Abstract
Escherichia coli SOS functions constitute a multifaceted response to DNA damage. We undertook to study the role of yafP, a SOS gene with unknown function. yafP is part of an operon also containing the dinB gene coding for DNA Polymerase IV (PolIV). Our phylogenetic analysis showed that the gene content of this operon is variable but that the dinB and the yafP genes are conserved in the majority of E. coli natural isolates. Therefore, we studied if these proteins are functionally linked. Using a murine septicaemia model, we showed that YafP activity reduced the bacterial fitness in the absence of PolIV. Similarly, YafP increased cytotoxicity of two DNA damaging nitroaromatic compounds, 4-nitroquinoline-1-oxide (NQO) and nitrofurazone, in the absence of PolIV. The fact that PolIV counterbalances YafP-induced cytotoxicity could explain why these two genes are transcriptionally linked. We also studied the involvement of YafP in genotoxic-stress induced mutagenesis and found that PolIV and YafP reduced NQO-induced mutagenicity. The YafP antimutator activity was independent of the PolIV activity. Given that YafP was annotated as a putative acetyltransferase, it could be that YafP participates in the metabolic transformation of genotoxic compounds, hence modulating the balance between their mutagenicity and cytotoxicity.
INTRODUCTION
Living organisms possess DNA repair enzymes capable of repairing practically all types of DNA lesions they encounter in natural environments. Some genes coding for DNA repair enzymes are constitutively expressed, while some are expressed only when the amount of a particular DNA lesion exceeds a certain threshold. A factor determining such a threshold is for example, the amount of unprotected single-stranded (ss)DNA. During DNA replication and transcription ssDNA is transiently present at low concentrations and the proteins involved in those processes usually protect it. An increase in the concentration of unprotected ssDNA is frequently a secondary consequence of the DNA lesions caused by cellular metabolites or environmental insults. Detection of unprotected ssDNA leads to the activation of the DNA damage response pathway in order to remove the lesions and restore genome integrity. One such inducible DNA repair pathway is the SOS system in bacteria (1). The SOS system is regulated by the RecA and LexA proteins. The persistent contact with ssDNA activates co-protease activity of the RecA protein, which promotes the self-cleavage of the transcriptional repressor LexA, thus inducing the SOS response. Escherichia coli SOS response induces the transcription of at least 40 genes of which many code for DNA maintenance functions, e.g. nucleotide excision repair, translesion-synthesis and homologous recombination (2,3).
Translesion-synthesis is not a DNA repair pathway because it does not remove lesions from DNA. However, this process is critical for cell survival as it allows complete genome replication in the presence of DNA lesions that block replicative DNA polymerases. Translesion-synthesis requires specialized DNA polymerases, most of which belong to the Y-family of DNA polymerases. E. coli possesses two Y-family DNA polymerases capable of bypassing lesions: Pol IV and Pol V, encoded by the dinB and umuDC genes, respectively (4). Pol IV belongs to the most ubiquitous branch of the Y-family DNA polymerases that are found in prokaryotes, eukaryotes and archaea (5). In vivo, Pol IV can proficiently and accurately perform DNA synthesis across a variety of replication blocking DNA base modifications, for example those induced by alkylating agents, benzo[a]pyrene, 4-nitroquinoline N-oxide, 4-nitroquinoline-1-oxide (NQO), nitrofurazone (NFZ) and methylglyoxal (6–9). The nature of the replication-blocking DNA adducts generated by these DNA damaging agents, i.e. N3-Adenine, N2-Guanine and N3-Guanine, suggests that the minor-groove replication blocking DNA lesions are cognate substrates for the Pol IV polymerase. Similar substrate specificity is conserved also for its mammalian homologue Pol κ (6,10). These DNA lesions are probably continuously generated in E. coli genome as suggested by a high amount of Pol IV polymerase, i.e. 250 molecules, in unstressed cells (11). Upon SOS induction, the amount of Pol IV increases 10-fold. The expression of the dinB gene was also shown to be controlled by RpoS, a sigma subunit of RNA polymerase, which acts as general stress response and stationary phase modulator (12).
In the standard laboratory E. coli strain K-12, the dinB gene was shown to be the first gene of an operon composed of four genes, dinB, yafN, yafO and yafP (13). Despite extensive study on the biochemical activity and the biological role of Pol IV DNA polymerase, the knowledge about the biological functions of yafN, yafO and yafP genes is still scarce. Recently, it was shown that YafO is a toxin that acts as a ribosome-dependent mRNA interferase inhibiting protein synthesis, while YafN is its associated antitoxin (14,15). Because the SOS response requires protein synthesis, it is unclear what is the role of this toxin–antitoxin system in cellular recovery from a genotoxic stress. yafP gene product has been annotated as putative N-acetyltransferase with acyl-CoA binding domain and it has been shown not to be a partner of the YafN/YafO toxin–antitoxin system (14). Deletion of yafP gene has no effect on growth under standard laboratory conditions. It neither has an affect on Pol IV catalyzed stress-induced mutagenesis (14) nor on Pol IV catalyzed error-free bypass of cytotoxic alkylating agents (9).
Here, we undertook to study the biological role of the YafP protein. Phylogenetic analysis of dinB operon showed that dinB and yafP genes are well conserved within E. coli species unlike yafN and yafO. Therefore, instead of using the E. coli MG1655 K-12 strain that has a four-gene operon, we decided to use the E. coli CFT073 uropathogenic strain because it has only dinB and yafP genes in this operon. By using a murine septicemia model, we showed that, in the absence of Pol IV, YafP activity is deleterious for bacterial fitness. We obtained the same effect using NQO and NFZ, two mutagenic and carcinogenic compounds, which generate cognate DNA lesions for Pol IV (6). However, we also observed that YafP reduces NQO mutagenicity. Taken together, our results strongly suggest that YafP protein participates in metabolic transformation of DNA damaging compounds. We suggest that, by acetylating such compounds, or their derivatives, YafP modifies the nature of adducts they form, hence modulating the balance between their mutagenicity and cytotoxicity.
MATERIALS AND METHODS
Bacterial strains
E. coli strains used in this study were the E. coli CFT073 uropathogenic strain belonging to B2 phylogenetic group (16) and the 72 strains from ECOR collection of natural isolates representative of E. coli species genetic diversity (17). Low copy number plasmid pGB2 and pGB2 carrying functional dinB gene under the control of its own promoter were kindly provided by R. Fuchs (18).
Phylogenetic analysis of the dinB operon structure
dinB operon structure from sequenced genomes of E. coli strains was analyzed using NCBI genome database (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html) and Genoscope MAGE database (https://www.genoscope.cns.fr/agc/microscope/home/index.php) (19). Structure of the dinB operon in genomes of the strains from ECOR collection was determined by PCR, using one primer matching 3′-extremity of the dinB gene (5′-cgtaaaacctgggatgaacg) and another matching 5′-extremity of yafP gene (5′ttcgtcaatctgcgcccagg). Size of the PCR products was determined using agarose gel electrophoresis. In operons for which this analysis indicated that they contain only two genes, PCR products were sequenced and aligned using clustal W software. Hence, the consensus sequence of the fusion between dinB and yafP genes was established. Four strains were excluded from this analysis: ECOR 9, which has an insertion sequence in the dinB genes leading to a non-functional operon and ECOR 5, 64, 47 strains. For the latter three strains, no PCR amplification was obtained suggesting that dinB operon region could be rearranged or deleted in genomes of these strains. ECOR strain phylogeny used in this study is from (20). Briefly, sequences of six housekeeping genes (trpA, trpB, pabB, putP, icd and polB) obtained by multilocus sequence typing (MLST) were concatenated in a 5919 bp sequence, then maximum-likelihood tree has been constructed using PHYML program and the GTR + G + I model. Bootstrap values correspond to 1000 resampling trees.
Media and chemicals
Overnight bacterial cultures were grown in 5 ml Luria Broth (LB, Difco) medium at 37°C with agitation at 200 r.p.m. Colony forming units (CFU) were scored on LB agar 15 g/l (Difco) plates after 24 h incubation at 37°C. When needed, antibiotics (all purchased from Sigma Aldrich) were added at the following concentrations: chloramphenicol (Cm) 30 µg/ml, ampicillin 100 µg/ml, spectinomycin (Spec) 75 µg/ml, streptomycin (Str) 50 µg/ml. DNA damaging agents: NQO, NFZ, methyl methanesulfonate (MMS) and mitomycine C (MMC) were all purchased from Sigma Aldrich.
Construction of dinB operon mutants
Sequence of CFT073 dinB yafP operon has been recovered from MAGE database (19). E. coli CFT073 dinB operon mutants were constructed using classical gene replacement method (21). The pKD3 vector served as template to amplify chloramphenicol resistance (CmR) cassette flanked by FRT sites. Primers used for deletions of dinB and yafP genes have been chosen inside genes in order not to disturb promoter or ATGA sequence. ATGA is an overlap sequence of dinB and yafP genes. The sequences of the primer used are as follow: Pdinb1: gtggagatgcgcgacaatcccgccctgcgcgatatccctattgctattggcggtgtaggctggagctgcttc; Pdinb2: ggcgagcaatcagtaaatcaggttttacctttgccagacggcgttcaagttccatatgaatatcctccttag; Pyafp1: ctatcagcctggtgattttcagcaactatgcgctattttcgtgtaggctggagctgcttc; Pyafp2: gcgcacatactagacgggcgttattttcattgcaagctggcatatgaatatcctccttag. We showed that the replacement of dinB gene by CmR cassette did not prevent expression of the yafP gene using quantitative PCR (data not shown). Two different full deletions of the whole operon have been constructed. One by deleting both genes simultaneously using Pdinb1 and Pyafp2 primers (dinB yafP I) and the second by deleting of yafP gene in the dinB mutant using Pyafp2 and Pyafp1 primers (dinB yafP II). This second mutant (dinB yafP II) was used only in the septicemia assay; all the other experiments were done using the first mutant (dinB yafP I). When needed, CmR cassette was removed by expressing the FRT recombinase from the pCP20 vector as described in (21). The removal of the CmR cassette has been verified by PCR. Complementation of dinB gene function has been obtained by using the low copy number vector pGB2 carrying functional dinB gene under the control of its own promoter.
Construction of the YafP::YFP translational fusion
The C-terminal translational fusion between YafP and YFP venus proteins was constructed using classical gene replacement method as described in (21). Linear DNA including YFP, a CmR cassette flanked by FRT sites, and the sequence with homology for the recombination was obtained by PCR from Pr4 plasmid (22). Primer DNA sequences used for this amplification have as 5′-extension, an overhang coding for the last 40 nt of yafP gene without the stop codon, in frame with the sequence coding for a six amino acid linker (DPPVAT), and as 3′-extension, an overhang having identity with the 40 nt downstream yafP stop codon. The sequences of the primers are as follow: PYY1: cgtggtttactaatttttatatgcgatataaaccgcaacatgatccgccggtggcgaccatggctagcaaaggagaaga; PYY2: gcacataccaggcgggcgttattttcattgcaagctggatcctccttagttcctattccg. The linear amplicon has been transformed in electrocompetent CFT073 cell expressing the red gam recombinase via pkD46 vector.
Fluorescence activated cell sorting analysis
YafP::YFP production was monitored using BD fortessa cytometer with a 488 nm argon laser and 515–545 nm emission filter at a maximum of 2500 event per second. A minimum of 100 000 cells were analyzed per time point. Overnight cultures were diluted 1/500 v/v and grown in 15 ml LB medium at 37°C with agitation of 200 r.p.m. For measurements of spontaneous induction, samples were taken from untreated cultures every 30 min during 10 h and analyzed. SOS response was induced by addition of 1 µg/ml of MMC 30 min after dilution of an overnight culture. For measuring YafP::YFP production after MMC treatment, samples were taken immediately after addition of the drug and every 15 min subsequently, for a maximum of 45 min to avoid the presence of large filamentous cells. For all time points, approximately 106 cells were collected, washed and resuspended in sterile 1× PBS pH 7.2 prior to analysis. Flowdjo software (treestar) was used for data analysis.
In vitro competition assays
To determine the relative fitness of different bacterial strains we performed batch culture competitions. Fresh overnight cultures of competing strains were diluted at 1 : 1000 v/v into 5 ml of LB medium, at a 1 : 1 ratio, and incubated at 37°C at 200 r.p.m. Every day, for 5 days, overnight cultures were diluted 1 : 1000 v/v into fresh LB medium allowing growth of about 10 generations per day. CFU counts were determined by serial dilution spread on LB agar plates with (LBCm) or without chloramphenicol. Repicking LB colonies on LBCm plates served as control for CmSensitive/CmR ratio. Competitive index was calculated as follows: CI = [(LBt+1 − LBCmt+1)/LBCmt+1]/[(LBt0 − LBCmt0)/LBt0].
To measure differential long-term survival of different strains, LB medium was inoculated and incubated as described above. After 5 days of incubation, the number of CFUs was assayed by serial dilution spread on LB or LBCm plates.
In vivo competition assay
The mouse model of extraintestinal virulence used for E. coli CFT073 was previously described in (23). Animal experiments were performed in compliance with the recommendations of the French Ministry of Agriculture and approved by the French Veterinary Services (accreditation A 75-18-05). 200 µl of a suspension of about 109 bacteria/ml in physiological serum was inoculated subcutaneously in 14–16 g female mice (Charles River OF1). In this model, CFT073 killed all of the inoculated mice in around 24 h, whereas E. coli K-12 MG1655 did not have effect on mice survival (23). To avoid post-mortem bacterial proliferation, mice were sacrificed 20 h after inoculation. At this time point, mice presented clinical symptoms of severe septicemia. The animals were prostrated with anorexia symptom and had erected hairs. Animals were sacrificed and bacteria were recovered from spleen and plated on LB and LBCm plates. When plasmids pGB2 and pGB2 carrying functional dinB gene were used, bacterial cells were plated on LBSpec Str and LBCm Spec Str. The data from, at least 15 mice, inoculated in at least three independent experiments per competition were used for this analysis. Competitive index was calculated as follows: CI = [(LB20 h post inoculation − LBCm20 h post inoculation)/LBCm20 h post inoculation]/[(LBt0 − LBCmt0)/LBt0]. The cost of CmR cassette was tested by competing yafP::FRT and yafP::CmR and dinB yafP::FRT I and dinB yafP::CmR I mutants. We found that CmR is neutral in our assay.
Growth curves
Cells from fresh overnight cultures were diluted 1/500 v/v and inoculated in 96-well microtiter plates containing LB or M9 minimal medium supplemented with different carbon sources: 0.4% glucose or 0.2% casamino acids or in LB medium supplemented with different concentrations of NQO. Microtiter plates were incubated at 37°C in microplate reader incubator (IEMS reader system from Labsystems). Optical density at 600 nm was measured every 5 min for 12 h. Plates were vigorously shaken for 1 min prior to each measurement.
Spot tests
Fresh overnight cultures were serially diluted in MgSO4 10−2M. 7 µl of each dilution were spotted onto LBSpec Str agar plate as control and on LB agar plates containing different concentration of DNA damaging agents: 10 µM NFZ, 10 µM NQO and 1 µM MMS. Spec and Str were used in the solid medium in order to select cells carrying the pGB2 vector. NFZ and NQO were always freshly prepared in DMF avoiding light exposure. Because of the extreme light sensitivity of NFZ, spot assays were conducted in dark room and the plates were incubated in dark. Growth of spots was first determined 12 h after inoculation and then followed every hour for the next 10 h.
Mutagenesis assay
Cells from fresh overnight cultures (diluted 1/1000 v/v), were inoculated into LB medium containing or not 10 µM NQO, and incubated for 20 h at 37°C at 200 r.p.m in the dark. 200 µl of each culture was plated on LB agar plate with 100 µg/ml rifampicine (Rif). Total number of CFUs was determined on LB agar plate. Growing colonies have been scored after 2 days of incubation at 37°C.
RESULTS
Phylogenetic analysis of the dinB, yafN, yafO and yafP operon within E. coli species
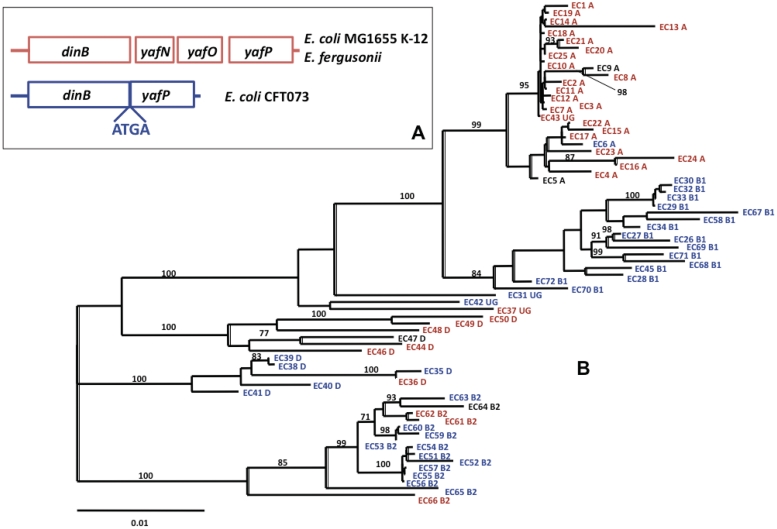
The genome sequences of 98 E. coli (including eight Shigella) strains are currently available at the NCBI genome database (http://www.ncbi.nlm.nih.gov/genomes). Database searches showed that dinB, yafN, yafO and yafP operon structure found in standard laboratory E. coli K-12 strain is not conserved across E. coli species. While, 60% of strains have the four-gene operon, 40% have only dinB and yafP genes. In this latter case, dinB and yafP genes are always fused together and overlap by 4 nt, i.e. ATGA (Figure 1A). The termination codon of dinB overlaps with the initiation codon of yafP. This structure of two-gene operon probably resulted from the recombination between two ATGA sequences present in the four-gene operon resulting in deletion of yafN and yafO genes.
Figure 1.
Phylogenetic analysis of the dinB, yafN, yafO and yafP operon within the E. coli species. (A) Schematic representation of the four-gene operon as found in E. coli K-12 MG1655 and E. fergusonii and the two-gene operon as found in E. coli CFT073. In the latter case, dinB and yafP genes are fused together and overlap by four nucleotides, i.e., ATGA (B) Structure of the dinB operon in genomes of the strains from ECOR collection representative of the E. coli strain phylogeny. The strain phylogenic tree was reconstructed from MLST data (20). Numbers at nodes represent bootstrap values superior to 70%. Strains labelled in red or blue possess either the four-genes operons or the two-genes operons, respectively. The four strains labelled in black correspond to strains whose operons were not analyzed for following reasons: ECOR 9 strain has an insertion sequence in the dinB genes leading to a non-functional operon, and ECOR 5, 64, 47 strains for which no PCR amplification was obtained suggesting that dinB operon region could be rearranged or deleted.
To extend this observation to more strains and to have a better overview of different E. coli phylogenetic groups, we performed PCR analysis of this operon in 72 E. coli strains from the ECOR collection of E. coli natural isolates using primers flanking yafN and yafO genes. The resulting PCR product was sequenced and analyzed (Figure 1B). We used MLST phylogenic tree representation of the ECOR collection to have a view of variation in operon composition among E. coli strain phylogenetic history. We found that in 50% of the strains, this operon does not have yafN and yafO genes. In those strains, dinB and yafP are always fused with ATGA sequence overlap, as observed previously. Clearly this pattern has a strong phylogenetic inertia. We noticed that almost all of the strains from phylogenetic group A have four-gene operon, while all B1 strains have two-gene operon. Strains from phylogenetic groups B2 and D, as well as ungrouped strains have both types of operons. Escherichia fergusonii, which is the most closely related species to E. coli possesses a four-gene operon. Taken together, our data show that dinB and yafP are present in nearly all E. coli studied strains, while yafN and yafO genes are not. The most parsimonious scenario is that yafN and yafO have been lost at least three times, once in the B2 group, once in the D group and once in the B1 group. This observation suggests the association of dinB with yafP and of yafN with yafO and, on the other hand the independence of both pairs of genes.
YafP protein synthesis
In order to study the biological role of the YafP protein, we used E. coli CFT073 uropathogenic strain as a model organism. This strain, referred as wild-type (WT) in the text, has only dinB and yafP genes in the operon, which eliminates possible confounding effects of the yafN and yafO genes. The genomic rearrangement event that deleted the yafN and yafO genes resulted in the dinB yafP gene fusion, which is transcribed from the dinB gene promoter producing a single messenger RNA. As this event puts yafP out of frame, and given that we were not able to detect a Shine Dalgarno sequence, the translation of the YafP protein may require translational frameshifting.
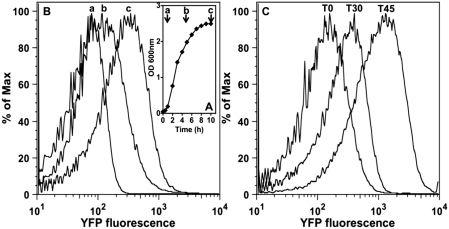
In order to study the conditions under which YafP is translated, we constructed a translational fusion between the yafP gene and the yellow fluorescent protein (YFP) and used fluorescence activated cell sorting (FACS) to monitor the fluorescence of cells. We followed cell fluorescence at different cell growth phases in the absence of SOS-inducing treatment (Figure 2A and B). Cell fluorescence was low during logarithmic growth phase, but it became much stronger upon entry to stationary phase. Exponentially growing cells treated with the SOS-inducing DNA damaging agent MMC produced a large amount of YafP–YFP protein fusion (Figure 2C). This is in accordance with the observation that the expression of E. coli K-12 dinB operon, including yafP, increases upon UV-induced SOS response (3,13). Therefore, it can be concluded that YafP protein is synthesized in CFT073 cells in spite of the fact that it requires translational frameshifting and that the regulation of CFT073 dinB operon appears to be similar to that one in K-12.
Figure 2.
Synthesis of the YafP–YFP protein fusion. A translational fusion between yafP gene and the yellow fluorescent protein (YFP) was used to monitor the synthesis of the YafP–YFP protein fusion in cells. (A) Growth curve of untreated cells. a, b and c represent three different time points at which the cells were sampled for (B) measurement of fluorescence using FACS analysis. (C) Cells sampled at b time point were treated with 1 µg/ml of the SOS-inducing DNA damaging agent Mitomycine C (MMC) and fluorescence was measured 30 min and 45 min after. Experiments were repeated three times. Representative curves are shown.
Fitness of dinB yafP operon mutants
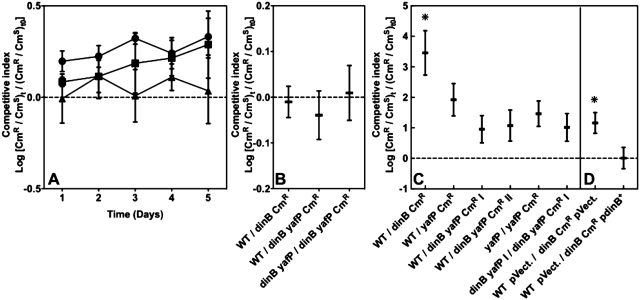
In order to find a phenotype associated with the YafP protein, we constructed a series of mutant derivatives of the CFT073 strain: dinB, yafP and dinB yafP. First, we assessed the growth rates of these mutants relative to the CFT073 strain in different media (LB, M9 minimal medium with different carbon sources) and found no differences. CmR cassette used for gene deletion has no effect on growth. Because competitions allow measurements of small differences in growth rates and survival, we performed in vitro batch culture competitions in LB between WT strain and dinB or dinB yafP mutants (Figure 3A). We found that there was no difference in fitness between these strains during five cycles, where each cycled allowed about 10 generations of growth. In addition, we monitored survival of different strains in mixed stationary phase cultures for 5 days (Figure 3B). There was no difference between the WT strain and dinB or dinB yafP mutants in this condition either. Hence, the phenotype of the yafP gene mutant cannot be detected under standard laboratory in vitro growth conditions.
Figure 3.
Fitness of CFT073 dinB yafP operon mutants. (A and B) Survival and fitness of dinB yafP operon mutants during in vitro competitions. (A) Competitive index of different mutants during batch culture competitions in LB: wild type (WT) and dinB (filled diamond); WT and dinB yafP (filled square); dinB yafP::FRT and dinB yafP::CmR (filled circle). The last competition was used as an evaluation of the cost of the CmR cassette. (B) Competitive index of different strains co-inoculated and incubated in LB for 5 days. Each point represents the mean (±SE) values from 3 independent experiments. (C and D) fitness of dinB yafP operon mutants during septicemia in the mouse model. (C) Competitive index of the WT and the different mutants, as well as measurements of CmR cassette cost during the competition. The latter was performed by competing yafP::FRT with yafP::CmR, and dinB yafP::FRT I with dinB yafP::CmR I mutants. (D) Competitive index of the WT and the dinB strain, which carried the pGB2 plasmid or the pGB2 bearing the functional dinB gene. Each point represents the mean (±SE) values from, at least 15 mice inoculated in at least three independent experiments. Asterisks show the competition for which the competitive index was significantly different (Wilcoxon test P < 0.05).
Because E. coli CFT073 is an extraintestinal pathogenic strain isolated from blood and urine of a woman with acute pyelonephritis (16), we decided to assess the fitness of different mutant strains by performing competition experiments using a mouse sepsis model. Twenty hours after inoculation, when animals presented the clinical symptoms of severe septicemia, animals were sacrificed and the number of live bacterial cells in spleen was established. We tested the cost of the CmR cassette by competing mutants yafP versus yafP CmR and dinB yafP I versus dinB yafP I CmR. We observed that the CmR cassette had no effect in our assay. The dinB mutant showed ∼1000-fold reduced fitness relative to the WT (Wilcoxon test, P < 0.05; Figure 3C). No other mutants showed significantly different fitness relative to the WT strain.
Interestingly, the inactivation of the yafP gene in the dinB mutant background restored fitness equivalent to that of WT strain. Similarly, when both genes (dinB and yafP) were deleted simultaneously as opposed to sequentially as in the previous case, the fitness of the resulting strain was equivalent to the WT strain. To ascertain that the dinB mutant did not carry an additional mutation responsible for the observed phenotype, we complemented dinB function using a low copy number plasmid carrying dinB controlled by its own promoter (Figure 3D). The presence of an empty plasmid had an important effect on competition, probably due to the cost incurred to the host cells by the plasmid. However, dinB mutant still showed significant, 10-fold reduction of fitness relative to WT strain (Wilcoxon test, P < 0.05). The expression of a functional dinB gene in trans restored the fitness of the dinB mutant back to the level of the WT strain. These data suggested that, during sepsis in mice the activity of YafP is toxic for cells in the absence of the Pol IV.
YafP protein mediates toxicity of genotoxic chemicals
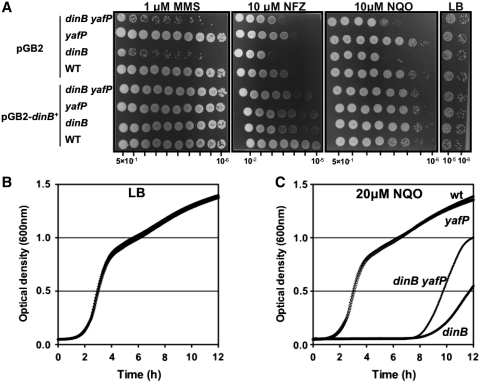
Because we observed that the presence of YafP is deleterious for bacteria in the absence of Pol IV in an in vivo stressful condition (Figure 3C), we hypothesized that YafP activity could be involved in the generation or in the processing of DNA lesions that are substrates for Pol IV. Thus, we tested the toxicity of several well known DNA damaging agents by spotting serial dilutions of the different CFT073 mutants having either an empty plasmid or plasmid expressing the functional dinB on LB plates containing DNA damaging agents (Figure 4A). Because we showed that YafP protein is synthesized upon treatment of cells with SOS inducing MMC, we tested resistance of WT strain and the different mutants to this DNA damaging agent and found no difference (data not shown). This suggests that neither YafP protein nor DinB are involved, directly or indirectly, in SOS induction via MMC or in the repair of DNA crosslinks.
Figure 4.
Sensitivity of dinB yafP operon mutants to different DNA damaging agents. (A) Sensitivity to 1 µM MMS, 10 µM NFZ and 10 µM NQO was estimated by spotting 7 µl of 5-fold serial dilutions of overnight cultures of the WT, dinB, yafP and dinB yafP strains onto LB plates containing DNA damaging agents. Strains carried the pGB2 plasmid or the pGB2 coding for the functional dinB gene. Experiments were repeated three times. Representative results are shown. (B and C) Effect of NQO on growth of WT, dinB, yafP and dinB yafP strains in liquid LB (B) or LB with 20 µM NQO (C). Experiments were repeated three times. Representative growth curves are shown.
We then tested the resistance to MMS because it was previously shown that K-12 strain Pol IV contributes to the tolerance of cytotoxic alkylating DNA lesions (9). We found that CFT073 Pol IV also counteracts the cytotoxic effects of MMS, but also that there is no contribution of YafP in this process. This suggests that YafP does not modulate, directly or indirectly, Pol IV activity. Next, we tested the resistance to NQO and NFZ. These two DNA damaging agents generate replication-blocking N2-guanine adducts, which can be bypassed efficiently by K-12 Pol IV polymerase in an error free manner (6). We found that, in the absence of Pol IV YafP increases the toxicity of NQO and NFZ about 5-fold. This effect was completely abolished by the trans-complementation using functional dinB carried on the pGB2 plasmid. To support these results, we also tested the effect of NQO on the growth rates of the WT strain and the different mutants in liquid LB supplemented with 20 µM NQO (Figure 4B and C). We found that the growth of WT and yafP strains was not affected by NQO. However, the growth of dinB and dinB yafP mutants was retarded in the presence of NQO. As in spot tests, the toxicity of NQO to dinB defective cells was reduced by the inactivation of the yafP gene.
Antimutator activity of the YafP protein
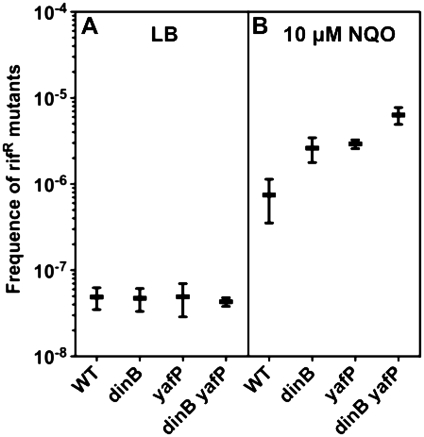
NQO is a DNA damaging agent that reacts with DNA only after metabolic activation (6). Because acetylation changes the reactivity of NQO with DNA (24), and because YafP is a putative N-acetyltransferase, we tested whether YafP plays a role in NQO mutagenicity (Figure 5). It was previously shown that the inactivation of the dinB gene increases NQO-induced mutagenesis (6), but the role of YafP was not tested. Strains were treated with 10 µM NQO. Mutagenic effect was estimated by scoring of rifampicin resistant (RifR) mutants. While the RifR mutant frequency was identical for all strains in the absence of the NQO treatment, all tested strains showed significant NQO-induced mutagenesis (Figure 5). The ratio of median values of NQO-induced RifR mutant frequencies and RifR mutant frequencies of untreated controls were: 9-, 57-, 94- and 113-fold for parental, dinB, yafP and dinB yafP strains, respectively. dinB, yafP and dinB yafP strains had significantly higher NQO-induced mutant frequency relative to the WT strain (Mann–Whitney P < 0.05). dinB yafP strain had significantly higher RifR mutant frequency compared to the dinB strain. yafP strain did not show significantly different RifR mutant frequency compared to either dinB or dinB yafP strain. In conclusion, these data show an epistatic effect of Pol IV and YafP proteins on reduction of NQO mutagenicity.
Figure 5.
Effect of Pol IV and YafP on NQO-induced mutagenesis. Frequency of spontaneous (A) and NQO-induced (B) rifampicin resistant (RifR) mutants in WT, dinB, yafP and dinB yafP strains. Each point represents the mean (±SE) values from at least five replicates in three independent experiments. The median values for WT, dinB, yafP, dinB yafP RifR frequency mutants are respectively: 3.30 × 10−8, 3.04 × 10−8, 2.71 × 10−8, 4.21 × 10−8 in LB and 2.86 × 10−7, 1.74 × 10−6, 2.54 × 10−6, 4.74 × 10−6 in LB with 10 μM NQO.
DISCUSSION
Operons are widespread genetic structures in bacteria, which often encode functionally linked proteins. Phylogenetic linkage of operon members often indicates importance of their functional interactions for the host. We found that E. coli K-12 strain dinB, yafN, yafO and yafP operon organization is not conserved within the species. yafN and yafO are absent from many strains. In contrast, dinB and yafP are present and transcriptionally linked in 57% of strains analyzed in this study. Because the four-gene operon was found in the genome of E. fergusonii, which is the species phylogenetically closest to E. coli, we hypothesized that the ancestral form of this operon in Escherichia contains four genes. Therefore, the deletions of the yafN yafO module, leading to the fusion of dinB and yafP genes, most probably occurred after separation from the common Escherichia ancestor.
The deletion of yafN and yafO occurred in half of the studied strains producing always the same partial overlap of dinB and yafP genes. This genomic configuration that includes translational overlaps is not unusual in bacteria (25). The overlap according an ATGA sequence is the most predominant in overlapping sequences in the E. coli genome (26). Hence, the synthesis of YafP probably requires translational frameshifting. In E. coli, translational frameshifting was found to be involved in the synthesis of several proteins, e.g., protein release factor 2 or TrpR repressor (27,28). The frequency of translation frame shifting depends on many factors including the levels of translation initiation and ribosome pausing. A regulatory sequence called downstream box (DB) was also implicated in the initiation of translation independent of the Shine Dalgarno sequence (29). The yafP gene sequence presents a high identity with the consensus sequence of DB (11/15 nt at the position −2 from the ATG). Consequently, the relative ratio of proteins can vary in different conditions. In the case of the dinB yafP operon, the requirement of translational frameshifting could assure that YafP protein is present in lower amounts than Pol IV when not needed and that YafP is in no case present without the DinB protein.
The 1000-fold loss of fitness of the dinB mutant strain that has a functional yafP gene during sepsis in mice shows that YafP activity is deleterious in vivo in the natural host of E. coli in the absence of Pol IV. No such phenotype was observed under the standard laboratory growth conditions. YafP protein has been annotated as putative a N-acetyltransferase with acyl-CoA binding domain. N-acetyltransferases catalyze the transfer of an acetyl group from acetyl coenzyme A to the nitrogen of primary arylamines, hydrazines and their N-hydroxylated derivatives (30). These enzymes play an important role in detoxification and/or metabolic activation of numerous xenobiotics (31). Given the difficulty of identifying the natural substrate of YafP in host of E. coli, and based on the putative function of this protein, we decided to test the effect of YafP on genotoxicity (i.e. cytotoxicity and mutagenicity) of some well characterized chemical compounds.
The DNA damaging properties of nitroaromatic compound depend on their metabolic activation, which requires reduction or acetylation by cellular enzymes (32). In this study, we used two such chemicals, NQO and NFZ, because it was previously shown that Pol IV reduces their cytotoxicity in E. coli (6). Both chemicals produce replication blocking adducts at the N2 position of the guanine, which are accurately bypassed by Pol IV (6). NFZ is a synthetic antibacterial compound used for the treatment of burns and skin graft patients (33). The catabolism of NFZ by cellular enzymes was shown to generate many different byproducts (34). In E. coli, this process starts by a reduction of NFZ by a nitroreductase that generates hydroxylamine derivatives that are highly reactive with proteins and DNA (35). It has been reported that in vitro acetylation of NFZ derivatives greatly increases their stability (36). NQO is a carcinogenic compound whose ultimate carcinogenic form is acetyl-4-hydroxyamino-quinoline-1-oxide (AcHAQO) (37), indicating the importance of acetyltransferases for its activation in vivo. We observed that YafP contributes to the toxicity of NQO and NFZ in the absence of Pol IV. This suggests that YafP may participate in the metabolic activation of these compounds. The deleterious effect of YafP on the cells resistance to NQO and NFZ is ∼5-fold. The observed YafP-mediated cytotoxicity is readily neutralized by Pol IV activity, which could explain why those two genes are transcriptionally linked. Consequently, YafP activity does not constitute a major handicap for the WT cells.
YafP also has antimutator activity. It reduces the NQO-induced mutagenicity by ∼90% (9-fold mutation frequency induction for WT strain versus 94-fold mutation frequency induction for yafP strain). How can these two different phenotypes, i.e. enhancement of cytotoxicity and antimutator activity, be explained? By changing the acetylation status of DNA damaging metabolites YafP may modify the amount and/or the nature of the DNA adducts produced. As a consequence, this modification may change the propriety of the DNA lesion, hence modulating its mutagenicity. For example, it was found in vitro that NQO metabolite, 4-hydroxyamino-quinoline 1-oxide (HAQO), can be either mono- or di-acetylated (38). Both forms generate adducts at the same positions of purine nucleotides, but not with the same efficiency. Monoacetylated form of HAQO is much more reactive with DNA than the diacetylated HAQO derivative (38). YafP could favour diacetylation of HAQO, and therefore, reduce the amount of DNA lesions. However, diacetylation of HAQO might increase replication-blocking capacity of the generated DNA adduct. These adducts, bypassed by the Pol IV in an error free manner, could be responsible for the observed increase in cytotoxicity in the dinB deficient yafP proficient strain. However, this hypothesis remains to be demonstrated biochemically.
Concluding remarks
Stress-response regulons, like SOS, provide coordinated gene expression that is important for the adaptation and survival in natural environments. Conservation of regulon composition can vary as a function of specific environmental pressures. We found that dinB and yafP genes are conserved in the majority of E. coli strains, while yafN and yafO genes are not. This suggests that dinB and yafP functions are frequently solicited in E. coli natural environments. Our data indicate that YafP protein participates in metabolic transformation of some DNA damaging agents. For example, it reduces NQO-induced mutagenicity, but also increases NQO cytotoxicity. As Pol IV counterbalances this cytotoxicity, Pol IV and YafP proteins can be considered as a peculiar toxin–antitoxin system. This could explain why dinB and yafP genes are in the same operon with translational overlap. YafP antimutagenic activity fits well with the functions of SOS, which coordinately act to assure recovery from diverse genotoxic shocks with as little loss of integrity of genetic information as possible.
FUNDING
FP7-HEALTH-F3-2010-241476; French Ministry of Science and Education (to A.G.). Funding for open access charge: Inserm laboratory subvention.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank M. Selva and L. Garry for technical assistance.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. ASM Press: Washington, DC; 2006. [Google Scholar]
- 2.Erill I, Campoy S, Barbe J. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 2007;31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 3.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rattray AJ, Strathern JN. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- 5.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 6.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 7.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu P, Oum L, Geacintov NE, Broyde S. Nucleotide selectivity opposite a benzo[a]pyrene-derived N2-dG adduct in a Y-family DNA polymerase: a 5′-slippage mechanism. Biochemistry. 2008;47:2701–2709. doi: 10.1021/bi701839q. [DOI] [PubMed] [Google Scholar]
- 9.Bjedov I, Dasgupta CN, Slade D, Le Blastier S, Selva M, Matic I. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics. 2007;176:1431–1440. doi: 10.1534/genetics.107.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, Woodgate R. Eukaryotic Y-family polymerases bypass a 3-methyl-2′-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic Acids Res. 2008;36:2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu. Rev. Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- 12.Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenzie GJ, Magner DB, Lee PL, Rosenberg SM. The dinB operon and spontaneous mutation in Escherichia coli. J. Bacteriol. 2003;185:3972–3977. doi: 10.1128/JB.185.13.3972-3977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singletary LA, Gibson JL, Tanner EJ, McKenzie GJ, Lee PL, Gonzalez C, Rosenberg SM. An SOS-regulated type 2 toxin-antitoxin system. J. Bacteriol. 2009;191:7456–7465. doi: 10.1128/JB.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Yamaguchi Y, Inouye M. Characterization of YafO, an Escherichia coli toxin. J. Biol. Chem. 2009;284:25522–25531. doi: 10.1074/jbc.M109.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–49. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Medigue C. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, Weinert K, Tenaillon O, Matic I, Denamur E. Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog. 2009;5:e1000257. doi: 10.1371/journal.ppat.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elez M, Radman M, Matic I. The frequency and structure of recombinant products is determined by the cellular level of MutL. Proc. Natl Acad. Sci. USA. 2007;104:8935–8940. doi: 10.1073/pnas.0610149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 2006;194:1141–1150. doi: 10.1086/507305. [DOI] [PubMed] [Google Scholar]
- 24.Galiegue-Zouitina S, Bailleul B, Loucheux-Lefebvre MH. Adducts from in vivo action of the carcinogen 4-hydroxyaminoquinoline 1-oxide in rats and from in vitro reaction of 4-acetoxyaminoquinoline 1-oxide with DNA and polynucleotides. Cancer Res. 1985;45:520–525. [PubMed] [Google Scholar]
- 25.Normark S, Bergstrom S, Edlund T, Grundstrom T, Jaurin B, Lindberg FP, Olsson O. Overlapping genes. Annu. Rev. Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- 26.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl Acad. Sci. USA. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sipley J, Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc. Natl Acad. Sci. USA. 1993;90:2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benhar I, Miller C, Engelberg-Kulka H. Frameshifting in the expression of the Escherichia coli trpR gene is modulated by translation initiation. J. Bacteriol. 1993;175:3204–3207. doi: 10.1128/jb.175.10.3204-3207.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sprengart ML, Fatscher HP, Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sim E, Walters K, Boukouvala S. Arylamine N-acetyltransferases: from structure to function. Drug Metab. Rev. 2008;40:479–510. doi: 10.1080/03602530802186603. [DOI] [PubMed] [Google Scholar]
- 32.Diaz E, Ferrandez A, Prieto MA, Garcia JL. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 2001;65:523–569, table of contents. doi: 10.1128/MMBR.65.4.523-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCalla DR, Reuvers A, Kaiser C. Mode of action of nitrofurazone. J. Bacteriol. 1970;104:1126–1134. doi: 10.1128/jb.104.3.1126-1134.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCalla DR. Mutagenicity of nitrofuran derivatives: review. Environ. Mutagen. 1983;5:745–765. doi: 10.1002/em.2860050512. [DOI] [PubMed] [Google Scholar]
- 35.Race PR, Lovering AL, Green RM, Ossor A, White SA, Searle PF, Wrighton CJ, Hyde EI. Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme. J. Biol. Chem. 2005;280:13256–13264. doi: 10.1074/jbc.M409652200. [DOI] [PubMed] [Google Scholar]
- 36.Ebetino FF, Carroll JJ, Gever G. Reduction of Nitrofurans. I. Aminofurans. J. Med. Pharm. Chem. 1962;91:513–524. doi: 10.1021/jm01238a011. [DOI] [PubMed] [Google Scholar]
- 37.Bailleul B, Daubersies P, Galiegue-Zouitina S, Loucheux-Lefebvre MH. Molecular basis of 4-nitroquinoline 1-oxide carcinogenesis. Jpn J. Cancer Res. 1989;80:691–697. doi: 10.1111/j.1349-7006.1989.tb01698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailleul B, Galiegue S, Loucheux-Lefebvre MH. Adducts from the reaction of O,O’-diacetyl or O-acetyl derivatives of the carcinogen 4-hydroxyaminoquinoline 1-oxide with purine nucleosides. Cancer Res. 1981;41:4559–4565. [PubMed] [Google Scholar]