Abstract
Nuclear speckles are known to be the storage sites of mRNA splicing regulators. We report here the identification and characterization of a novel speckle protein, referred to as NSrp70, based on its subcellular localization and apparent molecular weight. This protein was first identified as CCDC55 by the National Institutes of Health Mammalian Gene Collection, although its function has not been assigned. NSrp70 was colocalized and physically interacted with SC35 and ASF/SF2 in speckles. NSrp70 has a putative RNA recognition motif, the RS-like region, and two coiled-coil domains, suggesting a role in RNA processing. Accordingly, using CD44, Tra2β1 and Fas constructs as splicing reporter minigenes, we found that NSrp70 modulated alternative splice site selection in vivo. The C-terminal 10 amino acids (531–540), including 536RD537, were identified as a novel nuclear localization signal, and the region spanning 290–471 amino acids was critical for speckle localization and binding to SC35 and ASF/SF2. The N-terminal region (107–161) was essential for the pre-mRNA splicing activity. Finally, we found that knockout of NSrp70 gene in mice led to a lack of progeny, including fetal embryos. Collectively, we demonstrate that NSrp70 is a novel splicing regulator and essentially required early stage of embryonic development.
INTRODUCTION
Nuclear speckles are subnuclear structures that appear as irregular, punctuate structures which vary in size and shape when analyzed under a fluorescence microscope. They are located in the interchromatin regions of the nucleoplasm of mammalian cells. These particles occur in localized ‘clouds’, and cytochemical analysis indicates that they contain RNA (1). Using antibodies specific to splicing factors such as small nuclear ribonucleoprotein particles (snRNPs), the connection between nuclear speckles and pre-mRNA splicing was initially determined (2). It is now clear that nuclear speckles serve as storage sites of the components of the pre-mRNA splicing machinery, including snRNPs, spliceosome subunits and other non-snRNP protein splicing factors, and therefore play an important role in alternative splicing.
Alternative pre-mRNA splicing is a critical step in gene expression and proteomic diversity in all metazoans. Purification and analysis of the multiple proteins in speckles show that these can be roughly separated into two classes: one class consists of relatively widely expressed proteins, which seem to have wide-ranging roles in mRNA biogenesis. These proteins can be classified into two groups: SR proteins and hnRNP proteins. SR proteins tend to promote exon inclusion while hnRNP proteins usually have the opposite effect (3). The other class consists of factors with restricted expression patterns that are responsible for regulating tissue-specific alternative splicing events. These proteins, including Nova-1/2 and Hu proteins, have been identified by a variety of approaches and each of them shares a salient feature: each contains an RNA-binding domain of the K homology or RNA recognition motif (RRM) type (3).
Members of the SR family include ASF/SF2, SC35 and SRp20. Each of these proteins is required for constitutive splicing and can regulate alternative splicing. SR proteins are essential splicing factors since they can complement splicing-deficient S100 extracts (4). They contain one or more RRMs and a C-terminal arginine/serine (RS)-rich domain that contains serine residues that can be phosphorylated (5,6). RRMs mediate sequence-specific binding to the RNA, whereas the RS-rich domain is mainly involved in protein–protein interactions that are thought to be essential for the recruitment of the splicing machinery, for splice-site pairing (7,8), and for nuclear localization signaling (NLS) to the speckles (9). Notably, deletion of two SR proteins, ASF/SF2 or SC35, in the germ line of mice leads to embryonic lethality before day 7.5 (10–13).
Another class of splicing factors is SR-related proteins, which contain RS domains of varying lengths. Unlike SR proteins, SR-related proteins may or may not contain a RRM, and instead may contain other domains such as a DEXD/H box or a zinc finger. U2AF35, U1-70K and SRm160 are all examples of SR-related proteins (14), and many other proteins have recently been shown to belong to this family.
Recently, we unexpectedly found, through microarray analysis, a gene that can potentially affect T-cell motility. This gene was first identified and named as coiled-coil domain containing 55 (CCDC55) by the National Institutes of Health Mammalian Gene Collection program because it contains a coiled-coil domain in its sequence. This gene was significantly up-regulated in motile cells, but down-regulated in non-motile cells. However, both ectopic expression and knockdown experiments revealed no specific connection of CCDC55 protein with T-cell motility. As identification of CCDC55 in the nuclear speckle domain provided us with new insights into the splicing machinery (15), we compared the sequence of CCDC55 with other known splicing-related proteins. Interestingly, we found that CCDC55 had putative RRMs in the N terminus, an RS-like region in the C terminus, and two coiled-coil domains in both the N- and C-termini, thereby suggesting a role in RNA processing. Thus, based on its subcellular localization and molecular weight on SDS–PAGE, we renamed this protein as nuclear speckle-related protein 70 (NSrp70) and further explored its biological functions. We found that NSrp70 interacted and colocalized with SC35 and ASF/SF2 in nuclear speckles and modulated alternative splice site selection in vivo as determined by CD44, Tra2β1 and Fas minigenes. We determined the regions for the NLS sequence, for speckle localization and binding to SC35 and ASF/SF2, and for the pre-mRNA splicing activity. Finally, based on the NSrp70 knockout mice approach, we found that NSrp70 is an essential gene during early embryonic development.
MATERIALS AND METHODS
Reagents and antibodies
Phorbol 12-myristate 13-acetate (PMA), A23187, fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG, phalloidin-TRITC, and rabbit polyclonal anti-human NSrp70 (CCDC55) antibody were purchased from Sigma Chemical Co. (St. Louis, MO). 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) was purchased from Molecular Probes (Eugene, OR). DirectPCR Lysis Reagent (Tail) was purchased from VIAGEN Biotech (Wilshire Boulevard, LA, USA). Recombinant Human SDF-1α was purchased from R&D Systems (Minneapolis, MN, USA). Welprep™ Total RNA Isolation Reagent was purchased from JBI (Join Bio Innovation, Korea). Reverse transcript PCR premix and conventional PCR premix were purchased from iNtRON (iNtRON Biotechnology, Korea). Magna RIPTM Kit was purchased from Millipore (Billerica, MA, USA). Goat polyclonal anti-human β-actin, mouse monoclonal anti-myc-tag (9B11), rabbit polyclonal anti-human acetyl-histone H2A (Lys5) and rabbit polycolonal anti-human PARP antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Rabbit polyclonal anti-human I-κB antibody was purchased from Santa Cruz biotechnology (Santa Cruz, CA, USA). Mouse and rabbit polyclonal anti-human SC35 antibody was purchased from BD Pharmingen (San Jose, CA, USA). Rabbit polyclonal anti-GFP antibody was developed in rabbit using purified recombinant full-length GFP protein. [α-32P]dCTP was purchased from Perkin Elmer (Waltham, MA, USA). OmicsLinkTM shRNA expression clone and siRNA against NSrp70 were purchased from GeneCopoeiaTM (Germantown, MD, USA) and Dharmacon Inc. (Chicago, IL, USA), respectively.
Cell cultures and transfections
COS-7 and HEK293T cells were maintained in DMEM (Gibco-BRL, Gaithersburg, MD, USA) supplemented with 10% FBS (Giboco-BRL), penicillin and streptomycin (100 µg/ml; Invitrogen, Carlsbad, CA, USA). Constructs were transfected by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in HEK293T or COS-7 cells according to the manufacturer’s instructions. At 24 h after transfection, cells were collected for RNA purification or biochemical analysis. Transfection in Jurkat T cells was performed by Nucleofector Kit V (Amaxa, Koeln, Germany) according to the manufacturer’s instructions.
Plasmids
The mammalian expression vector of pEGFP-C1/ASF/SF2 and pETv5/CD44, and pCR3.1/MGTra minigene were generous gifts from Dr Brain J. Morris (University of Sydney, NSW2006, Australia). Fas minigene was a gift from Dr Michael Sattler (GSF-National Research Center for Environment and Health, Neuherberg, Germany). The pEGFP-C3/SP100, Daxx and PML3 were obtained from Dr David J. Picketts (University of Pennsylvania School of Medicine, Philadelphia, PA, USA). The cDNA of SC35 was obtained by RT–PCR using total RNA prepared from Jurkat T cells as a template. These cDNAs were then inserted into pCS4-3Myc, pEGFP-C1 and pmCherry-C1 by in-frame fusion. To generate the pEGFP/NSrp70 construct, the human NSrp70 clone coding the full-length open reading frame of NSrp70 (CCDC55) was purchased from imaGenes ORF Expression Clone (imaGenes GmbH, Germany). DNA fragments encoding NSrp70 or deletion or substitution mutants were generated by PCR from full-length NSrp70 ORF clone and inserted into pEGFP-C1 or pmCherry-C1. The expression vectors for chimeric NSrp70 proteins were as follows: M16 (Δ290C_fNLS), which is 1–289 amino acid of NSrp70 in-frame fused with NLS of NUCKS (nuclear casein kinase and cyclin-dependent kinase substrate); M17 (Δ 105N_Δ 290C_fNLS), which is 106–289 amino acids of NSrp70 in-frame fused with NLS of NUCKS. All the NSrp70 deletion mutants, chimeric muatants and SR proteins contained in-frame tagged GFP, mCherry or Myc at the N-terminus, and their sequences were verified by automatic sequencing.
Immunofluorescence staining and confocal imaging analysis
For visualization of fluorescence, cells were cultured on 18-mm non-coated round coverslips (Nalge Nunc International, Denmark) and fixed with 3.7% formaldehyde for 15 min. After blocking with either 5% BSA in phosphate-buffered saline (PBS) for 1 h, the primary antibodies were applied for overnight at 4°C, followed by the secondary antibodies for 1 h. In some case, cells were stained with 300 nM 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes, Eugene, OR, USA) before being mounted with gelatine/glycerol in PBS, 50:50 vol./vol. (Sigma Chemical Co., St Louis, MO, USA). The samples were examined with a FV1000 confocal laser scanning microscope (Olympus, Japan) equipped with 40×, 60× and 100× objectives. For the live-cell imaging, cells were directly mounted in a PC-R-10 bath flow chamber (Live Cell Instruments, Seoul, Korea) and observed by FV1000 confocal microscopy.
Immunoprecipitation and western blotting
Cell extracts from HEK293T or COS-7 cells (1 × 107 cells) which were transfected with indicated constructs were prepared by lysis buffer (1% Triton X-100, 150 mM NaCl, 20 mM Tris pH 7.5). Approximately 1 mg of the extract was mixed with 50 µl of pre-anti-GFP or anti-NSrp70 antibody-conjugated Sepharose 4B (GE Healthcare, Sweden). The immunocomplexes were incubated overnight at 4°C with frequent mixing, washed two times with washing buffer I (1% Triton X-100, 150 mM NaCl, 20 mM Tris–HCl pH 7.5) and buffer II (150 mM NaCl, 10 mM Tris–HCl pH 7.5) for each and resolved by 12% SDS–PAGE. Proteins were electroblotted on to a PVDF membrane (Perkin Elmer) by means of a Trans-Blot SD semidry transfer cell (Bio-Rad, Hercules, CA, USA). The membrane was blocked in 5% skim milk (1 h), rinsed, and incubated with the indicated antibodies in TBS containing 0.1% Tween-20 (TBS-T) and 3% skim milk overnight. Excess primary antibody was removed by washing the membrane four times in TBS-T. The membrane was then incubated with 0.1 µg/ml peroxidase-labeled secondary antibody (against rabbit or mouse) for 2 h. After three washes in TBS-T, bands were visualized by ECL western blotting detection (iNtRON Biotechnology Inc.). RNA immunoprecipitation experiments were performed according to the protocol provided by the kit (Millipore; catalog 17–701).
Nuclear fractionation
HEK293T or COS-7 cells (1 × 107 cells) were washed in ice-cold PBS and resuspended in 1 ml of hypotonic digitonin extraction buffer (5 mM Tris pH 7.5, 10 mM NaCl, 0.5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 40 µg/ml digitonin, 5 µg/ml aprotinin, 10 µg/ml leupeptin, 1 mM PMSF and 1 mM Na3VO4). After 15 min on ice, the cells were pelleted for 5 min at 1000 g, and the suspernatant was collected and assayed for cytosol protein content. The insoluble pellet was washed with PBS, and then re-extracted with 0.5 ml of NP-40 lysis buffer (10 µg/ml Tris pH 7.5, 40 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.2% NP-40, 5 µg/ml aprotinin, 10 µg/ml leupeptin, 1 mM PMSF and 1 mM Na3VO4). The samples were vortexed and incubated on ice for 10 min. Cell lysates were cleared of insoluble material by centrifugation at 18 000 g and soluble proteins were quantitated. Aliquots of the digitonin- and NP-40-extractable fractions from each sample were resolved by SDS–PAGE and analyzed by western blot.
In vivo splicing assays
In vivo splicing assays were performed essentially as described (16,17). Briefly, a splicing reporter minigene was co-transfected with an increasing amount of NSrp70 expression construct (pEGFP/NSrp70) in HEK293T cells. Empty pEGFP-C1 plasmids were added to ensure that the same amount of DNA was transfected. After 24 h of transfection, RNA was isolated using Welprep™ Total RNA Isolation Reagent. RT–PCR for the CD44 minigene was performed using primers N3INS (5′-CTCCCGGGCCACCTCCAGTGCC-3′) and N5INS (5′-GAGGGATCCGCTTCCTGCCCC-3′) and a program involving 30 cycles of 94°C for 20 s, 68°C for 30 s and 72°C for 40 s. For Tra2β1, we used primer pCR3.1-RT-Rev (5′-GCCCTCTAGACTCGAGCTCGA-3′) for the RT and primers MGTra-Bam (5′-GGGCCAGTTGGGCGACCGGCGCGTCGTGCGCGGGG-3′) and MGTra-R-Xho (5′-GGGCTCGAGTACCCGATTCCCAACATGACG-3′) for the PCR consisting of 35 cycles of 94°C for 20 s, 65°C for 20 s and 72°C for 40 s. For Fas, we used primers PT1 (5′-GTCGACGACACTTGCTCAAC-3′) and PT2 (5′-AAGCTTGCATCGAATCAGTAG-3′) for the PCR consisting of 33 cycles of 94°C for 20 s, 60°C for 10 s and 72°C for 20 s. The PCR products were analyzed with a 1% agarose gel and the splicing pattern was quantified using ImageJ 1.44d (ImageJ is a public domain Java image processing program inspired by National Institutes of Health Image).
Knockout mouse
To obtain knockout mice, we searched Gene-Trap embryonic stem (ES) cell repositories for NSrp70 (CCDC55) insertions. A BLAST search using the mouse cDNA sequence identified line Ayu21-T93. C57B6 NSrp70+/GT heterozygous mouse was generated as follows. Briefly, exchangeable gene trap pU-21T vector was used for random gene trap mutagenesis. The mutant construct contains a splice acceptor sequence linked to the β-geo reporter gene, and its integration site is concentrated in the 5′-end of a trapped gene. pU-21T (20–40 µg) was transfected by electroporation (800 V, 3 µF with Bio-Rad Gene Pulser) into the feeder-free ES cell line KTPU8 (F1 of B6 and CBA). Thereafter, G418-resistant clones were selected and expanded. Genomic DNA were prepared from clones and examined by PCR and Southern blotting for single-copy integration and for the existence of lox71–lox2272 sites, which are indispensable for site-specific recombination. To identify the trapped gene, 5′-RACE was performed. We found that pU-21T was inserted at 1141 bp downstream of the exon 1 of NSrp70 gene. Furthermore, we found that the 5′-end 569 bp of trap vector was deleted. Clones that had trap gene insertion in NSrp70 were used to generate chimeric mice. ES cells were aggregated with morulae from ICR (Imprinting Control Region) mice. We used 125 morulae per line and transferred them into five foster mothers. Chimeric mice were mated with C57BL/6 females. Genomic DNAs of F1 progenies and original ES cells were subjected to Southern blotting to confirm that the integration pattern of the vector is identical between the mouse line and the original ES clone.
PCR genotyping assay of embryos and progenies
Tails of embryos or progenies were lysed in 1.5 ml tubes containing tail lysis buffer (DirectPCR Lysis Reagent, VIAGEN Biotech), and 2 µg lysate was used in for PCR genotyping. The PCR primers for the wild-type allele were located in exon 1 (21T 93-1: 5′-ATATACACGTCGGCGTCAGC-3′) and exon-2 (21T93-3R: 5′-CAAAATAAGCCCATACCTGCGTAA-3′), and the PCR primers for the trap allele were located between exon 1 and exon 2 (21T93-9: 5′-GAAGAGAGGCCCATTGGTTG-3′, SA-5AS: 5′-GGGCAAGAACATAAAGTGACC-3′). Primers 21T93-1 and 21T93-3R were used in PCR of 1737 base pairs of nature intron 1 (35 cycles of 1 min at 94°C, 1 min at 58°C, and 2 min at 72°C), and primers 21T93-9 and SA-5AS were used in PCR of 597 base pairs that consist of the partial trap vector and the intron 1 sequence (35 cycles of 1 min at 94°C, 15 s at 56°C, and 30 s at 72°C).
Lenti-viral infection
Lenti-viral vector (2 µg of non-target psiLv-H1 or NSrp70 target psiLv-H1) with the appropriate insert (10 µg of packaging plasmid) was transfected into HEK293T cells using lipofectamine 2000 (Invitrogen). The culture supernatants were collected 48 h after transfection. The supernatants were pooled, concentrated by ultracentrifugation, and stored at −80°C until use. For lenti-viral infection, virus particles were mixed with Jurkat T cells (2 × 105/500 µl) in a 15-ml centrifuge tube in the presence of 8 µg/ml polybrene. Following centrifugation at 1341 g for 1 h, the cells were transferred into a 60-mm culture dish and infection and knockout efficiency were checked after 48 h of infection by FACs analysis and RT–PCR.
Cell migration assay
Cell migration assay was performed using transwell chemotaxis system (CHEMOTX system, 3–5 µm pores, Neuro Probe Inc., Gaithersburg, MD, USA). Lymphocytes (5 × 105) obtained from wild-type or NSrp70+/GT were placed on the upper wells (50 µl), and media containing 50 ng/ml SDF-1α were added to the lower wells (30 µl). The plates were incubated for 2.5 h at 37°C in a humidified CO2 incubator. After incubation, the inserts were removed, and the cells that had migrated through the filter to the lower wells were counted by using ImageJ 1.44d. Three different fields for each well were selected for imaging and the cells within the microscope field were counted using ImageJ 1.44d.
RESULTS
NSrp70 is a novel nuclear speckle protein
In this study, we initially identified NSrp70 as a migration-related gene in T cells. As shown in Supplementary Figure S1A, the expression of NSrp70 was ∼4-fold higher in motile T cells than in parent T cells. In silico analysis based on BioGPS showed that NSrp70 is highly expressed in immune function-related cells, including dendritic cells, T cells, B cells and natural killer cells (Supplementary Figure S1B). Northern blot analysis consistently revealed that NSrp70 is significantly expressed in secondary lymphoid organs such as spleen; and mesenteric, axillary and brachial lymph nodes (Supplementary Figure S1C). However, both RNA interference or ectopic overexpression of NSrp70 showed little effect on Jurkat T cells in terms of their migration (chemokinesis) and activation (IL-2 expression) in response to SDF-1α and PMA+A23187, respectively (Supplementary Figure S1D and E).
The absence of a connection between NSrp70 and T-cell migration and activation led us to track the localization of NSrp70 in various cell types, as a previous report demonstrated that NSrp70 formed speckle structures in the nucleus (15). We consistently observed typical nuclear speckled patterns when green fluorescent protein (GFP)-tagged NSrp70 cDNA was transfected into COS-7 cells (Figure 1A). High-resolution confocal microscopic analysis revealed either diffused or doughnut-shaped distribution of GFP-tagged NSrp70 in many cells (Supplementary Figure S2A). However, a colocalization study revealed that NSrp70 shows a precise overlap with splicing factor SC35, confirming its localization in nuclear speckles (Supplementary Figure S2B). Endogenous NSrp70 was also detected in the nucleus, but mostly in a diffuse pattern in fixed HEK293T cells (Figure 1B, left). To clarify whether the slightly different pattern of GFP-tagged NSrp70 compared to the endogenous NSrp70 in terms of nuclear localization pattern is due to the artifact of tagged protein, we further used Myc-tagged NSrp70 cDNA in 293T cells, and then co-localization study was performed with SC35. myc-tagged NSrp70 also showed similar pattern as endogenous NSrp70, ranging from the diffuse pattern to the speckle pattern, and was unambiguously overlapped with SC35, therefore suggesting that tagged protein had little effect on NSrp70 localization. Instead, it might be a difference resulted by the direct visualization (GFP) versus indirect visualization after antibody staining. Localization of NSrp70 was further confirmed by nuclear fractionation assay with COS-7 and HEK293T cells (Figure 1A and B, right). Sequence alignment of NSrp70 orthologs demonstrated a highly conserved homology with other species (Supplementary Figure S3).
Figure 1.
Exogenous and endogenous NSrp70 are localized within the nucleus as speckles or small punctuate forms. (A, left): COS-7 cells were transfected for 24 h with 2 µg of pEGFP or pEGFP/NSrp70 cDNAs. The cells were stained with DAPI (blue) and phalloidin-TRITC (red), and were observed under a confocal microscope equipped with a 100× objective. (B) Endogenous NSrp70 (green) was detected with a rabbit polyclonal anti-NSrp70 antibody and visualized with an FITC-conjugated anti-rabbit antibody in HEK293T cells. (A and B) The subcellular localization of NSrp70 was confirmed by nuclear fractionation assay of the COS-7 (A, right) and HEK293T (B, right) cell lines. I-κB and PARP (or SC35) were used as cytosolic and nucleoplasmic fraction markers.
NSrp70 is a novel SR-related protein and influences alternative splice site selection
The speckle pattern of GFP_NSrp70 protein may correspond to perichromatin fibrils (PFs) and interchromatin granule clusters (IGCs), which can be detected by electron microscopy when cells are stained with antibodies against components of the splicing machinery (18). To further characterize the potential roles of NSrp70, we performed alignment of full-length NSrp70 amino acids with other speckle proteins according to the needle method of EMBOSS Pairwise Alignment Algorithms. We also searched the conserved domains in BLAST and Protein Knowledgebase Database as described in ‘Materials and Methods’ section. Through sequence analysis, however, we found that NSrp70 has low-sequence similarity and identity with classic SR proteins or SR-related proteins (data not shown).
As we thought that the low percentage of similarity and identity obtained through the needle method was due to the whole-length sequence that has many inserted gaps, we selected known RRM and RS domain sequences from different SR or SR-related protein, and then aligned them with the total NSrp70 sequence according to the water method of EMBOSS Pairwise Alignment Algorithms to find the best region of similarity between the two sequences. Interestingly, two N-terminal regions (20–85 and 170–289 amino acids) were observed to be in a similar region as RRMs (Table 1). A signature sequence, ‘RDAEDA’, which appeared in known RRM sequences of SR proteins, was also found between 20 and 85 amino acids of NSrp70 (Supplementary Figure S3A). We also found an RS-like region at the C terminus (290–551 amino acids), while its predicted RS-like region sequences were interrupted by aspartic acid (D) or glutamic acid (E) after arginine (R). To understand how far back in evolution does this protein go, we analyzed potential orthologs proteins by ClustalW analysis. The constructed phylogenetic tree revealed that human NSrp70 shares 82% sequence identity with bovine NSrp70, 14% identity with human U2-associated, and 15% identity with SC35 (Supplementary Figure S3B). The analysis of orthologs together with the fact that NSrp70 is a nuclear speckle protein strongly suggests that NSrp70 is an SR-related protein.
Table 1.
Identification of RRMs and RS-like region in NSrp70 and their percent similarity/identity as compared to other SR or SR-related proteins
| Protein name | RRMa |
RS domain |
Conserved regions in NSrp70 |
|||
|---|---|---|---|---|---|---|
| Region | Similarity/Identity (%) | Region | Similarity/identity (%) | RRM | RS-like | |
| Classical SR proteins | ||||||
| SRp75 | 2–72 | 43.2/ 29.5 | 179–494 | 39.9/23.6 | 233–292 | 282–552 |
| 104–163 | 40.9/20.5 | 335–370 | ||||
| SRp55 | 1–72 | 45.5/25 | 184–343 | 37.1/20.5 | 233–274 | 282–491 |
| 110–183 | 43/19 | 199–274 | ||||
| SC35 | 14–92 | 35.9/21.8 | 117–221 | 43.5/23.5 | 192–269 | 335–419 |
| SRp40 | 4–74 | 37.3/24.1 | 182–267 | 51.1/22.7 | 188–264 | 445–531 |
| 108–181 | 30.9/18.6 | 178–274 | ||||
| 9G8 | 11–84 | 30.9/18.6 | 121–238 | 38.3/24.1 | 178–274 | 281–410 |
| Additional SR proteins | ||||||
| SRrp86 | 66–142 | 66.7/55.6 | 194–261 | 26.6/536 | 256–264 | 320–398 |
| 346–430 | 29.4/50.6 | 259–342 | ||||
| U1-70K | 103–181 | 43.1/27.6 | 230–305 | 33.3/54.8 | 285–339 | 363–455 |
| 348–390 | 34.8/58.7 | 363–408 | ||||
| p54 | 33–113 | 35.8/17 | 247–353 | 26.8/44.7 | 39–88 | 281–376 |
| XE7 | 147–256 | 33.8/25 | 642–691 | 52.8/34 | 502–554 | 303–352 |
| SRp46 | 14–92 | 55/40 | 83–205 | 32.5/38.8 | 303–319 | 281–359 |
| hTra2β | 118–196 | 57.1/10.7 | 31–113 | 45.7/34.6 | 58–85 | 277–355 |
| 231–287 | 41.5/20.8 | 294–346 | ||||
| hTra2α | 119–197 | 45.8/41.7 | 30–112 | 45.7/33 | 448–471 | 262–355 |
| 226–282 | 40.4/28.8 | 312–358 | ||||
| U2AF65 | 149–231 | 61.1/27.8 | 27–62 | 57.1/37.1 | 61–78 | 364–398 |
| 259–337 | 75/62 | 149–159 | ||||
| 385–466 | 45.5/21.8 | 34–80 | ||||
| RNA binding SR-related factors | ||||||
| U2-associated protein SR140 | 274–355 | 40/28.3 | 922–1001 | 38.9/27.4 | 127–182 | 281–374 |
| RBM23 | 166–243 | 58.8/47.1 | 366–382 | |||
| 263–341 | 43.9/26.8 | 470–502 | ||||
| RBM39 | 153–230 | 58.8/47.1 | 41–90 | 46.3/29.9 | 366–382 | 371–437 |
| 250–328 | 46.4/35.7 | 36–57 | ||||
| 445–508 | 43.3/25 | 173–228 | ||||
| RBM5 | 98–178 | 48.6/28.3 | 206–235 | |||
| 231–315 | 62.5/37.5 | 473–488 | ||||
| SFRS15 | 508–582 | 46.2/40.8 | 501–550 | |||
aRRM and RS domain sequences of each SR and SR-related protein were collected from The Nuclear Protein Database and Protein Knowledgebase UniPortKB, and then each sequence was aligned with NSrp70 full-length sequence by using water method of EMBOSS Pairwise Alignment Algorithms to find the best region of similarity between two sequences. The URLs used in this study was described in Supplementary Data.
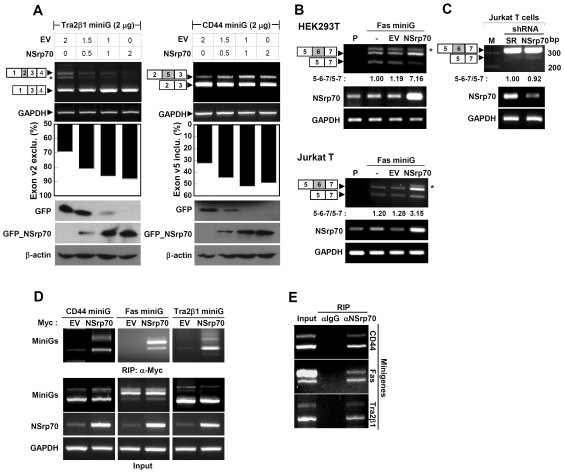
Based on the results obtained from sequence alignment with other SR or SR-related proteins, we next conducted in vivo splicing assays using minigenes to test whether NSrp70 behaves like a typical SR protein i.e. whether it can activate pre-mRNA splicing or regulate alternative splicing in a concentration-dependent manner (5,19). To this end, Tra2β1 or CD44 minigene was transfected into HEK293T cells with increasing amounts of pEGFP/NSrp70. RT–PCR was performed with primers directed at sequences located in the flanking constitutive exons. As shown in Figure 2A, we found that addition of NSrp70 in small increment from 0.5 to 2 µg resulted in an increased exon v2 exclusion from 69 to 92% in Tra2β1 minigene and exon v5 inclusion from 39 to 65% in CD44 minigene. As a control, cotransfection of Tra2β1 or CD44 with an empty vector, pEGFP-C1, did not change the splicing pattern, demonstrating that the splicing reaction and involvement of NSrp70 in splicing modulation were not an artifact. Western blot data represent the protein amounts after transfection of GFP and GFP_NSrp70 in HEK293T cells (Figure 2A).
Figure 2.
NSrp70 influences alternative splice site selection of several minigenes in different cell types. (A) Promotion of exon v2 exclusion on Tra2β1 and exon v5 inclusion on CD44 minigene by NSrp70. HEK293T cells were cotransfected with increasing amounts of pEGFP/NSrp70 (0–2 µg) and 2 µg of the minigene. Parental vector (0–2 µg) was added to ensure that similar amounts of cDNAs were transfected. Exon inclusion or exclusion was determined by RT–PCR (top), as described in ‘Materials and Methods’ section. The ratio of exclusion or inclusion of Tra2β1 or CD44 minigene was shown as a histogram (middle). Dose-dependent expression of GFP_NSrp70 and GFP was confirmed by western blotting (bottom). (B) NSrp70 promotes exon v6 inclusion on Fas minigene in HEK293T or Jurkat T cells. Two micrograms of Fas minigene was transfected with 2 µg of pEGFP/NSrp70 or pEGFP into HEK293T or Jurkat T cells. Exon inclusion or exclusion was determined by RT–PCR. Note: P, parent; EV, pEGFP vector; NSrp70, pEGFP/NSrp70. Asterisks denote an uncharacterized PCR product (A and B). (C) Interference of NSrp70 by shRNA decreases exon v6 inclusion in Jurkat T cells. psiLv-H1/scramble or psiLV-H1/shNSrp70 (CCDC55) vector was infected to Jurkat T cells by the lentiviral system. Exon inclusion or exclusion was determined by RT–PCR. Note: M, 100 bp DNA marker; SR, psiLv-H1/scramble, NSrp70, psiLV-H1/shNSrp70 infection. The ratio of exon inclusion or exclusion splicing forms was analyzed by ImageJ. Each experiment was repeated at least three times to confirm the reproducibility of data. (D and E) mRNA of minigenes interacts to NSrp70. HEK293T cells were transfected with (D) or without (E) plasmid expressing myc_NSrp70 and indicated minigene and then immunoprecipitated with anti-Myc (D) or anti-NSrp70 antibody (E). The RNAs in the immunoprecipitates were then analyzed by RT–PCR using primers specific for the CD44, Fas or Tra2β1 transcripts of minigenes. Ten microliter of the supernatant of total lysate was used for total input sample.
To further confirm that NSrp70 is indeed engaged in alternative splicing modulation, we utilized another splicing reporter minigene, Fas, to test the efficiency of NSrp70 in an in vivo splicing assay. As shown in Figure 2B, NSrp70 transfection caused ∼3–7-fold increase in exon v6 inclusion of Fas minigene in both HEK293T and Jurkat T cells. Importantly, we also tested whether depletion of NSrp70 could oppositely affect exon v6 inclusion of endogenous Fas in Jurkat T cells. To obtain Jurkat T cells with stable depletion of NSrp70, we utilized a lentiviral system that contains shRNA targeted for NSrp70. As shown in Figure 2C, depletion of NSrp70 resulted in increased exon v6 exclusion of endogenous Fas. Taken together, these results strongly demonstrate that NSrp70 can influence the alternative splice site selection of various pre-mRNAs, including Tra2β1, CD44 and Fas.
An important issue raised at this point was whether NSrp70 directly interacts with the RNAs used in the study. To this end, transfected or endogenous NSrp70 was immunoprecipitated with antibodies against myc (Figure 2D) or NSrp70 (Figure 2E), and then the precipitates were analyzed for the presence of each minigene by RT–PCR. All three minigenes were immunoprecipitated by myc-tagged NSrp70 or endogenous NSrp70, suggesting that NSrp70 physically interacts with RNAs, including Tra2β1, CD44 and Fas.
Nuclear localization is required for NSrp70-mediated alternative splicing
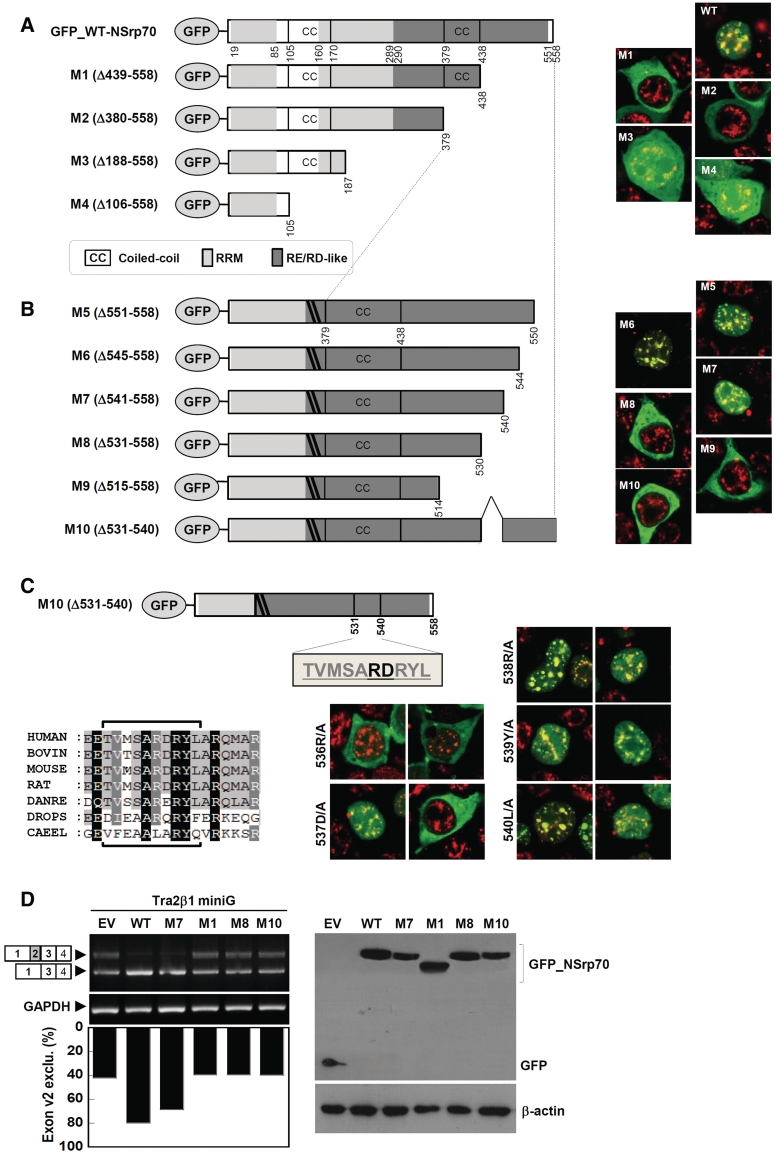
The RS domain of SR or SR-related proteins is thought to be required for protein–protein interaction with each other or with other components of the splicing machineries (5,7,20) as well as for subcellular localization (21,22). As we found an RS-like region in NSrp70, which resembles the RS domain in other SR proteins, we hypothesized that this region may contain the NLS sequence. To this end, we generated a series of GFP-tagged deletion mutants for probing NLS sequence in the putative RS-like region of NSrp70. After transient transfection, the localization of GFP-tagged NSrp70 mutants was analyzed by confocal microscopy. To better monitor the localization, the fixed cells were also counter stained with endogenous SC35. In contrast to the wild-type (GFP_NSrp70), deletion of amino acids spanning the 439–558 (M1) region appeared to be localized in the cytosol, suggesting that this region included the NLS sequence (Figure 3A). To find the exact NLS sequence, we therefore designed more NSrp70 mutant constructs (Figure 3B). Interestingly, we found that mutant M10 (Δ531–540) localized in the cytosol, suggesting that this region belongs to the NLS sequence of NSrp70. Further point mutations at 536R/A or 537D/A demonstrated that these two amino acids are critical residues for nuclear localization and target the speckle compartment (Figure 3C). To extend these findings with regard to the functional role, we performed in vivo splicing assay with Tra2β1 minigene. M1 (Δ439–558), M8 (Δ531–558) and M10 (Δ531–540), which localize in the cytosol, did not induce alternative splicing (Figure 3D). Interestingly, however, M7 (Δ541–558), which localizes in the nucleus, affected the alternative splice site selection and is comparable to that of the wild-type (Figure 3D). These results demonstrate that nuclear localization by the NLS sequence (531–540), including 536RD537, is important for the functional role of NSrp70. Further, we found that this sequence does not match the known NLS sequence, suggesting that this is a novel NLS sequence.
Figure 3.
Nuclear localization is mediated by a novel NLS sequence and is required for NSrp70-mediated alternative splicing. (A–C) Schematic diagram of deletion or substitution mutants of NSrp70 (left) and their subcellular localization in HEK293T cells (right). HEK293T cells were transfected with pEGFP vectors (1 µg) that contain wild-type or mutant NSrp70. After 24 h of transfection, cells were fixed, stained with anti-SC35 antibody followed by TRITC-conjugated anti-mouse antibody, and visualized by confocal microscopy (right). Sequence alignment of the NLS sequence with other species, as analyzed by Clustal X and GeneDoc, is depicted in (C) (left bottom). (D) Nuclear localization is required for NSrp70-mediated exon 2 exclusion of Tra2β1 minigene. HEK293T cells were cotransfected with Tra2β1 minigene (2 µg) and the indicated NSrp70 constructs (2 µg). After 24 h of transfection, exon v2 exclusion was determined by RT–PCR (left). The ratio of exclusion or inclusion of Tra2β1 or CD44 minigene was shown as a histogram (left bottom). GAPDH and β-actin are shown as loading controls. The protein levels of GFP and GFP_NSrp70 mutants were confirmed by western blotting (bottom). Experiment was repeated at least three times to confirm the reproducibility of data.
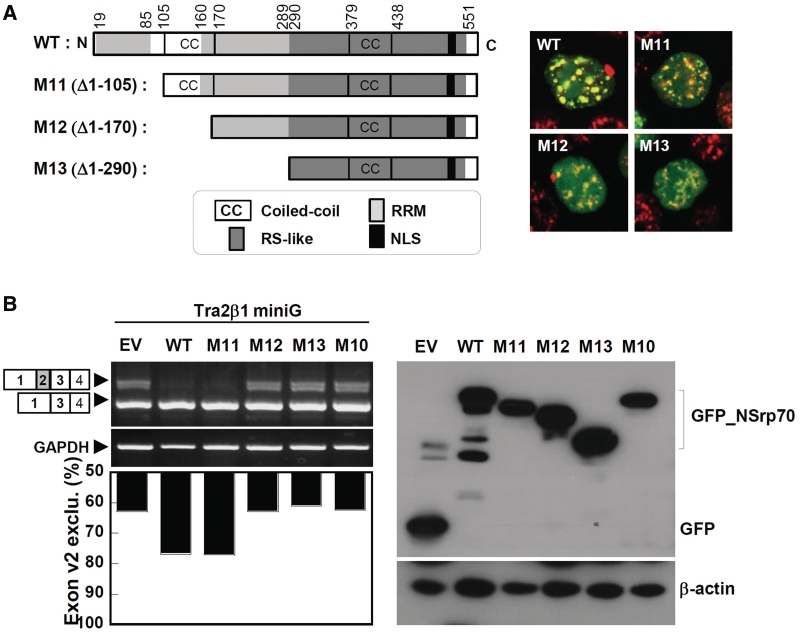
Coiled-coil domain of the N-terminus is critical for alternative splicing activity
SR proteins contain one or two RRMs containing the signature sequences RDAEDA, RDADDA and SWQDLKD (23,24). Splice-site selection is determined by the nature of the RRM (25). As NSrp70 also contains the highly conserved RDAEDA sequence between amino acids 47 and 70 at the N terminus (Supplementary Figure S3), we investigated the role of this sequence in terms of alternative splicing activity. As the N terminus also contains a coiled-coil domain in amino acids 106–161, we also tested whether this region corresponds to the splicing activity of NSrp70. To this end, we designed a series of N-terminal deletion constructs and investigated their splicing activities for Tra2β1 minigene in HEK293T cells (Figure 4A). Interestingly, although all of the deletion mutants (M11–M13) showed nuclear speckle localization, M12 (Δ1–170), a deletion mutant of the coiled-coil domain, but not M11 (Δ1–105), completely lost alternative splice site selection of pre-mRNA of Tra2β1 (Figure 4B), suggesting that the coiled-coil domain (106–170 amino acids) is critical for alternative splicing activity of NSrp70. All expressed proteins accumulated to similar levels in transfected HEK293T cells, as verified by western blot analysis of whole cell lysates (Figure 4B, right).
Figure 4.
Deletion of the coiled-coil domain results in loss of alternative splicing activity. (A) Schematic diagram of deletion mutants of NSrp70 (left) and their subcellular localization with SC35 in HEK293T cells (right). (B) HEK293T cells were cotransfected with Tra2β1 minigene (2 µg) and the indicated NSrp70 constructs (2 µg). After 24 h of transfection, exon v2 exclusion was determined by RT–PCR (left). The ratio of exclusion or inclusion of Tra2β1 or CD44 minigene was shown as a histogram (left bottom). GAPDH and β-actin are shown as loading controls. The protein levels of GFP and GFP_NSrp70 mutants were confirmed by western blotting (bottom). Experiment was repeated at least three times to confirm the reproducibility of data.
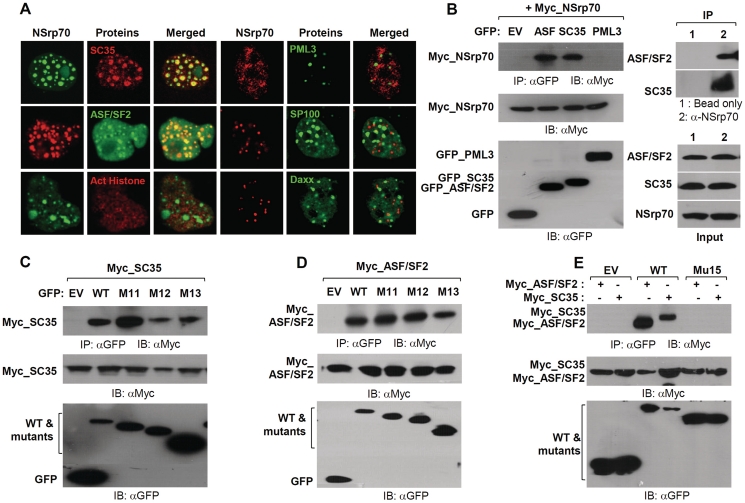
NSrp70 interacts with SC35 and ASF/SF2 through the C-terminal RS-like region
As we found that NSrp70 colocalized with SC35, a spliceosomal component (Supplementary Figure S2B), we extended the experiment to test whether NSrp70 is colocalized with other spliceosomal proteins such as ASF/SF2 or non-spliceosomal proteins such as acetylated-histone3, PML3, Sp100 and Daxx. We found that NSrp70 was significantly overlapped with SC35 and ASF/SF2 but not with acetylated-histone3, PML3, Sp100 and Daxx, strongly suggesting that NSrp70 is a new spliceosomal protein (Figure 5A). To prove the physical interactions between NSrp70 and ASF/SF2 or SC35, we generated Myc-tagged NSrp70 and GFP-tagged ASF/SF2 or SC35 and then cotransfected them into HEK293T cells. As shown in Figure 5B (left), immunoprecipitation results revealed that NSrp70 could interact with both ASF/SF2 and SC35 but not with PML3. To test whether native proteins are also physically interacted, endogenous NSrp70 was immunoprecipitated with anti-NSrp70 antibody, and then immunoblotted with anti-ASF/SF2 or anti-SC35 antibodies. As shown in Figure 5B (right), endogenous NSrp70 were also physically interacted with native forms of ASF/SF2 and SC35.
Figure 5.
NSrp70 interacts with ASF/SF2 and SC35 through the RS-like region of NSrp70. (A) HEK293T cells were cotransfected with pEGFP/NSrp70, pmCherry/NSrp70, pmCherry/SC35, pEGFP/ASF/SF2, pEGFP/PML3, pEGFP/SP100 or pEGFP/Daxx. Each transfectant was visualized in the live chamber by using a confocal microscope equipped with a 100× objective. For staining of the endogenous acetylated histone (Act-His), pEGFP/NSrp70-transfected cells were cultured on an 18-mm cover glass for 24 h, and then stained with rabbit polyconal anti-human acetyl-histone H2A (Lys5) antibody followed by TRITC-conjugated anti-rabbit antibody. (B) Coimmunoprecipitation of NSrp70 with ASF/SF2 or SC35. Left: HEK293T cells were transfected with pCS4-3Myc/NSrp70 and the indicated constructs, and then the cell extracts (1 mg) were immunoprecipitated with 50 µl of anti-GFP antibody-conjugated Sepharose 4B slush. Immune complexes were resolved on an SDS–PAGE gel and blotted. The presence of NSrp70 was determined by using an anti-Myc antibody. The expression of indicated constructs was shown by western blotting. Right: HEK293T cells were immunoprecipitated with anti-NSrp70 antibody-conjugated Sepharose 4B slush. Immune complexes were resolved as described above and blotted with an anti-ASF/SF2 or SC35 antibodies. (C–E) HEK293T cells were transfected with pCS4-3Myc/SC35 or pCS4-3Myc/ASF/SF2 and the indicated mutant constructs of NSrp70 (M11, M12, M13, or M15). Immunoprecipitation was performed as described above.
We next questioned whether the physical interaction between NSrp70 and SC35 or ASF/SF2 is connected for the splicing activity of NSrp70. As shown in Figure 5C and D, although mutants M12 and M13 had no alternative splicing activity (Figure 4A and B), they could still interact with SC35 or ASF/SF2, demonstrating that the physical interaction of NSrp70 with these two spliceosomal proteins is not directly connected with its activity and further suggests that the C-terminal RS-like region may include the interaction site. In fact, the RS domain is known as a region that mediates protein–protein interactions. For example, upon deletion of its RS domain, SC35 fails to associate with Tra, Tra2 and other SR proteins (7). In accordance with the previous report, we found that upon partial deletion of the RS-like region (Δ290–471) in NSrp70, the protein failed to interact with ASF/SF2 or SC35 (Figure 5E).
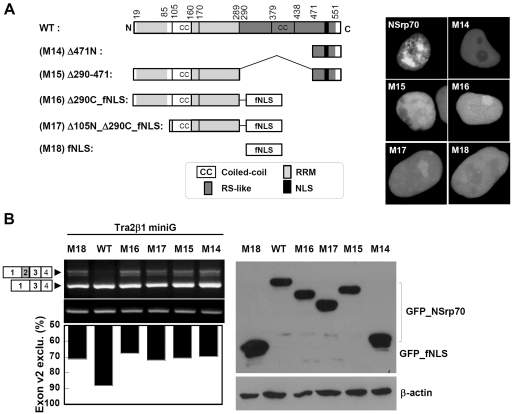
Speckle localization through the RS-like region is also important for NSrp70-mediated splicing activity
Our results thus far demonstrated the requirement for both the N-terminal coiled-coil domain and C-terminal NLS for proper activity of NSrp70, but did not establish the function of the RS-like region through which SC35 and ASF/SF2 are associated. We therefore examined whether the RS-like region is also involved in NSrp70-mediated splicing function. Interestingly, we found that partial deletion of this region (M15; Δ290–471) caused the protein to completely lose its localization in the nuclear speckles and revealed its diffuse pattern in the nucleus, demonstrating that this region contained the sequence for both speckle localization (Figure 6A) and SC35 and ASF/SF2 association (Figure 5E). To further confirm whether the N-terminal region alone is enough to induce splicing activity, the RS-like region was replaced with the foreign sequence containing an 18 amino acid NLS sequence taken from NUCKS (26). Figure 6A shows a schematic map of mutants and their nuclear localization. Interestingly, all mutants showed a similar pattern as M15, suggesting that the RS-like region is important for speckle localization. Consequently, these mutants showed no activity for Tra2β1 exon v2 exclusion in HEK293T cells (Figure 6B). Taken together, although we did not further determine the exact sequence for speckle localization, the region for SC35 and ASF/SF2 association might be overlapping with that for speckle localization. Therefore, our current result suggests that speckle localization is important for the functional role of NSrp70.
Figure 6.
Speckle localization through the RD/RE-rich domain is important in NSrp70-mediated alternative splicing. (A) Schematic diagram of deletion mutants of NSrp70 (left). HEK293T cells were transfected with the indicated construct (1 µg). After 24 h of transfection, cells were visualized in the live chamber by using a confocal microscope equipped with a 100× objective. (B) HEK293T cells were cotransfected with Tra2β1 minigene (2 µg) and the indicated construct (1 µg). After 24 h of transfection, exon v2 exclusion was determined by RT–PCR (left top). The ratio of exclusion of Tra2β1 minigene was shown as a histogram (left bottom). GAPDH and β-actin are shown as loading controls. The protein levels of GFP and GFP_NSrp70 mutants were confirmed by western blotting (bottom). Experiment was repeated at least three times to confirm the reproducibility of data.
NSrp70 has a recessive phenotype and knockout of NSrp70 leads to early embryonic lethality
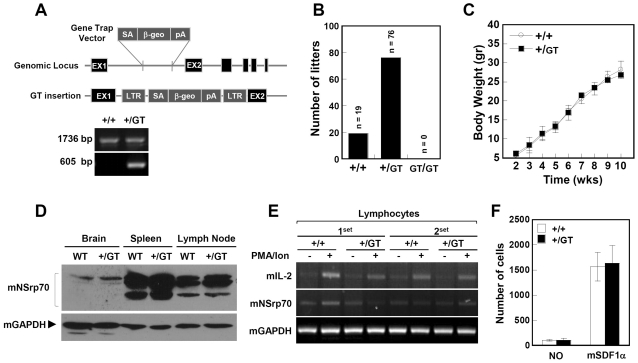
Because human NSrp70 has high similarity (75.6%) with its mouse ortholog, as depicted in Supplementary Figure S3, we investigated the role of NSrp70 in a mouse system generated in a large-scale Gene-Trap project (27). By sequence analysis of DNA isolated from NSrp70+/GT (Gene-Trap) heterozygous mice, we confirmed that the retroviral Gene-Trap vector was inserted in intron 1 of the mouse NSrp70 gene on chromosome 11, downstream of the exon encoding the initiation methionine (Figure 7A). To generate NSrp70GT/GT mice, heterozygous mutant mice were intercrossed and the genotypes of the offspring were analyzed by PCR. Among more than 95 offspring investigated, only NSrp70+/GT (∼80%) and NSrp70+/+ (∼20%) mice, but not viable NSrp70GT/GT mice, were detected (Fig. 7B), leading to the assumption that knockout of NSrp70 in vivo results in embryonic lethality. To determine in which developmental stage knockout of NSrp70 induces lethality, we carried out genotyping of embryos from embryonic day (E) 6.5 to 19.5. However, no homozygous embryo was observed (data not shown), suggesting that homozygous mutation leads to embryonic lethality shortly after implantation. In contrast, heterozygous mice (NSrp70+/GT) showed no differences to wild-type (NSrp70+/+) with regard to body weight and expression levels of NSrp70 protein (Figure 7C and D). In addition, no differences were observed in terms of IL-2 response in splenocytes, as evaluated by mitogen stimulation and migration of isolated T cells by SDF-1α (Figure 7E and F), suggesting that NSrp70 has a recessive phenotype in mice.
Figure 7.
NSrp70-deficient homozygous mouse exhibits early embryonic lethality. (A) Schematic diagram of the exchangeable Gene-Trap system. Note: numbered boxes, exons; lines, introns; diagonal lines, regions of homology where crossover can take place; β-geo, encoding a fusion protein with both β-galactosidase and neomycin phosphotransferase activity; SA, splicing acceptor; pA, polyadenine. Representative gel data revealed the PCR products of NSrp70+/+ and NSrp70+/GT. Primers used target the region from EX1 to EX2 (1736 bp) and the trap vector (605 bp), respectively. (B) Intercrossing the heterozygous mouse (NSrp70+/GT) produced no offspring homozygous for the allele containing the Gene-Trap vector (GT). The bar graph represents the number of offspring litters from the intercrossed NSrp70+/GT mice. (C) Body weight changes in NSrp70+/+ and NSrp70+/GT mice. Mice were weighed every week through the entire experimental period indicated. Data represent mean ± SEM (n = 8). (D) Western blot analysis was performed using rabbit polyclonal anti-NSrp70 antibody with mouse tissues from NSrp70+/+ and NSrp70+/GT (50 µg for each lane). mGAPDH was used as a loading control. (E) Lymphocytes from NSrp70+/+ and NSrp70+/GT mouse were stimulated with PMA (200 nM) and A23187 (1 µM) for 24 h, and then the expression of mIL-2 and mNSrp70 mRNA was determined by RT–PCR. mGAPDH was used as a loading control. (F) Lymphocytes (5 × 105) from NSrp70+/+ or NSrp70+/GT mice were allowed to migrate to the SDF-α (50 ng/ml)-containing lower well for 2.5 h at 37°C. The bar graph represents quantitative determinations of migrated cells obtained from three different fields of lower compartment.
DISCUSSION
We report here the identification and characterization of a novel SR-related and nuclear speckle protein NSrp70 whose expression is high in the testis, kidney and immune tissues such as spleen and lymph nodes. Endogenous NSrp70 protein contained novel sequences for NLS and speckle localization and was found to be colocalized with SC35 and ASF/SF2 in nuclear speckles. In addition, NSrp70 had a putative RRM in the N terminus followed by an RS-like region in the C terminus, suggesting that NSrp70 is a novel SR-related protein that may be involved in pathways related to RNA processing. Accordingly, functional splicing assay for NSrp70 with reporter minigenes demonstrated that NSrp70 could modulate alternative splice site selection in vivo. Particularly, the fact that NSrp70 knockout mice produced no progeny strongly suggests that NSrp70 is an essential gene in early embryonic development.
Alternative splicing is considered to be the most important source of protein diversity in vertebrates (28,29). To date, hundreds of splicing factors have been identified (30); however, attempts to explain genome-wide alternative splicing patterns will be impossible without a complete list of alternative splicing regulators. Although we unexpectedly found NSrp70 in rapidly migrating T cells, our results unambiguously demonstrated that NSrp70 is a novel protein localized in nuclear speckles that functions as a splicing regulator. This finding was substantiated by the fact that NSrp70 was detected as an uncharacterized protein in the nucleus by the systemic approach that aimed to define the subcellular localization of novel proteins identified by large-scale cDNA sequencing (15).
Many nuclear factors show a punctuate staining pattern when observed by indirect immunofluorescence microscopy, and are localized in distinct structures, such as speckles, paraspeckles, nucleoli, cajal bodies, GEMS and PML bodies (31). The obvious interaction of NSrp70 with speckle proteins such as ASF/SF2 or SC35, but not with PML bodies such as PML3, Daxx or SP100, demonstrates that NSrp70 is strictly located in speckles. As many pre-mRNA splicing factors are enriched in nuclear speckle, it is natural to question whether NSrp70 is a member of the SR protein or SR-related protein family (4). The following two factors are considered. First, SR proteins are characterized by the presence of RRM and the SR domain, which consists largely of serine/arginine repeats (4). The RRM has a consensus sequence of ∼90 amino acidscontaining a central sequence of eight or six conserved residues that are mainly aromatic and positively charged (32–34). SR proteins also contain signature sequences such as RDAEDA, RDADDA and SWQDLKD (23,24). Second, SR proteins contain the RS domain at the C terminus after one or two copies of RRM at the N terminus. However, the fact that the first homology search based on the conserved domain of the proteins resulted in no obvious RRM, while a second-round alignment with the selected RRM sequences from the known SR or SR-related proteins found putative RRM sites at two regions in the N terminus (20–85 and 161–290 amino acids), suggests that NSrp70 is not a typical member of SR proteins.
No clear RS domain that is conserved in other SR proteins was found in NSrp70. However, the sequences that cover the 290–551 amino acids region revealed consecutive RS, SR, RD and RE repeats, thereby representing an RS-like region in NSrp70. It is becoming increasingly clear that the RS domain mediates protein–protein interactions and, in some cases, acts as NLS (9,21,22). In an agreement with this observation, we found that the RS-like region in NSrp70 could not only bind to ASF/SF2 or SC35 but also contains an NLS sequence. Furthermore, the RS-like region might contain active sites of NSrp70 as it has been reported that replacement of the RS residue in ASF/SF2 to RD/RE, but not RG, could induce splicing of pre-mRNA in vitro (35,36). The RS-like region also affects nuclear speckle formation because partial deletion of this region (290–471 amino acids) disabled the formation of the nuclear speckle. Collectively, the presence of RRM and the RS-like region containing NLS and nuclear speckle sequences strongly suggest that NSrp70 is an SR-related protein.
Based on the prediction by UniProtKB/Swiss-Prot, we identified two potential coiled-coil domains at the regions spanning 106–170 and 379–438 amino acids. The first region might be critical for the splicing activity as deletion of this region (106–170 amino acids) led to complete loss of the alternative splicing activity of the protein. As the coiled-coil region can mediate homo- or heterodimeric interaction, we determined whether this region is the site for binding to ASF/SF2 and SC35 or, otherwise, mediates self-interaction (i.e. homodimerization). However, the result that NSrp70 still binds to either ASF/SF2 or SC35 upon deletion of 1–129 amino acids (M13: Δ1–129) suggests that this site does not have a role in the interaction with other splicing regulators. Although further studies are needed to verify its role(s), we recently obtained evidence that this site is required for homodimeric interaction (data not shown). There is also another possibility that this coiled-coil domain can function as an RRM. For example, it has been reported that RNG105, a novel RNA-binding protein in neuronal RNA granules, directly binds to the mRNA through its coiled-coil domain (37). However, as this site is predicted to be located between two potential RRMs in the N terminus, it is also plausible that this site can stabilize RNA binding mediated by two RRMs, presumably through homodimeric interaction. Therefore, further studies are now under way to identify the relationship between the coiled-coil domain and two RRMs in NSrp70.
The interaction of NSrp70 with SC35 and/or ASF/SF2 through the RS-like region led us to investigate whether NSrp70 could affect the function of SC35 or ASF/SF2. However, we did not see any difference in terms of alternative splicing activity and speckle localization when NSrp70 expression was down-regulated by siRNA (data not shown). Functional redundancy could be one of the possible reasons. For example, ASF/SF2 and SC35 were shown to be functionally interchangeable in in vitro splicing assays; both activate constitutive splicing in SR protein-depleted extracts (also called S100 extracts) and induce the same splice site selection in alternatively spliceable model pre-mRNAs (38). It is also possible that NSrp70 is not a major protein that is involved in mRNA splicing. However, the lack of progeny of NSrp70 knockout mouse suggests that this protein is essential for development, although the exact mechanism behind embryonic lethality due to NSrp70 knockout is not currently defined. Since we did not see any embryo that shows a homozygotic genotype, we believe that this gene is involved in very early stages of embryonic development. Therefore, we need to examine the effect of NSrp70 in early stages, including blastocyst and morula. Similarly, it has been demonstrated that SRp20, a splicing factor belonging to the highly conserved family of SR proteins, is essential for early embryonic development, and that mutant preimplantation embryos fails to form blastocytes and died at the morula stage (39). Knockout of some individual SR genes in mice is also known to result in early embryonic lethality (10,12,39). Taken together, these in vivo results suggest that SR proteins have their own distinctive role in vivo rather than a mere functional redundancy. Identification of distinct DNA sequences in alternatively spliced exons that are responsive to specific SR proteins also reveals the importance of each SR protein among redundant SR proteins in vivo (40).
We have initially identified NSrp70 gene by microarray as a candidate gene that may affect T-cell migration. We confirmed that NSrp70 is highly expressed in motile Jurkat T cells compared to non-motile Jurkat T cells (Supplementary Figure S1A). In addition, in silico analysis based on BioGPS demonstrated that NSrp70 was highly expressed in some immune-related cells, and this result was supported by Northern blot analysis (Supplementary Figure S1B and C). Nevertheless, overexpression or knockdown of NSrp70 did not show any effect in terms of chemokinesis by hSDF-1α and T-cell activation by PMA and A23187 (Supplementary Figure S1D and E). Interestingly, however, a recent report demonstrated that this gene is 2.4-fold highly expressed in two invasive and metastatic subpopulations of tumor cells isolated from two different mammary tumor types (PyMT and MTLn3) (data are available at the Cell Migration Gateway consortium website; http://cmckb.cellmigration.org). More interestingly, according to the WormBase database (http://www.wormbase.org/), there were variations in the migration pattern of distal tip cells compared to control animals when NSrp70 expression was disrupted by RNAi. These findings support the idea that NSrp70 could still be involved in cell migration. Further studies are needed to verify the role of NSrp70 in immune cell migration.
In conclusion, we identified a novel protein, NSrp70, with structural features similar to certain SR-related proteins, and characterized its subcellular localization in nuclear speckles. We further delineated its function with regard to modulation of pre-mRNA alternative splicing in vivo based on its subnuclear speckled distribution pattern and physical interaction with SC35 and ASF/SF2. In addition, we showed that this protein is essential for early embryonic development. Further study, in addition to the current results, of this novel splicing regulator may provide us with new information on RNA biogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDINGS
Funding for open access charge: Cell Dynamics Research Center (2010-0001621); Mid-career Researcher Program of the National Research Foundation of Korea/Ministry of Education, Science and 65 Technology (2008-0060838); Korean government, Korea Research Foundation Grant (KRF-2008-314-C00265); BioImaging Research Center at GIST; Korean government, Korea Research 70 Foundation Grant (KRF-2008-314-C00265).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Brian J. Morris for the Tra2β1, CD44 and Srp20 minigenes, and the pEGFP-ASF/SF2 plasmid; Dr Michael Sattler for the Fas minigene; and Dr M. Andrea Markus for discussion and suggestions throughout the study. We also wish to thank Dr Michelle Tallquist for advice on the mouse genotyping experiment. We are grateful to the members of CARD in Kumamoto University for providing the Ayu21-T93 mouse line.
REFERENCES
- 1.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 2.Huang S, Spector DL. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc. Natl Acad. Sci. USA. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–285. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 5.Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 6.Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl Acad. Sci. USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 8.Tacke R, Manley JL. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 1999;11:358–362. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 9.Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang HY, Xu X, Ding JH, Bermingham JR, Jr, Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Fu XD. Conditional knockout mice to study alternative splicing in vivo. Methods. 2005;37:387–392. doi: 10.1016/j.ymeth.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr, Ye Z, Liu F, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Moroy T, Heyd F. The impact of alternative splicing in vivo: mouse models show the way. RNA. 2007;13:1155–1171. doi: 10.1261/rna.554607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 15.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoss O, Stoilov P, Hartmann AM, Nayler O, Stamm S. The in vivo minigene approach to analyze tissue-specific splicing. Brain Res. 1999;4:383–394. doi: 10.1016/s1385-299x(99)00043-4. [DOI] [PubMed] [Google Scholar]
- 17.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. The EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 20.Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5'-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 21.Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- 23.Escobar AJ, Arenas AF, Gomez-Marin JE. Molecular evolution of serine/arginine splicing factors family (SR) by positive selection. In Silico Biol. 2006;6:347–350. [PubMed] [Google Scholar]
- 24.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. The FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 25.van Der Houven Van Oordt W, Newton K, Screaton GR, Caceres JF. Role of SR protein modular domains in alternative splicing specificity in vivo. Nucleic Acids Res. 2000;28:4822–4831. doi: 10.1093/nar/28.24.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundt K, Haga IV, Huitfeldt HS, Ostvold AC. Identification and characterization of two putative nuclear localization signals (NLS) in the DNA-binding protein NUCKS. Biochim. Biophys. Acta. 2007;1773:1398–1406. doi: 10.1016/j.bbamcr.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Araki M, Araki K, Yamamura K. International Gene Trap Project: towards gene-driven saturation mutagenesis in mice. Curr. Pharm. Biotechnol. 2009;10:221–229. doi: 10.2174/138920109787315006. [DOI] [PubMed] [Google Scholar]
- 28.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 30.Peres Lopes GM, de Souza SJ. Dissecting the human spliceosome through bioinformatics and proteomics approaches. J. Bioinform. Comput. Biol. 2004;1:743–750. doi: 10.1142/s0219720004000405. [DOI] [PubMed] [Google Scholar]
- 31.Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 32.Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson MS, Nakagawa TY, LeVan K, Dreyfuss G. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol. Cell. Biol. 1987;7:1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J, Krainer AR. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiina N, Shinkura K, Tokunaga M. A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 2005;25:4420–4434. doi: 10.1523/JNEUROSCI.0382-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu XD, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc. Natl Sci. Acad. USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 40.Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]