Abstract
Mitochondrial genome diversity in closely related species provides an excellent platform for investigation of chromosome architecture and its evolution by means of comparative genomics. In this study, we determined the complete mitochondrial DNA sequences of eight Candida species and analyzed their molecular architectures. Our survey revealed a puzzling variability of genome architecture, including circular- and linear-mapping and multipartite linear forms. We propose that the arrangement of large inverted repeats identified in these genomes plays a crucial role in alterations of their molecular architectures. In specific arrangements, the inverted repeats appear to function as resolution elements, allowing genome conversion among different topologies, eventually leading to genome fragmentation into multiple linear DNA molecules. We suggest that molecular transactions generating linear mitochondrial DNA molecules with defined telomeric structures may parallel the evolutionary emergence of linear chromosomes and multipartite genomes in general and may provide clues for the origin of telomeres and pathways implicated in their maintenance.
INTRODUCTION
Genome fragmented into multiple linear chromosomes terminating with telomeric arrays is a hallmark of the eukaryotic cell. In contrast, molecular architectures of genomes in prokaryotes and organelles vary substantially (1). For instance, certain animal mitochondrial DNAs (mtDNAs) are monomeric circles (2), kinetoplastid protists have networks of catenated circles (3,4), and most plants and fungal mitochondria contain linear (circularly permuted) concatemers that are heterogeneous in size (termed polydisperse linear DNA) (5–7). Finally, uniform linear mtDNAs terminating with defined terminal structures (mitochondrial telomeres) are found in a number of phylogenetically diverse taxa (8–28). In addition, mitochondria of numerous organisms contain multipartite genome; i.e. fragmented into multiple (from few to several hundred) circular or linear chromosomes (29–39).
The predominant genome architecture may even differ in closely related organisms, i.e. conceptually different (monomeric linear versus circular-mapping and linear polydisperse) or containing varying proportions of topologically different mtDNA molecules. For example, mitochondria of Candida glabrata and Saccharomyces cerevisiae contain polydisperse linear DNA molecules, with a minor fraction of circles and lariat structures generated by rolling circle replication (40). In contrast, a recent study revealed that mitochondria of C. albicans lack significant amounts of circular mtDNA molecules, containing predominantly a network of branched DNA structures with linear polydisperse mtDNA molecules—interpreted as recombination-driven replication (41,42). An alternative interpretation would be mitochondrial replication just like in S. cerevisiae, and a reduced level of circular replicative DNA molecules, due to more effective recombination that is also responsible for branched structures. Whatever the replication mechanism, physical mapping approaches such as restriction mapping, DNA sequencing or PCR amplification indicate that any of these populations of linear polydisperse mtDNA molecules have predominantly circularly permuted sequences (i.e. can be reasonably well represented as single sequence records as deposited in GenBank, but should not be labeled circular as enforced by the database management). Such genome architectures are in most cases illustrated as circular maps. In contrast, species containing linear mtDNA molecules terminating with specific telomeric structures are best depicted in linear maps (43). Accordingly, we term mitochondrial genomes as circular- or linear-mapping.
However, the picture is more complex in some yeast species with multiple forms of mitochondrial genomes. In Pichia pijperi and Williopsis mrakii, the linear mtDNA molecules terminating with telomeric hairpins (t-hairpins) coexist with monomeric and dimeric circles and linear polydisperse mtDNAs (11,44,45). Two types of DNA replicons occur in mitochondria of C. parapsilosis. Namely, the linear mtDNA molecules with telomeric arrays (t-arrays) of tandem repeats and multimeric minicircles derived exclusively from the sequence of mitochondrial telomeres (telomeric circles, t-circles) implicated in the telomere maintenance pathway (16,20,46–48).
At present, little is known about the biological roles of different mtDNA forms and molecular mechanisms leading to architectural alterations of the mitochondrial genomes. Our ambition is to identify these mechanisms and to uncover their role in the evolution of linear chromosomes and corresponding telomeric structures. Therefore, we initiated a large-scale comparative study of the mitochondrial genomes in yeast species closely related to C. parapsilosis, whose mitochondrial telomeres share a number of structural features with their counterparts at the ends of nuclear chromosomes (49,50). In previous reports, we described that strains of C. metapsilosis and C. orthopsilosis possess either linear-mapping mitochondrial genome, with similar architecture as found in C. parapsilosis, or a circularized (mutant) form of the genome (51,52). Moreover, we found that C. subhashii contains yet another type of linear mitochondrial genome, which does not come with any detectable circular or concatemeric form. Instead, its linear mtDNA terminates with invertron-like telomeres, with a protein covalently bound to both 5′ termini (10). The four Candida species containing linear mitochondrial genomes are classified within the monophyletic ‘CTG clade’ of Hemiascomycetes (53,54). The same phylogenetic group also contains species with circular-mapping mitochondrial genomes such as C. albicans (55), Debaryomyces hansenii (56) and Pichia sorbitophila (57). The occurrence of closely related organisms or even strains of the same species with different mitochondrial genome architecture is in line with the hypothesis that linear- and circular-mapping mitochondrial genomes do not exhibit a radical difference, but that the genome forms may sporadically interconvert via currently unknown molecular mechanism(s) (11,52).
In this study, we analyze the complete mtDNA sequences of eight additional Candida species. Our survey reveals that their molecular forms vary dramatically providing a unique opportunity for identification of structural elements and molecular mechanisms affecting the genome architecture. At the same time, our analysis provides an insight on the evolution of linear chromosomes and their telomeric structures.
MATERIALS AND METHODS
Yeast strains and cultivation
Yeast strains analyzed in this study are listed in Table 1. Yeasts were grown in liquid YPDG media (1% (w/v) yeast extract, 1% (w/v) peptone, 0.5% (w/v) glucose and 3% (v/v) glycerol), with constant shaking at 25–30°C until the late exponential phase.
Table 1.
Summary of the mitochondrial genome mapping in yeast species investigated in this study
| Species | Strain | Mitochondrial genome |
|||||
|---|---|---|---|---|---|---|---|
| Genome forma | Mitochondrial telomeresb | Genome sizea (bp) | % G + C | GenBank Acc. No. | Referencec | ||
| Candida alai | NRRL Y-27739T | Cd,e | 30 368e | 20.9 | HQ267968 | This study | |
| Candida albicans | CBS 562NT | Cd | This study | ||||
| SC 5314 | Ce | 40 420e | 32.2 | AF285261 | (85) | ||
| WO-1 | n.d. | 55 284 (unfinished)e | 32.6 | Broad institute | |||
| Candida blackwellae | CBS 10843T (AS2.3639T) | Cd | This study | ||||
| Candida bohiensis | NRRL Y-27737T | Cd | This study | ||||
| Candida buenavistaensis | NRRL Y-27734T | Cd | This study | ||||
| Candida chauliodes | NRRL Y-27909T | Cd | This study | ||||
| Candida corydali | NRRL Y-27910T | Cd | This study | ||||
| Candida dubliniensis | CD36T (CBS 7987T) | Cd,e | 34 732e | 30.6 | Sanger Institute, this study | ||
| Candida frijolesensis | NRRL Y-48060T | 3xL1d,e | t-hairpins | 37 215 (master chromosome)e | 21.6 | HM594866 | This study |
| Candida gigantensis | NRRL Y-27736T | Cd | This study | ||||
| Candida guilliermondii (Pichia guilliermondii) | ATCC6260T (CBS 566T)f | n.d. | 23 890 (unfinished)e | 25.7 | Broad Institute | ||
| Candida jiufengensis | CBS 10846T (AS2.3688T) | Cd,e | 29 672e | 28.4 | GU136397 | This study | |
| Candida labiduridarum | NRRL Y-27940T | 3xL1d | n.d. | ∼38 000 (master chromosome)d | This study | ||
| Candida maltosa | CBS 5611T | Cd,e | 62 949b | 22.0 | This study | ||
| Candida metapsilosis | MCO 448T | L2d,e | t-arrays + t-circles | 23 062 + 2nx 620e | 25.1 | AY962591 | (51,52) |
| PL 448 | Cd,e | 22 175e | 25.8 | AY391853 | (52,60) | ||
| Candida neerlandica | NRRL Y-27057T (CBS434T) | Cd,e | 32 141e | 24.4 | EU334437 | (60), this study | |
| Candida orthopsilosis | MCO 456 | Cd,e | 22 528e | 25.0 | AY962590 | (51,52) | |
| MCO 457T | Cd | ∼25 000d | (52) | ||||
| MCO 471 | L2d,e | t-arrays + t-circles | 24 697 + 2nx 777e | 25.0 | DQ026513 | (52,60) | |
| Candida parapsilosis | CLIB 214T (CBS 604T) | L2d,e | t-arrays + t-circles | 30 928 + 2nx 738e | 23.8 | DQ376035 | (60) |
| SR 23 (CBS 7157) | L2d,e | t-arrays + t-circles | 30 923 + 2nx 738e | 23.8 | X74411 | (20,46) | |
| Candida pseudojiufengensis | CBS 10847T (AS2.3693T) | Cd | This study | ||||
| Candida salmanticensis | CBS 5121T | L2d,e | t-arrays + t-circles | 25 718 + 2nx 104e | 20.5 | HQ267969 | (20), this study |
| Candida sojae | CBS 7871T | Cd,e | 39 415e | 29.1 | EF468347 | This study | |
| Candida subhashii | FR-392-06T (CBS 10753T) | L3d,e | t-proteins | 29 795e | 52.7 | GU126492 | (10), this study |
| Candida tetrigidarum | NRRL Y-48142T | Cd | This study | ||||
| Candida tropicalis | CBS 94T | Cd,e | This study | ||||
| MYA-3404 | Ce | 50 304e | 37.3 | Broad institute | |||
| Candida viswanathii | CBS 1924g | C ↔ L1d | palindrome/t-hairpins | ∼39 000 | This study | ||
| CBS 4024T | C ↔ L1d,e | palindrome/t-hairpins | 39 242e | 26.2 | EF536359 | This study | |
| Clavispora (Candida) lusitaniae | CBS 6936T (ATCC42720T) | Cd | 18 942 (unfinished)e | 27.4 | Broad Institute, this study | ||
| Debaryomyces hansenii (Candida famata) | CBS 767T | Cd,e | 29 462e | 27.0 | DQ508940 | (56), this study | |
| Lodderomyces elongisporus | NRRL YB-4239T (CBS 2605T) | Cd,e | 35 601e | 28.8 | Broad Institute, (52) | ||
| Pichia guilliermondii (Candida guilliermondii) | CBS 2030T (ATCC46036T)f | Cd | This study | ||||
aGenome form and size were deduced from PFGE analysis and DNA sequencing (see also Figures 1–3 and Supplementary Figure S1 for more details). Note that the terminal nucleotides of the shortest linear mtDNA molecules were not precisely mapped in the case of C. salmanticensis; C, circular-mapping; L1, L2, L3, linear-mapping, the types of linear-mapping genomes are defined according to their terminal structures (10,11,20); 3xL1, tripartite type I linear-mapping.
bMitochondrial telomeres classified according to Tomaska et al. (50).
cmtDNA mapping or sequence data were determined in this study, published previously or downloaded from public databases of the Broad Institute (http://www.broad.mit.edu/) and the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/).
dPFGE analysis.
eDNA sequencing.
fCandida guilliermodnii ATCC6260 is the anamorphic strain of Pichia guilliermondii.
gCBS 1924 is the type strain of Candida lodderae, its mtDNA displays similar restriction enzyme profile as the mtDNA from C. viswanathii CBS4024T.
n.d.—not done.
T and NT indicate the type and the neotype strain of the species, respectively.
Pulsed field gel electrophoresis
Screening for linear mitochondrial genomes was performed by pulsed field gel electrophoresis (PFGE) approach (10,11). Briefly, whole-cell DNA samples were prepared in agarose blocks, and separated in a 1.5% (w/v) agarose gel using a CHEF Mapper XA Chiller System (Biorad) with pulse switching set at 5–20 s (linear ramping and 120° angle) for 42 h at 5 V/cm. All separations were performed in 0.5× TBE buffer (45 mM Tris–borate, 1 mM EDTA, pH 8.0) at 10°C.
DNA sequencing and mitochondrial genome annotation
The mtDNA used for DNA sequencing was purified from isolated mitochondria. Procedures for mtDNA preparation, DNA sequence analysis and contig assembly were described previously (10,46,51,58–60). Genome annotations were performed using the MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl), and manually adjusted according to the sequence alignments of deduced protein products with their homologs from closely related yeast species. Intron sequences were identified using MFannot, RNAweasel (61) and Rfam (62). The precise boundaries were confirmed by alignments of the corresponding sequence with an intron-less gene from a related species. Putative protein products encoded by intronic open reading frames were identified by searching the Pfam database (63) and classified accordingly.
Phylogenetic analysis
For phylogenetic analysis, we have used amino acid sequences of a protein set (Atp6–Atp8–Atp9–Cob–Cox1–Cox2–Cox3–Nad1–Nad2–Nad3–Nad4–Nad4L–Nad5–Nad6) encoded by 23 mitochondrial genomes. The sequences were translated using translation table 4 (mold, protozoan and coelenterate mitochondrial code), except for S. cerevisiae, where translation table 3 (yeast mitochondrial code) applies. The multiple alignments were performed by MUSCLE (64) and concatenated to one alignment. Alignment columns with >50% of gaps were filtered out, resulting in an alignment with 3932 sites. The phylogenetic tree was built with three different programs: PhyloBayes with the CAT substitution model (65), MrBayes (66) with JTT model of amino acid substitution (67) and γ-distributed rate variation between sites, and PhyML (68) with JTT model. Application of all three programs gives the same tree topology. The only differences occur within C. metapsilosis–C. orthopsilosis–C. parapsilosis clade due to high sequence similarity among these species. In the rest of the tree, all branches are highly supported by posterior probabilities (above 0.9 in MrBayes and PhyloBayes), and most branches have bootstrap value of 100 in PhyML. Placement of C. alai in the tree has high posterior probability in both Bayesian programs, but low bootstrap values in PhyML.
Gene order comparison
To infer possible ancestral gene order, we used protein coding, rRNA and tRNA genes of 16 mitochondrial genomes from the ‘CTG clade’. Non-conserved genes that occur only in some species (i.e. trnM3 in D. hansenii and P. sorbitophila; dpoBa, dpoBb and orf756 in C. subhashii) were omitted in this analysis. We have reconstructed a possible history of rearrangements using a simple double cut and join model (DCJ) (69). The DCJ model is based on parsimony and includes commonly considered rearrangement operations, such as reversal, translocation, chromosome circularization, linearization, fusion and fission. There is an efficient algorithm to compute the parsimonious distance of two genomes in DCJ model (69); however, an exact algorithm for inferring ancestral gene orders for a given set of present day genomes is not known. To infer the ancestral gene order under DCJ, we have implemented a local optimization procedure that in each iteration attempts to improve the solution by choosing new ancestral gene orders from a local neighborhood using a dynamic programming algorithm (70). The DCJ model does not handle genomes with duplicated genes. To resolve recent duplications in some of the genomes (C. albicans, C. maltosa, C. sojae, C. viswanathii), we removed duplicated genes, and included both possible forms of the genomes as alternatives in the corresponding leaves. Similarly, both isomers are allowed in the genomes that include long inverted repeats (C. alai, C. albicans, C. maltosa, C. neerlandica, C. sojae, L. elongisporus). In each leaf, one of the alternative orders is chosen as a representative, so as to minimize the overall parsimony cost. Finally, we penalize occurrence of multiple circular chromosomes, or combinations of linear and circular chromosomes in ancestral genomes.
Enzymatic mapping of termini
Approximately 1 µg mtDNA aliquots were treated with exonuclease III (ExoIII; New England Biolabs) or BAL-31 nuclease (New England Biolabs) according to the manufacturer’s instructions, for increasing time periods. After enzyme inactivation (ExoIII for 20 min at 70°C; BAL-31 for 10 min at 65°C in the presence of EDTA), the mtDNA was precipitated with ethanol, dissolved in water, digested with a restriction endonuclease and electrophoretically separated in a 0.9% agarose gel. The labeling of mtDNA termini with T4 polynucleotide kinase has been performed essentially as described previously (20).
DNA hybridization probes
Southern hybridization of PFGE separated yeast DNA samples (Figure 1) was performed with a probe containing an equimolar mixture of PCR products derived from cox2 (345 bp) and nad4 (374 bp) of corresponding species. The following PCR primers were used: 5′-TAGATGTNCCWACWCCWTGAG-3′ and 5′-AYTCRTATTTTCAATATCATTG-3′ (cox2); 5′-AGGTATHWTGGTWAARACACC-3′ and 5′-CAGGWGAWACDAAWCCATG-3′ (nad4). For C. subhashii, the equivalent PCR primers were 5′-CGTCCCAACACCATGAGG-3′ and 5′-ACTCGTACTTCCAGTACCACTG-3′ (cox2); 5′-AGGGATCATGGTCAAGACG-3′ and 5′-CTGGTGAGACTAGCCCGTG-3′ (nad4). In subsequent experiments, we used the following probes: P-668 (668 bp fragment amplified by PCR from the C. frijolesensis mtDNA using primers 5′-ATAATGGGTCAGTGAGTT-3′ and 5′-ACGTTCTCTAGCAGTTGA-3′), EH-1350 (1350 bp EcoRV-HindIII fragment from C. frijolesensis mtDNA), H-1030 (1030 bp HindIII fragment from C. neerlandica mtDNA), and Oligo-32 (32 nt oligonucleotide 5′-AATGAGATGAGGAAGTAAAGGGATAAGGATAA-3′, corresponding to a palindrome sequence in C. viswanathii mtDNA).
Figure 1.
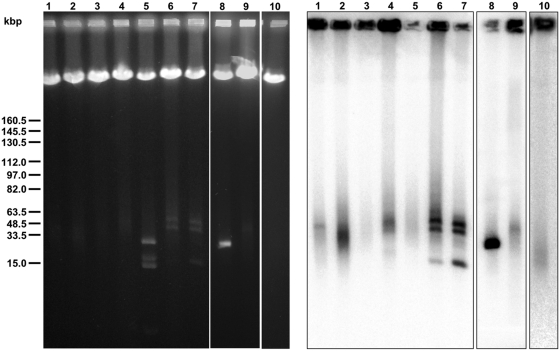
PFGE analysis of the yeast mtDNAs. The whole-cell DNA samples were separated by PFGE using a CHEF Mapper XA Chiller System (Biorad), blotted onto a nylon membrane and hybridized with mtDNA-derived probes as described in ‘Material and Methods’ section. Lane 1—C. viswanathii CBS 4024; lane 2—C. sojae CBS 7871; lane 3—C. maltosa CBS 5611; lane 4—C. neerlandica NRRL Y-27057; lane 5—C. alai NRRL Y-27739; lane 6—C. labiduridarum NRRL Y-27940; lane 7—C. frijolesensis NRRL Y-48060; lane 8—C. subhashii CBS 10753; lane 9—C. jiufengensis CBS 10846; lane 10—C. albicans CBS 562. Note that three discrete bands migrating in the region <50 kb represent three linear mitochondrial chromosomes in C. labiduridarum and C. frijolesensis (lanes 6 and 7). In contrast, four bands in C. alai (lane 5) do not hybridize with mtDNA probes and correspond to linear DNA plasmids (data not shown).
DNA sequence accession numbers
Mitochondrial DNA sequences described in this work were deposited in the GenBank nucleotide sequence data library under following accession numbers: HQ267968 (C. alai NRRL Y-27739), HM594866 (C. frijolesensis NRRL Y-48060), GU136397 (C. jiufengensis CBS10846), EU267175 (C. maltosa CBS5611), EU334437 (C. neerlandica NRRL Y-27057), EF468347 (C. sojae CBS7871), EF536359 (C. viswanathii CBS4024), HQ267969 (C. salmanticensis CBS5121).
RESULTS AND DISCUSSION
PFGE analysis of yeast mitochondrial genomes
We employed the PFGE approach (11,40) to distinguish between polydisperse and uniform linear mtDNA molecules in samples from 24 yeast species (Table 1 and Figure 1). In experimental conditions used for the screening (see ‘Materials and Methods’ section), the uniform linear mtDNA molecules from C. subhashii (10) migrate as a discrete band of ∼30 kb (Figure 1, lane 8). In contrast, a smear is typical for C. albicans [Figure 1, lane 10; (42)] and most other yeast species, with mtDNA-derived probes revealing a strong signal between ∼20 and 50 kb (Figure 1 and Table 1). This indicates that most examined species contain polydisperse linear mtDNAs. On average, their lengths are larger than the genome unit, apparently containing more than one genome equivalent per molecule. However, in C. labiduridarum and C. frijolesensis we detected three discrete bands migrating in the region between 15 and 50 kb (Figure 1, lanes 6–7). This points to a possibility that these species contain three uniform chromosomes in mitochondria (see below). Four distinct bands were also found in C. alai. However, these bands did not hybridize with the mtDNA probe (Figure 1, lane 5) and subsequent sequence analysis revealed that they correspond to linear DNA plasmids (to be described elsewhere).
Mitochondrial genome isomers in species with polydisperse mtDNA
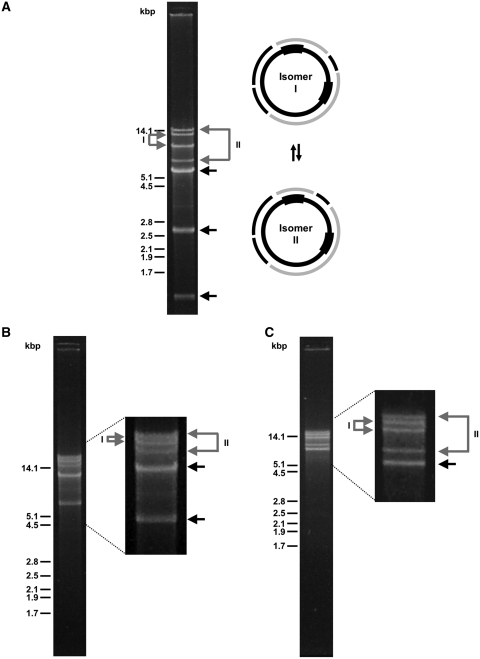
Since the mitochondrial genome of C. albicans occurs in two isomers (42,71), we examined the presence of genome isomers also in other species with polydisperse mtDNAs. Restriction enzyme analysis of the mtDNAs from C. maltosa, C. neerlandica and C. sojae identified four minor mtDNA fragments (e.g. ∼15, ∼13, ∼9 and ∼7 kb in the case of C. neerlandica mtDNA digested with BamHI and PvuII) indicating that they contain a circular-mapping genome with large repeated regions generating two isomers that are present in a stoichiometric ratio (Figure 2A–C). Subsequent sequence analysis confirmed that all three genomes contain large inverted repeats (LIRs) that could be involved in the flip-flop recombination generating genome isomers. This is in line with the observation that the LIRs represent recombination hotspots in C. albicans mtDNA (42). The LIRs were also detected in the C. alai mtDNA sequence, but the presence of contaminating linear plasmids rendered the identification of isomers by restriction enzyme analysis inconclusive.
Figure 2.
Restriction enzyme analysis reveals circular-mapping genome isomers. Candida neerlandica (A), C. maltosa (B) and C. sojae (C) mtDNAs were digested with the restriction enzyme combinations BamHI + PvuII, ApaLI + MluI and AgeI + ApaLI, respectively, and electrophoretically separated in 0.9% (w/v) agarose gel. Black arrows indicate the DNA fragments present in both isomers, grey arrows label the pair of fragments specific to the isomers I or II. Schemes illustrate both isomers with the position of inverted repeats (shown bold within the inner circle) and corresponding restriction enzyme fragments (the outer circle).
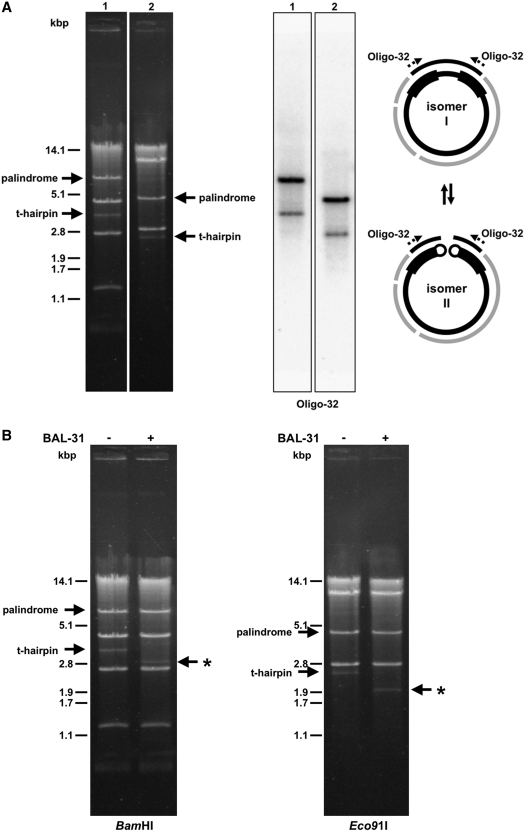
Physical mapping uncovered genome isomers also in mitochondria of C. viswanathii. However, in this case we observed two BamHI (∼7 and ∼3.5 kb) and Eco91I (∼5 and ∼2.5 kb) bands, with sizes corresponding to a monomer (lower faint band) and a dimer (upper band). Southern hybridization indicates that the two fragments have the same sequence (Figure 3A), and that the ratio between them varied in different preparations (data not shown). In this case, the complete mtDNA sequence of C. viswanathii contains LIRs arranged as a large palindrome (see below), suggesting that the palindrome (represented by the upper band) could be resolved into the smaller fragment (the lower band), i.e. a linear mtDNA with defined terminal sequences/structures. To support this idea we treated isolated mtDNA with BAL-31 nuclease prior to restriction enzyme analysis. This experiment demonstrated that the lower faint band was the only mtDNA fragment sensitive to BAL-31 nuclease activity (Figure 3B). On the other hand, this fragment seems to be refractory to both exonuclease III and T4 polynucleotide kinase (data not shown). This indicates that the termini of resolved linear mtDNA molecules are protected by a special arrangement. We presume that, similar to species from the genera Williopsis and Pichia (44), the linear mtDNA molecules terminate with single-stranded covalently closed telomeric hairpins (t-hairpins). In contrast, we did not identify genome isomers in C. jiufengensis, which lacks LIRs.
Figure 3.
Circular- and linear-mapping genome isomers in mitochondria of C. viswanathii. (A) The mtDNA samples were digested with BamHI (lane 1) or Eco91I (lane 2) and separated in 1% (w/v) agarose gel. The Southern blot was hybridized with radioactively labeled oligonucleotide probe Oligo-32 derived from the large palindrome (shown as dashed arrows). The solid arrows show positions of the palindrome and the presumed terminal fragments of resolved linear molecules capped with t-hairpins. Scheme shows presumed circular- (I) and linear-mapping (II) genome isomers. (B) Isolated mtDNA was treated or untreated with BAL-31 nuclease (0.2 U for 5 min). The mtDNA was then extracted from the reaction, digested with BamHI or Eco91I endonuclease, and electrophoretically separated. Note that the fragments containing presumed t-hairpins were sensitive to BAL-31 nuclease (indicated by asterisk).
Multipartite (fragmented) linear-mapping genomes
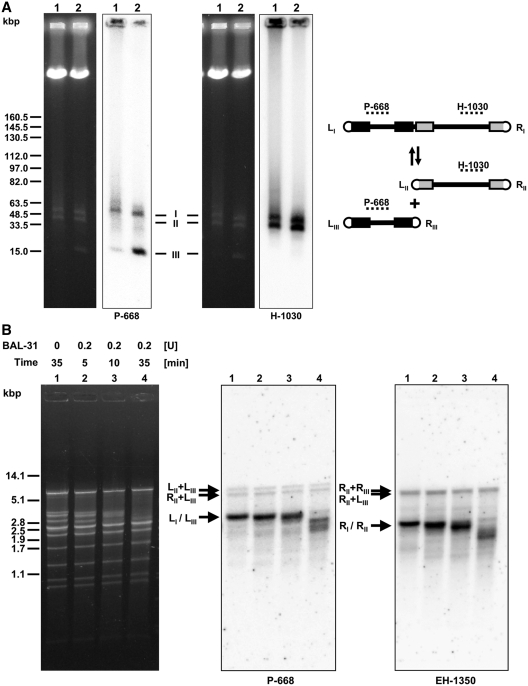
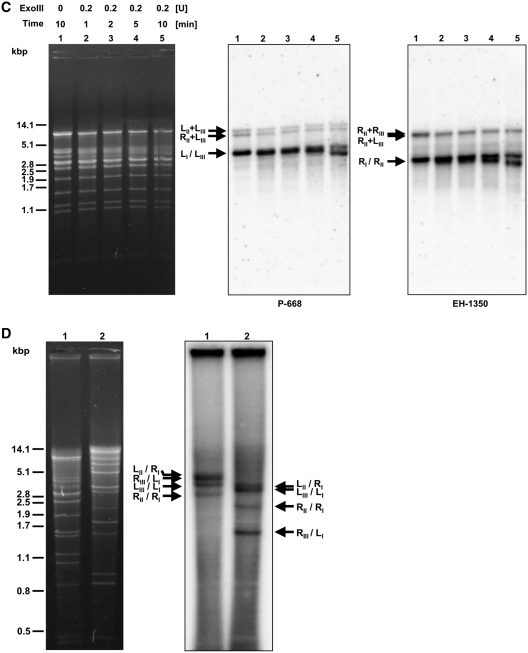
As mentioned above, PFGE analysis revealed the presence of three distinct mtDNA bands in C. labiduridarum and C. frijolesensis. The size of the largest band (chromosome I) corresponds approximately to the sum of the middle (chromosome II) and the smallest (chromosome III) bands (Figure 1, lane 6–7), suggesting that the longest molecules might represent a master chromosome split into two non-identical fragments. Therefore, we analyzed PFGE-separated mtDNA molecules of C. labiduridarum and C. frijolesensis by Southern hybridization using two probes derived from distant regions of chromosome I (Figure 4A). The probe P-668 hybridized with chromosomes I and III and also detected some mtDNA in wells and smears. The pattern detected by the probe H-1030 was similar, except that it hybridized with chromosome II instead of chromosome III. To confirm that all three chromosomes are linear, we treated C. frijolesensis mtDNA with BAL-31 nuclease, exonuclease III, and T4 polynucleotide kinase. We observed that the presumed terminal restriction enzyme fragments were all sensitive to BAL-31 nuclease (Figure 4B). Interestingly, the treatment with exonuclease III revealed two subpopulations of terminal fragments differing in their accessibility to the enzyme activity (Figure 4C). This indicates that the ends of the linear mtDNA molecules might adopt a covalently closed structure such as a t-hairpin, which in a fraction of molecules is opened and thus accessible to exonuclease III and T4 polynucleotide kinase (Figure 4D). Southern hybridization revealed four faint restriction fragments of the mtDNA (designated as LII + LIII, RII + RIII, LII + RIII, LIII + RII) that are refractory to all three enzymes (Figure 4B–D). The fragment sizes correspond to junctions between chromosomes II and III, and their presence shows that the master chromosome occurs in four flip-flop isomers (i.e. LIII − RIII − LII − RII, LIII − RIII − RII − LII, RIII − LIII − LII − RII and RIII − LIII − RII − LII). This suggests that fragmented linear-mapping genomes (i.e. uniform linear mtDNAs with resolved termini corresponding to chromosomes I–III) may coexist with circular-mapping genome forms (i.e. polydisperse linear mtDNAs lacking homogeneous terminal structures that correspond to the smear observed in PFGE).
Figure 4.
Multipartite linear-mapping genomes in C. labiduridarum and C. frijolesensis (A) PFGE separated samples of C. labiduridarum NRRL Y-27940 (lane 1) and C. frijolesensis NRRL Y-48060 (lane 2) were blotted onto a nylon membrane and hybridized with the radioactively labeled probes P-668 and H-1030 (regions hybridizing with both probes are shown as dashed lines). Presumed master (I) and two smaller chromosomes (II and III) are indicated. Note that the master chromosome occurs in four isomers (i.e. LIII − RIII − LII − RII (shown in the scheme), LIII − RIII − RII − LII, RIII − LIII − LII − RII and RIII − LIII − RII − LII. ‘L’ and ‘R’ indicate the left and the right telomere, respectively). The C. frijolesensis mtDNA (∼1 µg) was digested with BAL-31 nuclease (B) or exonuclease III (ExoIII) (C) as indicated. After nuclease inactivation, the DNA was digested with EcoRV, separated in 0.9% (w/v) agarose gel. The Southern blots were hybridized with the P-668 and EH-1350 probes specific for the left and the right arm of the master chromosome, respectively (see ‘Materials and Methods’ section). Arrows show the positions of the left (L) and right (R) terminal fragments and their fusions (R + R, R + L and L + L). Note that after ExoIII treatment the telomeric fragments form two subpopulations that differ in their sensitivity to the ExoIII treatment. This indicates that the linear mtDNA molecules possess an open structure with 5′ overhang or blunt end or covalently closed t-hairpin. (D) The C. frijolesensis mtDNA was treated with antarctic phosphatase and labeled with [γ32P]ATP and T4 polynucleotide kinase. The mtDNA was then digested with restriction endonuclease EcoRV (lane 1) or BglII (lane 2) and separated in 0.8% (w/v) agarose gel (left panel). The gel was fixed in 10% (v/v) methanol/10% (v/v) acetic acid for 30 min, dried overnight and autoradiographed (right panel). Arrows indicate the position of telomeric fragments containing the open structures accessible to terminal labeling.
The occurrence of multipartite genomes raises a question concerning the distribution of mtDNA molecules during cell division. Since we do not have any evidence for a specific segregation machinery analogous to the mitotic spindle ensuring the proper segregation of individual chromosomes in mitochondria, we propose that presumed circular-mapping genome and/or chromosome I may represent the ‘master copies’ playing a key role in the genome transmission.
Genetic organization of the mitochondrial genomes
With the aim to investigate the mitochondrial genome architecture in more detail and to identify sequence and/or structural features involved in the genome architecture alterations, we determined the complete mtDNA sequences of eight yeast species; C. alai, C. frijolesensis, C. jiufengensis, C. maltosa, C. neerlandica, C. salmanticensis, C. sojae and C. viswanathii (Supplementary Figure S1). The sizes of sequenced mtDNAs range from 25.7 (C. salmanticensis) to 62.9 kb (C. maltosa), and their G + C content varies from 20.5 (C. salmanticensis) to 29.1% (C. sojae) (Table 1). The genomes contain essentially the same set of conserved genes including the genes for subunits of ATP synthase (atp6,8,9), apocytochrome b (cob), cytochrome c oxidase (cox1,2,3), NADH:ubiquinone oxidoreductase (nad1,2,3,4,4L,5,6), large and small ribosomal RNA (rnl and rns) and a complete set of transfer RNAs (trn). The C. salmanticensis mtDNA has two additional genes: i.e. rps3/var1 coding for a subunit of the mitochondrial ribosome and rnpB/rpm1 for the RNA subunit of RNase P (Table 2). One or more genes are duplicated in C. maltosa, C. sojae and C. viswanathii mtDNAs as they are localized within the LIRs.
Table 2.
Genes present in the mitochondrial genomes sequenced in this study
| Species | Protein subunits of oxidative phosphorylation complexes |
Protein synthesis and RNA processing |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I |
III | IV |
V |
Ribosomal protein | rRNA |
tRNA | RNase P RNA | ||||||||||||
| nad1 | nad2 | nad3 | nad4 | nad4L | nad5 | nad6 | cob | cox1 | cox2 | cox3 | atp6 | atp8 | atp9 | rps3 | rns | rnl | trn | rnpB | |
| Candida alai | + | + | + | + | + | + | + | + | + | + | + | +a | +a | + | − | + | + | 24 + 1a | − |
| Candida frijolesensis | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | 24 | − |
| Candida jiufengensis | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | 24 | − |
| Candida maltosa | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | 24 + 2a | − |
| Candida neerlandica | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | 24 | − |
| Candida salmanticensis | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 24 | + |
| Candida sojae | + | +a | +a | + | + | + | + | +a | + | +a | + | + | + | +a | − | +a | + | 24 + 5a | − |
| Candida viswanathii | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | 24 + 1a | − |
aDuplicated within LIRs.
The presence of tRNATrp with an UCA anticodon, and the absence of an S. cerevisiae homolog of the abnormal tRNAThr1 with 8-nt in the anticodon loop (decoding CUN as threonine), indicate that UGA and CUN codons are recognized as tryptophan and leucine, respectively. The codon assignment was verified by multiple alignments of protein sequences, which led us to the conclusion that UGA(Trp) is the only deviation from the standard genetic code.
Introns were identified in cob, cox1, nad5 and rnl genes (Table 3). Their number and distribution among species vary from one (in C. alai) to 10 (in C. frijolesensis). The introns predominantly belong to group I and in many cases contain an open reading frame (ORF) coding for putative LAGLIDADG and GIY-YIG type endonucleases (group I) or reverse transcriptases/maturases (group II introns).
Table 3.
Identified intron sequences
| Species | Gene | Intron | Intron group | Intronic ORFs |
|---|---|---|---|---|
| Candida alai | cox1 | aI1 | IB | |
| Candida frijolesensis | cob | bI1 | I | |
| bI2 | IA | |||
| bI3 | IB | orf5 (LAGLIDADG1 endonuclease) | ||
| cox1 | aII1 | II (domainV) | orf1 (reverse transcriptase/maturase; HNHc domain) | |
| aI1 | IB | orf2 (LAGLIDADG1 endonuclease) | ||
| aI2 | IB | orf3 (LAGLIDADG1 endonuclease) | ||
| aI3 | I | orf4 (LAGLIDADG2 endonuclease) | ||
| aI4 | IB | |||
| nad5 | nd5I1 | I | orf6 (LAGLIDADG2 endonuclease) | |
| orf7 (LAGLIDADG2 endonuclease) | ||||
| rnl | rI1 | IA | ||
| Candida jiufengensis | cob | bI1 | ID | orf1 (GIY-YIG endonuclease) |
| cox1 | aI1 | IA (derived) | ||
| aI2 | ID | orf2 (LAGLIDADG1 endonuclease) | ||
| aI3 | IB | |||
| aI4 | IB | |||
| aI5 | IB | orf3 (LAGLIDADG endonuclease; truncated) | ||
| aI6 | IA (derived) | |||
| rnl | rI1 | IA (derived) | ||
| rI2 | IA (derived) | |||
| Candida maltosa | cob | bI1 | ID | orf2 (GIY-YIG endonuclease) |
| bI2 | IA (derived) | |||
| cox1 | aI1 | IB | orf1 (LAGLIDADG1 endonuclease) | |
| aI2 | IB2 (derived) | |||
| rnl | rI1 | IA | ||
| rI2 | IA (derived) | |||
| Candida neerlandica | cob | bI1 | I | |
| bI2 | IA | |||
| bI3 | IB | orf3 (LAGLIDADG1 endonuclease) | ||
| cox1 | aI1 | IB | orf1 (LAGLIDADG1 endonuclease) | |
| aI2 | IB1 (derived) | |||
| aI3 | IB1 (derived) | orf2 (LAGLIDADG1 endonuclease) | ||
| nad5 | nd5I1 | IB | orf4 (LAGLIDADG2 endonuclease) | |
| rnl | rI1 | IA | ||
| Candida salmanticensis | cob | bI1 | ID | orf1 (GIY-YIG endonuclease) |
| cox1 | aI1 | IB1 (derived) | ||
| nad5 | nd5I1 | IB2 (derived) | ||
| rnl | rI1 | IC2 | ||
| rI2 | I | |||
| Candida sojae | cox1 | aI1 | IB | orf1 (LAGLIDADG1 endonuclease) |
| aII1 | II (domainV) | orf2 (reverse transcriptase; HNHc domain) | ||
| Candida viswanathii | cob | bII1 | II (domainV) | orf3 (reverse transcriptase/maturase; HNHc domain) |
| cox1 | aI1 | IB (3′, partial) | orf1 (LAGLIDADG1 endonuclease) | |
| aI2 | IB | |||
| aI3 | IB | orf2 (LAGLIDADG1 endonuclease) | ||
| nad5 | nd5I1 | IB |
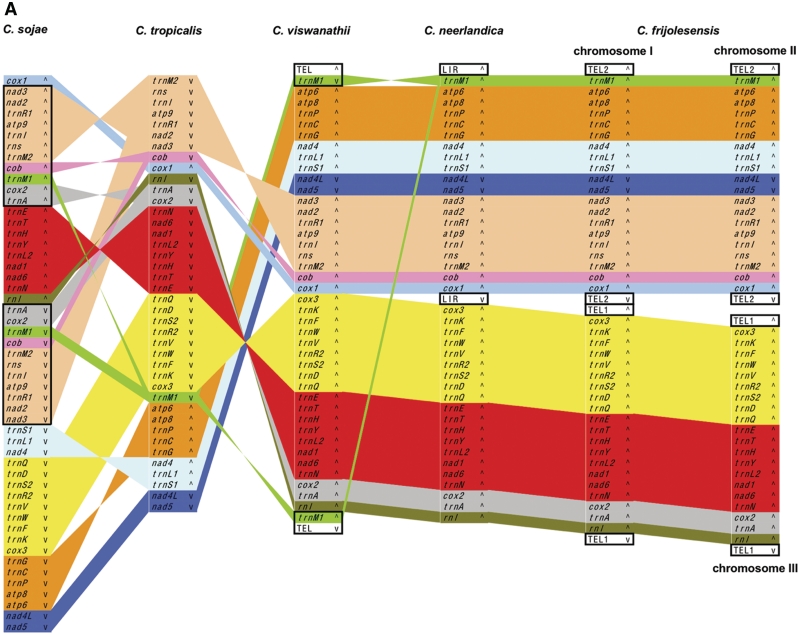
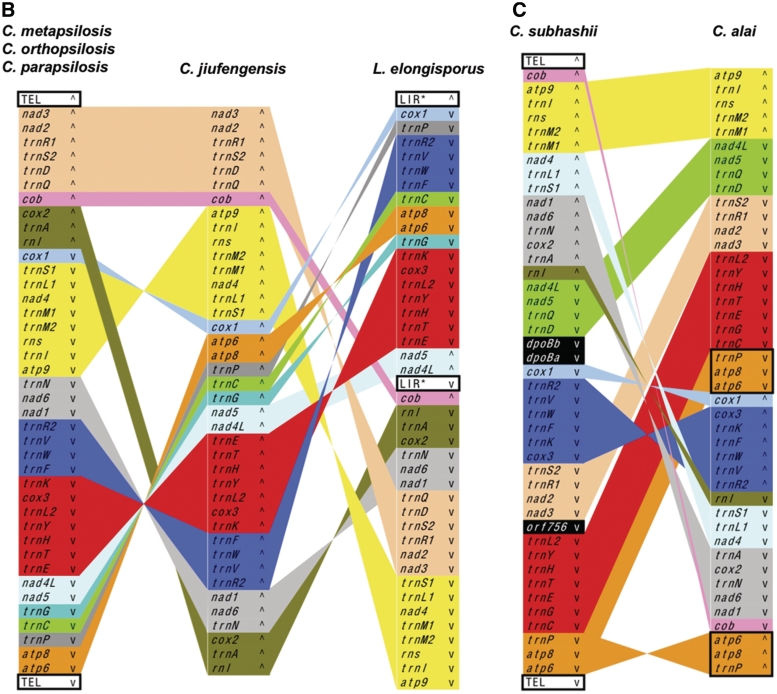
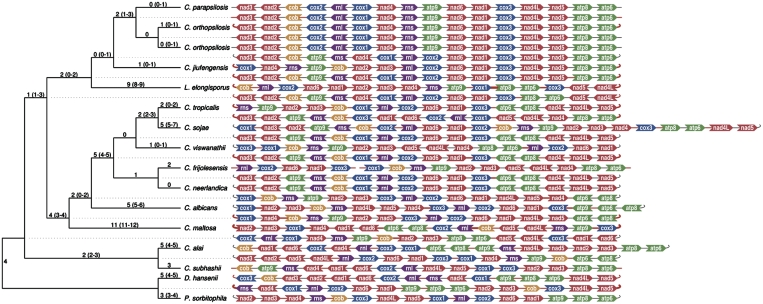
Our previous studies (46,51) revealed that C. metapsilosis, C. orthopsilosis and C. parapsilosis have the same gene order in their mtDNAs. Likewise, C. frijolesensis, C. neerlandica and C. viswanathii have the same genetic organization, except that trnM1 has been duplicated and inverted in the latter species (Figure 5A). All other species examined exhibit unique gene arrangements, with synteny reduced to four conserved gene clusters (i.e. trnN-atp6, cox1-atp9, cob-nad3 and rnl-cox2) in C. jiufengensis versus C. parapsilosis (Figure 5B).
Figure 5.
Comparison of mitochondrial gene orders among species from C. neerlandica–C. tropicalis (A), L. elongisporus–C. parapsilosis (B) and C. subhashii–C. alai (C) lineages. Individual genes and blocks with conserved gene order are shown by identical colors. Duplicated regions are framed. The symbols wedge and caret indicate the orientation of genes, TEL (telomeres) and LIR. In L. elongisporus, both LIRs (LIR*) consist of two regions of ∼4 kb separated by 574 and 95 bp-long unique sequences.
LIRs and the genome architecture
Analysis of the collected yeast mtDNA sequences reveals that the most prominent feature of the genome architecture is the presence of relatively long duplications, arranged as inverted repeats (LIRs). These elements are present in all but one (C. jiufengensis) mtDNA, and their lengths vary from 109 bp (present as the subterminal repeats in the linear mtDNA of C. salmanticensis) to 14 379 bp (in C. maltosa) thus substantially expanding the genome length (Supplementary Figure S1). In most cases, LIRs comprise non-coding sequences or contain only a few genes or gene fragments. In C. sojae, the 8658 bp LIRs represent a block duplication of 11 genes (i.e. trnA, cox2, trnM1, cob, trnM2, rns, trnI, atp9, trnR1, nad2, nad3). In most cases, the pairs of LIRs are separated by long unique regions. However, we noticed two special arrangements of LIRs: (i) in C. viswanathii identical copies of 4162 bp inverted repeats separated by a 798 bp non-coding sequence form a large palindrome and (ii) the two different inverted repeats (734 and 1229 bp) are separated by a 228 bp non-coding sequence. The second arrangement is present in the region of C. frijolesensis chromosome I, corresponding to the junction of chromosomes II and III. Since the two smaller chromosomes possess different LIRs at their termini, the chromosome I contains different sequences at the opposite ends.
As mentioned above, we demonstrate the presence of genome isomers in C. frijolesensis, C. labiduridarum, C. maltosa, C. neerlandica, C. sojae and C. viswanathii, but neither in C. jiufengensis lacking the LIRs nor in C. salmanticensis, which has the LIRs in subterminal regions of the linear mtDNA extended by t-arrays (2nx 104 bp).
Since the LIRs represent a suitable substrate for homologous recombination generating the genome isomers, the recombination transactions may be implicated in alterations of the genome architecture, which may in turn depend on LIR arrangements. We notice that the arrangement of LIRs in the mtDNA sequences correlates with the mitochondrial genome architecture. While C. maltosa, C. neerlandica and C. sojae have two circular-mapping genome isomers, C. viswanathii contains circular- and linear-mapping isomers, and C. frijolesensis possesses circular- and multipartite linear-mapping genome forms. This suggests that specifically arranged LIR copies (such as in C. viswanathii and C. frijolesensis) play a role as resolution elements, allowing interconversion between the circular- and linear-mapping genome forms (C. viswanathii), eventually leading to genome fragmentation into multiple linear chromosomes (C. frijolesensis).
The comparison of C. neerlandica, C. viswanathii and C. frijolesensis underlines the presumed role of LIRs in the genome architecture. All three species are phylogenetically closely related, with essentially the same mitochondrial genome organization. However, they differ in LIR arrangements and genome architecture (i.e. circular mapping; circular- and linear- or multipartite linear mapping).
Large palindromes are structural elements suitable for resolution of uniform linear molecules from circular and/or linear polydisperse mtDNAs. In general, such sequences are known hotspots of genomic instability due to their inherent ability to form cruciform or hairpin structures resulting in DNA replication stall sites (72–74). While in Escherichia coli, palindromic sequences cause double-stranded breaks induced by SbcCD complex (75), in the spirochete Borrelia the palindromes are processed by telomere resolvase (ResT) into t-hairpins (76). The latter mechanism parallels the palindrome resolution involved in the conversions of circular replication intermediates into linear-mapping mtDNAs in Williopsis and Pichia species (44) as well as in the formation of linear mtDNA monomers from linear and circular dimeric replication intermediates in the cilliate Paramecium (77) and the crustacean Armadillidium vulgare (78), respectively. Our results indicate that the palindrome in the C. viswanathii mtDNA is resolved into t-hairpins suggesting that linear mtDNA molecules with defined termini are generated during this process. On the basis of PFGE analysis (Figure 1, lane 1), we assume that the fraction of polydisperse mtDNA molecules fully processed into uniform mtDNA monomers is relatively low. In contrast, we detected three linear chromosomes, a smear of polydisperse mtDNA molecules and flip-flop isomers of chromosome I in C. frijolesensis samples. This indicates that circular-mapping genome forms are processed into chromosome I and further resolved into two smaller chromosomes.
Terminal inverted repeats appear to be a typical feature of linear-mapping mitochondrial genomes occurring in phylogenetically distant organisms (9,10,15,19–21,24,25,27,34,38,44,51,79,80) indicating that they arose by analogous evolutionary trajectories. These repeats usually consist of non-coding sequences, and sometimes a few genes. The linear-mapping mitochondrial genome of the stramenopile Proteromonas lacertae possesses even 15.6 kb-long terminal LIRs with about two-thirds of genes (21). Therefore, we assume that the terminal LIRs are remnants of resolution elements that emerge from segmental duplications of mitochondrial genome. Alternatively, the may derive from invertrons such as linear mitochondrial DNA plasmids that are known to integrate into mtDNAs (10).
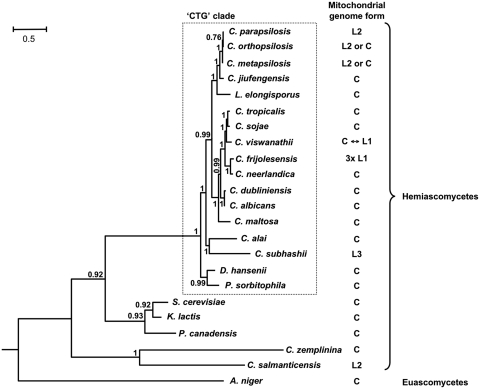
Phylogenetic analysis
We took advantage of the mtDNA-derived data and analyzed phylogenetic relationship of investigated yeast species by Bayesian and maximum likelihood methods. All three methods resulted essentially in the same tree topology. The tree calculated by PhyloBayes (Figure 6) is supported by high posterior probabilities on most branches and is consistent with the study of Fitzpatrick et al. (53) indicating that the monophyletic ‘CTG clade’ splits into two major lineages: the first represented by D. hansenii and P. sorbitophila, and the second by the C. albicans–C. parapsilosis group. Incorporation of additional recently described species (81–84) in the phylogenetic analysis revealed more detailed relationship among species in the latter lineage. This lineage splits into three subgroups (i.e. L. elongisporus–C. parapsilosis, C. maltosa–C. tropicalis and C. subhashii–C. alai) each containing species with circular- and linear-mapping mtDNAs. The occurrence of different types of mitochondrial telomeres (i.e. t-arrays in C. metapsilosis, C. orthopsilosis and C. parapsilosis; t-hairpins in C. viswanathii and C. frijolesensis; inverton-like telomeres with a t-protein in C. subhashii) in each subgroup is consistent with the tree topology. Similar to C. parapsilosis, the linear mitochondrial genome of C. salmanticensis terminates with t-arrays, although the sequence of its mitochondrial telomeres is different. Since C. salmanticensis belongs to early branching hemiascomycete lineages this linear mitochondrial genome emerged independently on linear mtDNAs in species from the ‘CTG clade’, presumably by employing similar molecular mechanism(s).
Figure 6.
Phylogenetic tree based on mtDNA encoded proteins. Phylogenetic tree was calculated from multiple sequence alignments of mitochondrial proteins by PhyloBayes (65). Posterior probabilities are shown at corresponding branches. The mitochondrial genome forms were reported elsewhere (10,20,22,51,55–57,60,86–89) or analyzed in this study. C—circular-mapping genome; L1, L2 and L3 indicate the type of linear-mapping genomes according to the telomeric structures, i.e. t-hairpins, t-arrays and invertron like with t-proteins, respectively; 3xL1—tripartite linear-mapping genome with t-hairpins (see Table 1 for details).
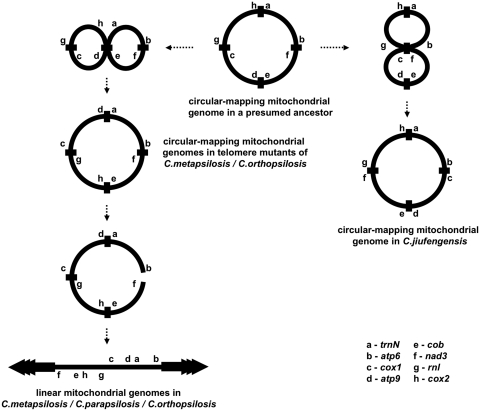
Reconstruction of ancestral mitochondrial genomes
Our previous reports (10,46) as well as the comparison of mtDNAs examined in this study revealed a number of conserved gene clusters. This prompted us to use the gene orders of 15 species from the ‘CTG clade’ for reconstruction of possible ancestral mitochondrial genomes in corresponding nodes of the phylogenetic tree (Figure 7), using the DCJ model (69) and local optimization procedures (70). For example, the presumed ancestor of C. parapsilosis and C. jiufengensis, which differ by the genome form, had a circular-mapping genome. We propose a simple evolutionary scenario leading to the linear-mapping mitochondrial genome (Figure 8). In this scenario, a resolution of a recombination transaction between the gene pairs cox2-trnN and cob-atp9 results in the formation of mtDNA with the gene order observed in circularized mutants of C. orthopsilosis and C. metapsilosis and its subsequent linearization between the genes nad3 and atp6 generates a linear mtDNA with genetic organization observed in the linear-mapping mitochondrial genomes of C. metapsilosis, C. orthopsilosis and C. parapsilosis. In contrast, recombination between the gene pairs rnl-cox1 and atp6-nad3 in the presumed ancestral genome leads to the identical arrangement of genes as is present in the C. jiufengensis mtDNA.
Figure 7.
Reconstruction of ancestral genomes. The figure shows possible ancestral gene orders and the number of events on each branch found by local optimization for the DCJ model. The intervals show range of numbers of events in equally parsimonious histories. Red connectors in the gene orders for present day and ancestral genomes represent inferred breakpoints on the branch to the nearest ancestor. Due to space constraints, the figure omits tRNA genes (even though the reconstructions were performed including tRNAs); full gene orders including tRNAs are shown in Supplementary Figure S2. The figure includes duplicated genes, which were restored after ancestral gene order reconstruction. Note that the linear and its circularized (mutant) mitochondrial genome forms of C. orthopsilosis were used in the analysis (51,60).
Figure 8.
A hypothetical pathway leading to mitochondrial genomes of C. parapsilosis and C. jiufengensis from the most recent common ancestor. We propose a simple scenario allowing delineation of the gene order found in both the circular-mapping genome of C. jiufengensis and the ‘true’ linear genome of C. parapsilosis from a reconstructed circular-mapping ancestor inferred by the analysis shown in Figure 7 and Supplementary Figure S2. The process includes reciprocal recombination events between the gene pairs (i) rnl/cox1 and nad3/atp6 or (ii) cox2/trnN and cob/atp9, followed by opening of the circular-mapping genome between the genes nad3 and atp6 in the latter case. Note that the circular-mapping genome intermediate prior its linearization has identical gene order as the mitochondrial telomere mutants of C. metapsilosis and C. orthopsilosis (51,52).
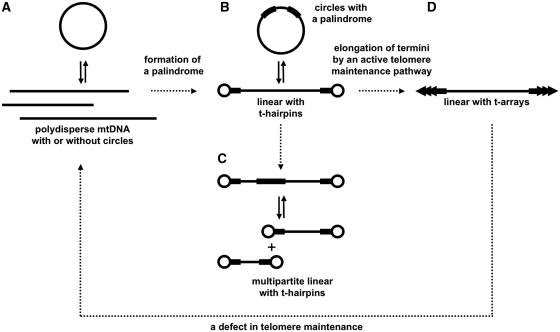
On the origin of ‘true’ linear mitochondrial genomes
In a number of species, replication of linear-mapping genomes relies on circular intermediates (monomers or dimers) generating linear concatemers via rolling circle and/or recombination-dependent replication mechanisms (44,45). In contrast, no genome-sized circles or genome concatemers were detected in mitochondria of C. parapsilosis and C. subhashii, which harbor uniform linear mtDNA molecules terminating with t-arrays and t-proteins, respectively (10,20,46). Hence, these linear-mapping genomes can be considered as ‘true’ linear genomes. This is further underlined by the presence of active telomere maintenance pathways ensuring their complete replication. We posit that linear-mapping genomes with terminal structures such as t-hairpins correspond to a transient state between circular mapping and ‘truly’ monomeric linear mitochondrial genomes. T-hairpins formed at linear mtDNA termini provide substrates for terminus elongation by an active telomere maintenance pathway [e.g. recombination-dependent mechanism operating in C. parapsilosis mitochondria (49,50)]. Once this pathway ensures the stability of a linear genome, circular replication intermediates and/or polydisperse mtDNAs become dispensable for the system. Conversely, a defect in the telomere-maintenance pathway may result in intramolecular end-to-end fusion, thus re-establishing the original circular-mapping mitochondrial genome architecture (Figure 9).
Figure 9.
A hypothesis on the origin of linear chromosomes in yeast mitochondria. (A) A circular-mapping genome represented by linear polydisperse mtDNAs with [e.g. C. glabrata, S. cerevisiae (40)] or potentially without [e.g. C. albicans (42)] a fraction of circular molecules. (B) Genome rearrangements may result in the formation of a palindrome allowing the resolution of a circular-mapping genome into linear chromosomes with defined terminal structures such as t-hairpins. Such genomes were observed in several species [e.g. P. pijperi and W. mrakii (11,44), C. viswanathii] containing uniform linear mtDNAs, with t-hairpins resolved from circular molecules (monomers and dimers) and/or linear polydisperse mtDNAs. (C) Multiple resolution elements (i.e. two types of LIRs) allow the genome fragmentation into multiple linear chromosomes (e.g. C. frijolesensis, C. labiduridarum). (D) The termini of the linear chromosomes may provide a substrate for further elongation via active maintenance mechanisms, such as the t-circle dependent pathway observed in ‘true’ linear genomes (e.g. C. metapsilosis, C. orthopsilosis, C. parapsilosis, C. salmanticensis) (49,50). Defects in the telomere maintenance result in the genome circularization via end-to-end fusion, as in mitochondrial telomere mutants of C. metapsilosis and C. orthopsilosis containing circular-mapping genomes (51,52).
ACCESSION NUMBERS
HQ267968, HM594866, GU136397, EU267175, EU334437, EF468347, EF536359, HQ267969.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Howard Hughes Medical Institute (grant HHMI 55005622 to J.N.); the Fogarty International Research Collaboration Award (2-R03-TW005654-04A1 to L.T.); European Community FP7 (IRG-224885 to T.V. and IRG-231025 to B.B.); Slovak Research and Development Agency (APVV 0024-07 and LPP-0164-06 to J.N., 20-001604 to L.T.); Canadian Research Chair program (to B.F.L.); Scientific Grant Agency of the Ministry of Education of Slovak republic (VEGA 1/0219/08 to J.N., 1/0132/09 to L.T., 1/0210/10 to T.V.); Comenius University (218/2009 to M.V.); Hungarian Scholarship Board based on the bilateral agreement between Hungary and Slovakia (workplan 0.9 b to Z.F.); The Canadian Research Chair Program (to B.F.L.). Funding for open access charge: Howard Hughes Medical Institute (HHMI 55005622).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Ladislav Kovac (Comenius University, Bratislava) for continuous support and discussion; Feng-Yan Bai (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China), Hiroshi Fukuhara (Institut Curie, Orsay, France), Cletus P. Kurtzman (National Center for Agricultural Utilization Research, Peoria, USA), Sung-Oui Suh and Meredith Blackwell (Louisiana State University, Baton Rouge, USA) for their gifts of yeast strains; and Serge Casaregola (Centre International de Ressources Microbiennes, Grignon, France) for providing us with the D. hansenii mtDNA sequence prior to publication.
REFERENCES
- 1.Bendich AJ. The size and form of chromosomes are constant in the nucleus, but highly variable in bacteria, mitochondria and chloroplasts. Bioessays. 2007;29:474–483. doi: 10.1002/bies.20576. [DOI] [PubMed] [Google Scholar]
- 2.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukes J, Guilbride DL, Votypka J, Zikova A, Benne R, Englund PT. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy J, Faktorova D, Lukes J, Burger G. Unusual mitochondrial genome structures throughout the Euglenozoa. Protist. 2007;158:385–396. doi: 10.1016/j.protis.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Bendich AJ. Reaching for the ring: the study of mitochondrial genome structure. Curr. Genet. 1993;24:279–290. doi: 10.1007/BF00336777. [DOI] [PubMed] [Google Scholar]
- 6.Backert S, Dorfel P, Lurz R, Borner T. Rolling-circle replication of mitochondrial DNA in the higher plant Chenopodium album (L.) Mol. Cell. Biol. 1996;16:6285–6294. doi: 10.1128/mcb.16.11.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson D. The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 2002;3:475–481. doi: 10.1038/nrg814. [DOI] [PubMed] [Google Scholar]
- 8.Coleman AW, Thompson W, Goff LJ. Identification of the mitochondrial genome in the chrysophyte alga Ochromonas danica. J. Eukaryotic Microbiol. 1991;38:129–135. [Google Scholar]
- 9.Forget L, Ustinova J, Wang Z, Huss VA, Lang BF. Hyaloraphidium curvatum: a linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol. Biol. Evol. 2002;19:310–319. doi: 10.1093/oxfordjournals.molbev.a004084. [DOI] [PubMed] [Google Scholar]
- 10.Fricova D, Valach M, Farkas Z, Pfeiffer I, Kucsera J, Tomaska L, Nosek J. The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology. 2010;156:2153–2163. doi: 10.1099/mic.0.038646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuhara H, Sor F, Drissi R, Dinouel N, Miyakawa I, Rousset S, Viola AM. Linear mitochondrial DNAs of yeasts: frequency of occurrence and general features. Mol. Cell. Biol. 1993;13:2309–2314. doi: 10.1128/mcb.13.4.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilson P, Waller R, McFadden G. Preliminary characterisation of chlorarachniophyte mitochondrial DNA. J. Eukaryot. Microbiol. 1995;42:696–701. doi: 10.1111/j.1550-7408.1995.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 13.Goddard JM, Cummings DJ. Structure and replication of mitochondrial DNA from Paramecium aurelia. J. Mol. Biol. 1975;97:593–609. doi: 10.1016/s0022-2836(75)80061-1. [DOI] [PubMed] [Google Scholar]
- 14.Kairo A, Fairlamb AH, Gobright E, Nene V. A 7.1 kb linear DNA molecule of Theileria parva has scrambled rDNA sequences and open reading frames for mitochondrially encoded proteins. EMBO J. 1994;13:898–905. doi: 10.1002/j.1460-2075.1994.tb06333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayal E, Lavrov DV. The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene. 2008;410:177–186. doi: 10.1016/j.gene.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Kovac L, Lazowska J, Slonimski PP. A yeast with linear molecules of mitochondrial DNA. Mol. Gen. Genet. 1984;197:420–424. doi: 10.1007/BF00329938. [DOI] [PubMed] [Google Scholar]
- 17.Martin FN. Linear mitochondrial genome organization in vivo in the genus Pythium. Curr Genet. 1995;28:225–234. doi: 10.1007/BF00309781. [DOI] [PubMed] [Google Scholar]
- 18.Moore LJ, Coleman AW. The linear 20 kb mitochondrial genome of Pandorina morum (Volvocaceae, Chlorophyta) Plant Mol. Biol. 1989;13:459–465. doi: 10.1007/BF00015557. [DOI] [PubMed] [Google Scholar]
- 19.Morin GB, Cech TR. Mitochondrial telomeres: surprising diversity of repeated telomeric DNA sequences among six species of Tetrahymena. Cell. 1988;52:367–374. doi: 10.1016/s0092-8674(88)80029-1. [DOI] [PubMed] [Google Scholar]
- 20.Nosek J, Dinouel N, Kovac L, Fukuhara H. Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol. Gen. Genet. 1995;247:61–72. doi: 10.1007/BF00425822. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Brocal V, Shahar-Golan R, Clark CG. A linear molecule with two large inverted repeats: the mitochondrial genome of the stramenopile Proteromonas lacertae. Genome Biol. Evol. 2010;2:257–266. doi: 10.1093/gbe/evq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pramateftaki PV, Kouvelis VN, Lanaridis P, Typas MA. The mitochondrial genome of the wine yeast Hanseniaspora uvarum: a unique genome organization among yeast/fungal counterparts. FEMS Yeast Res. 2006;6:77–90. doi: 10.1111/j.1567-1364.2005.00018.x. [DOI] [PubMed] [Google Scholar]
- 23.Sesterhenn TM, Slaven BE, Keely SP, Smulian AG, Lang BF, Cushion MT. Sequence and structure of the linear mitochondrial genome of Pneumocystis carinii. Mol. Genet. Genomics. 2010;283:63–72. doi: 10.1007/s00438-009-0498-7. [DOI] [PubMed] [Google Scholar]
- 24.Shao Z, Graf S, Chaga OY, Lavrov DV. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 2006;381:92–101. doi: 10.1016/j.gene.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 25.Smith DR, Lee RW. Mitochondrial genome of the colorless green alga Polytomella capuana: a linear molecule with an unprecedented GC content. Mol. Biol. Evol. 2008;25:487–496. doi: 10.1093/molbev/msm245. [DOI] [PubMed] [Google Scholar]
- 26.Suyama Y, Miura K. Size and structural variations of mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1968;60:235–242. doi: 10.1073/pnas.60.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G. Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Curr. Genet. 1993;24:241–247. doi: 10.1007/BF00351798. [DOI] [PubMed] [Google Scholar]
- 28.Wesolowski M, Fukuhara H. Linear mitochondrial deoxyribonucleic acid from the yeast Hansenula mrakii. Mol. Cell. Biol. 1981;1:387–393. doi: 10.1128/mcb.1.5.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer JD, Shields CR. Tripartite structure of the Brassica campestris mitochondrial genome. Nature. 1984;307:437–440. [Google Scholar]
- 30.Warrior R, Gall J. The mitochondrial DNA of Hydra attenuata and Hydra littoralis consists of two linear molecules. Arch. Sc. Geneve. 1985;38:439–445. [Google Scholar]
- 31.Laforest MJ, Roewer I, Lang BF. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res. 1997;25:626–632. doi: 10.1093/nar/25.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe KI, Bessho Y, Kawasaki M, Hori H. Mitochondrial genes are found on minicircle DNA molecules in the mesozoan animal Dicyema. J. Mol. Biol. 1999;286:645–650. doi: 10.1006/jmbi.1998.2523. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong MR, Blok VC, Phillips MS. A multipartite mitochondrial genome in the potato cyst nematode Globodera pallida. Genetics. 2000;154:181–192. doi: 10.1093/genetics/154.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan J, Lee RW. Mitochondrial genome of the colorless green alga Polytomella parva: two linear DNA molecules with homologous inverted repeat termini. Mol. Biol. Evol. 2002;19:999–1007. doi: 10.1093/oxfordjournals.molbev.a004180. [DOI] [PubMed] [Google Scholar]
- 35.Burger G, Forget L, Zhu Y, Gray MW, Lang BF. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl Acad. Sci. USA. 2003;100:892–897. doi: 10.1073/pnas.0336115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nash EA, Barbrook AC, Edwards-Stuart RK, Bernhardt K, Howe CJ, Nisbet RE. Organization of the mitochondrial genome in the dinoflagellate Amphidinium carterae. Mol. Biol. Evol. 2007;24:1528–1536. doi: 10.1093/molbev/msm074. [DOI] [PubMed] [Google Scholar]
- 37.Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J. Mol. Biol. 2007;372:356–368. doi: 10.1016/j.jmb.2007.06.085. [DOI] [PubMed] [Google Scholar]
- 38.Voigt O, Erpenbeck D, Worheide G. A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata. BMC Genomics. 2008;9:350. doi: 10.1186/1471-2164-9-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao R, Kirkness EF, Barker SC. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 2009;19:904–912. doi: 10.1101/gr.083188.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maleszka R, Skelly PJ, Clark-Walker GD. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 1991;10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bendich AJ. The end of the circle for yeast mitochondrial DNA. Mol. Cell. 2010;39:831–832. doi: 10.1016/j.molcel.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Gerhold JM, Aun A, Sedman T, Joers P, Sedman J. Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell. 2010;39:851–861. doi: 10.1016/j.molcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Nosek J, Tomaska L, Fukuhara H, Suyama Y, Kovac L. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 1998;14:184–188. doi: 10.1016/s0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- 44.Dinouel N, Drissi R, Miyakawa I, Sor F, Rousset S, Fukuhara H. Linear mitochondrial DNAs of yeasts: closed-loop structure of the termini and possible linear-circular conversion mechanisms. Mol. Cell. Biol. 1993;13:2315–2323. doi: 10.1128/mcb.13.4.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobs MA, Payne SR, Bendich AJ. Moving pictures and pulsed-field gel electrophoresis show only linear mitochondrial DNA molecules from yeasts with linear-mapping and circular-mapping mitochondrial genomes. Curr. Genet. 1996;30:3–11. doi: 10.1007/s002940050093. [DOI] [PubMed] [Google Scholar]
- 46.Nosek J, Novotna M, Hlavatovicova Z, Ussery DW, Fajkus J, Tomaska L. Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol. Genet. Genomics. 2004;272:173–180. doi: 10.1007/s00438-004-1046-0. [DOI] [PubMed] [Google Scholar]
- 47.Tomaska L, Nosek J, Makhov AM, Pastorakova A, Griffith JD. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 2000;28:4479–4487. doi: 10.1093/nar/28.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nosek J, Rycovska A, Makhov AM, Griffith JD, Tomaska L. Amplification of telomeric arrays via rolling-circle mechanism. J. Biol. Chem. 2005;280:10840–10845. doi: 10.1074/jbc.M409295200. [DOI] [PubMed] [Google Scholar]
- 49.Nosek J, Tomaska L. In: Origin and Evolution of Telomeres. Nosek J, Tomaska L, editors. Austin, Tx: Landes Bioscience; 2008. [Google Scholar]
- 50.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat. Struct. Mol. Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosa P, Valach M, Tomaska L, Wolfe KH, Nosek J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 2006;34:2472–2481. doi: 10.1093/nar/gkl327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rycovska A, Valach M, Tomaska L, Bolotin-Fukuhara M, Nosek J. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology. 2004;150:1571–1580. doi: 10.1099/mic.0.26988-0. [DOI] [PubMed] [Google Scholar]
- 53.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Massey SE, Moura G, Beltrao P, Almeida R, Garey JR, Tuite MF, Santos MA. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 2003;13:544–557. doi: 10.1101/gr.811003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wills JW, Troutman WB, Riggsby WS. Circular mitochondrial genome of Candida albicans contains a large inverted duplication. J. Bacteriol. 1985;164:7–13. doi: 10.1128/jb.164.1.7-13.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sacerdot C, Casaregola S, Lafontaine I, Tekaia F, Dujon B, Ozier-Kalogeropoulos O. Promiscuous DNA in the nuclear genomes of hemiascomycetous yeasts. FEMS Yeast Res. 2008;8:846–857. doi: 10.1111/j.1567-1364.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- 57.Jung PP, Schacherer J, Souciet J-L, Potier S, Wincker P, de Montigny J. The complete mitochondrial genome of the yeast Pichia sorbitophila. FEMS Yeast Res. 2009;9:903–910. doi: 10.1111/j.1567-1364.2009.00540.x. [DOI] [PubMed] [Google Scholar]
- 58.Burger G, Lavrov DV, Forget L, Lang BF. Sequencing complete mitochondrial and plastid genomes. Nat. Protoc. 2007;2:603–614. doi: 10.1038/nprot.2007.59. [DOI] [PubMed] [Google Scholar]
- 59.Lang BF, Burger G. Purification of mitochondrial and plastid DNA. Nat. Protoc. 2007;2:652–660. doi: 10.1038/nprot.2007.58. [DOI] [PubMed] [Google Scholar]
- 60.Valach M, Tomaska L, Nosek J. Preparation of yeast mitochondrial DNA for direct sequence analysis. Curr. Genet. 2008;54:105–109. doi: 10.1007/s00294-008-0200-3. [DOI] [PubMed] [Google Scholar]
- 61.Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 62.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 66.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 67.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 68.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 69.Bergeron A, Mixtacki J, Stoye J. Proceedings Sixth International Workshop Algs. in Bioinformatics (WABI’06), number 4175 in Lecture Notes in Computer Science. 2006. A unifying view of genome rearrangements; pp. 163–173. [Google Scholar]
- 70.Kovac J, Brejova B, Vinar T. A New Approach to the Small phylogeny problem. 2010. Technical Report. arXiv:1012.0935, arxiv.org. [Google Scholar]
- 71.Shaw JA, Troutman WB, Lasker BA, Mason MM, Riggsby WS. Characterization of the inverted duplication in the mitochondrial DNA of Candida albicans. J. Bacteriol. 1989;171:6353–6356. doi: 10.1128/jb.171.11.6353-6356.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 73.Lewis SM, Cote AG. Palindromes and genomic stress fractures: bracing and repairing the damage. DNA Repair. 2006;5:1146–1160. doi: 10.1016/j.dnarep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 74.Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl Acad. Sci. USA. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eykelenboom JK, Blackwood JK, Okely E, Leach DR. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol. Cell. 2008;29:644–651. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 76.Kobryn K, Briffotaux J, Karpov V. Holliday junction formation by the Borrelia burgdorferi telomere resolvase, ResT: implications for the origin of genome linearity. Mol. Microbiol. 2009;71:1117–1130. doi: 10.1111/j.1365-2958.2008.06584.x. [DOI] [PubMed] [Google Scholar]
- 77.Pritchard AE, Cummings DJ. Replication of linear mitochondrial DNA from Paramecium: sequence and structure of the initiation-end crosslink. Proc. Natl Acad. Sci. USA. 1981;78:7341–7345. doi: 10.1073/pnas.78.12.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marcade I, Cordaux R, Doublet V, Debenest C, Bouchon D, Raimond R. Structure and evolution of the atypical mitochondrial genome of Armadillidium vulgare (Isopoda, Crustacea) J. Mol. Evol. 2007;65:651–659. doi: 10.1007/s00239-007-9037-5. [DOI] [PubMed] [Google Scholar]
- 79.Burger G, Zhu Y, Littlejohn TG, Greenwood SJ, Schnare MN, Lang BF, Gray MW. Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J. Mol. Biol. 2000;297:365–380. doi: 10.1006/jmbi.2000.3529. [DOI] [PubMed] [Google Scholar]
- 80.Hikosaka K, Watanabe Y, Tsuji N, Kita K, Kishine H, Arisue N, Palacpac NM, Kawazu S, Sawai H, Horii T, et al. Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol. Biol. Evol. 2010;27:1107–1116. doi: 10.1093/molbev/msp320. [DOI] [PubMed] [Google Scholar]
- 81.Adam H, Groenewald M, Mohan S, Richardson S, Bunn U, Gibas CF, Poutanen S, Sigler L. Identification of a new species, Candida subhashii, as a cause of peritonitis. Med. Mycol. 2009;47:305–311. doi: 10.1080/13693780802380545. [DOI] [PubMed] [Google Scholar]
- 82.Ji ZH, Jia JH, Bai FY. Four novel Candida species in the Candida albicans/Lodderomyces elongisporus clade isolated from the gut of flower beetles. Antonie van Leeuwenhoek. 2009;95:23–32. doi: 10.1007/s10482-008-9282-7. [DOI] [PubMed] [Google Scholar]
- 83.Kurtzman CP, Robnett CJ, Yarrow D. Two new anamorphic yeasts: Candida germanica and Candida neerlandica. Antonie Van Leeuwenhoek. 2001;80:77–83. doi: 10.1023/a:1012218122038. [DOI] [PubMed] [Google Scholar]
- 84.Suh SO, Nguyen NH, Blackwell M. Yeasts isolated from plant-associated beetles and other insects: seven novel Candida species near Candida albicans. FEMS Yeast Res. 2008;8:88–102. doi: 10.1111/j.1567-1364.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 85.Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, et al. The diploid genome sequence of Candida albicans. Proc. Natl Acad. Sci. USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sekito T, Okamoto K, Kitano H, Yoshida K. The complete mitochondrial DNA sequence of Hansenula wingei reveals new characteristics of yeast mitochondria. Curr. Genet. 1995;28:39–53. doi: 10.1007/BF00311880. [DOI] [PubMed] [Google Scholar]
- 87.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 88.Zivanovic Y, Wincker P, Vacherie B, Bolotin-Fukuhara M, Fukuhara H. Complete nucleotide sequence of the mitochondrial DNA from Kluyveromyces lactis. FEMS Yeast Res. 2005;5:315–322. doi: 10.1016/j.femsyr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 89.Juhasz A, Pfeiffer I, Keszthelyi A, Kucsera J, Vagvolgyi C, Hamari Z. Comparative analysis of the complete mitochondrial genomes of Aspergillus niger mtDNA type 1a and Aspergillus tubingensis mtDNA type 2b. FEMS Microbiol. Lett. 2008;281:51–57. doi: 10.1111/j.1574-6968.2008.01077.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.