Abstract
The centromere is a highly specialized chromosomal element that is essential for chromosome segregation during mitosis. Centromere integrity must therefore be properly preserved and is strictly dependent upon the establishment and maintenance of surrounding chromatin structure. Here we identify WDHD1, a WD40-domain and HMG-domain containing protein, as a key regulator of centromere function. We show that WDHD1 associates with centromeres in a cell cycle-dependent manner, coinciding with mid-to-late S phase. WDHD1 down-regulation compromises HP1α localization to pericentric heterochromatin and leads to altered expression of epigenetic markers associated with this chromatin region. As a consequence, such reduced epigenetic silencing is manifested in disrupted heterochromatic state of the centromere and a defective mitosis. Moreover, we demonstrate that a possible underlying mechanism of WDHD1’s involvement lies in the proper generation of the small non-coding RNAs encoded by the centromeric satellite repeats. This role is mediated at the post-transcriptional level and likely through stabilizing Dicer association with centromeric RNA. Collectively, these findings suggest that WDHD1 may be a critical component of the RNA-dependent epigenetic control mechanism that sustains centromere integrity and genomic stability.
INTRODUCTION
The centromere is a distinctive chromosomal element upon which the kinetochore is anchored during mitosis (1,2). This highly compacted structure and its integrity are indispensable for mitotic chromosome alignment and segregation, and consequently the preservation of genomic information. DNA corresponding to the centromere (CT) and pericentromere (PCT) regions consists of extensive arrays of short tandem repeats, respectively, termed minor and major satellites, that have long been thought to be transcriptionally inert. However, research in the past decade has unequivocally demonstrated the expression of CT- and PCT-derived non-coding RNA transcripts across different eukaryotic species (3,4). Studies in the fission yeast, Schizosaccharomyces pombe, have provided the strongest evidence thus far that these RNA species serve as a critical determinant in the formation and maintenance of centromeres and associated chromatin structure (5–7). The centromeric silencing process involves an elaborate interplay between epigenetic and RNAi-dependent mechanisms, which entails a cascade of events leading ultimately to the generation of small RNA and establishment of a constitutive heterochromatic state.
While the possibility that the mammalian centromeres are governed by an analogous mechanism exists, it has not been addressed fully. As in the fission yeast, the constitutive heterochromatin state of the mammalian CT and PCT regions is specified and maintained by epigenetic mechanisms that constitute particular structural proteins (i.e. HP1) and distinct sets of histone tail modification (i.e. H3K9Me3 and H4K20Me3). Several chromatin modifiers, such as SUV39h1/2 (8,9), KDM2A (10) and Np95 (11–13), appear to be important for the regulation of CT/PCT transcription and structural stability. The key component of the RNAi pathway, Dicer, has also been implicated in the generation of small homologous dsRNA and mediating heterochromatin formation (6,14–16). Moreover, in line with a post-transcriptional mode of regulation, possible involvement of factors such as Vigilin and ADAR1 was previous documented (17,18). Therefore, despite the large differences among eukaryotic species in the centromeric DNA sequence and the structural organization of heterochromatin, the mechanism of its silencing may be highly conserved.
WDHD1 is an acidic nucleoplasmic DNA-binding protein that contains an amino-terminal WD40 domain and a carboxyl-terminal HMG-box motif (19). While recent reports have preliminarily linked human WDHD1 to S phase transaction and DNA damage response (20–23), much is not understood about the mammalian counterpart. Major clues about the function of this protein were derived mostly from previous functional studies on the yeast orthologs, Mcl1 and Ctf4, which have been regarded as DNA polymerase α accessory factors (24–26). Importantly, a role of Mcl1/Ctf4 in chromosome duplication and segregation was previously documented (26–31). Their functions in this regard are potentially exerted through the establishment of sister-chromatid cohesion and centromeric integrity. However, the molecular mechanism underlying the centromeric roles of Mcl1 as well as the functional link between human WDHD1 and centromeric silencing have not been substantiated. Conversely, despite extensive studies on dissecting the organization and regulation of yeast centromeric heterochromatin domains, the molecular pathway through which mammalian cells establish identity of this essential chromosomal element has not been fully explored.
In this study, we characterized WDHD1 as an integral centromeric heterochromatin-associated protein, and showed that WDHD1 plays a crucial role in CT/PCT silencing and associated heterochromatin stability. Our results further revealed that WDHD1 provides a fundamental function in generating the small RNA encoded by the satellite repeats, which may be linked to the Dicer-dependent, post-transcriptional RNA processing. Consequently, WDHD1 is required to sustain centromere integrity and mitotic fitness.
MATERIALS AND METHODS
Cell culture and transfection
HeLa, 293T and NIH-3T3 cells were grown as a monolayer in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (HeLa and 293T) or bovine calf serum (for NIH-3T3), and 100 U/ml penicillin and streptomycin, in a 5% CO2 humidified incubator at 37°C. Cells were transfected using Lipofectamine 2000 or Lipofectamine RNAiMAX (Invitrogen/Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. Transient transfection was done for 48–54 h prior to cell harvest, unless otherwise noted.
Cell cycle analysis and inhibitor treatment
Visualization of DNA replication sites by BrdU incorporation was done based on a previous report (32). G1/S junction synchronization of NIH-3T3 was performed as previously described (33). Briefly, NIH-3T3 cells were plated at low density and serum starved. After 48 h, cells were stimulated with normal growth medium containing 1 µg/ml aphidicolin (Sigma). After 16–18 h, cells were washed to remove aphidicolin, cultured in growth medium, and harvested at the indicated time points. Removal of soluble protein pools was achieved by extraction methods established elsewhere (8,34). Cells grown on coverslips were washed first with cytoskeleton (CSK) buffer (10 mM PIPES–KOH pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2), then incubated with CSK buffer supplemented with 0.2% Triton X-100 for 2 min on ice. For RNase A treatment, cells were pre-treated with 0.4% Triton X-100 for 30 s on ice to remove soluble proteins, followed by incubation with RNase A (1 mg/ml in PBS) for 3 min at room temperature. To inhibit RNA Pol II activity, NIH-3T3 cells at the G1/S junction were treated with 100 µM DRB for 2 h. For all the treatment experiments described above, cells were washed upon completion, fixed, and subjected to immunofluorescence analysis. Determination of the proliferation status of cell culture was done using BrdU Cell Proliferation Assay Kit (Chemicon; Billerica, MA, USA) based on the manufacturer’s instructions.
RNAi-mediated gene expression knockdown
NIH-3T3 cells were transfected with siRNAs with Lipofectamine RNAiMAX (Invitrogen) and harvested at 48 h after transfection. Twenty-five-nucleotide siRNA duplexes were designed to target the indicated mRNA, sequences of which are shown in the Supplementary Table.
Antibodies and reagents
Anti-WDHD1 antibody was raised in rabbit using a recombinant protein corresponding to amino acids 600–1123 of WDHD1, followed by antigen-specific purification. Mouse monoclonal antibody against HP1α and rabbit polyclonal antisera against H4K20me3, H3K27me1, AcH4 were obtained from Millipore (Temecula, CA, USA). Anti-CAF1 and anti-Vigilin goat polyclonal antibodies and mouse monoclonal antibodies against PCNA (PC10), ADAR1 and RNA Pol II (CTD4H8) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit antisera against Dicer and Drosha were from Bethyl Laboratories (Montgomery, TX, USA). An anti-Dicer monoclonal antibody (13D6) was from Abcam (Cambridge, MA, USA). Anti-phospho-histone H3 (Ser10; 6G3) monoclonal antibody was obtained from Cell Signaling Technology (Danvers, MA, USA). Monoclonal antibody against BrdU (BU33) was from Sigma (St Louis, MO, USA). Anti-DNA polymerase δ p125 (catalytic subunit) was purchased from MBL (Japan). Secondary antibodies used in the western blot assays were from Vector Laboratories (Burlingame, CA, USA), whereas those used in immunofluorescence analysis were obtained from Invitrogen. All chemicals were purchased from Sigma, except where otherwise indicated.
Indirect immunofluorescence and confocal microscopy
Immunofluorescence was modified from previously described methods (32). Cells seeded on slides were fixed in 2% formaldehyde solution and subsequently permeabilized with 0.5% Triton X-100 at room temperature. Cells were then blocked in blocking buffer (1% BSA in PBS) and probed with the indicated primary antibodies and species-specific secondary antibodies (Alexa 488- or 594-conjugated anti-mouse or anti-rabbit IgG). DNA or cell nuclei were counter-stained with DAPI (Sigma); F-actin was stained with phalloidin; condensed mitotic chromosomes were marked by the anti-phospho-H3 antibody. Images were captured on a Zeiss LSM 510 Meta confocal laser-scanning microscope (Carl-Zeiss, Feldbach, Switzerland), using a 63×/NA 1.4 oil immersion objective lens. Image overlays were colored by computer-assisted management of confocal microscopy data generated with Zeiss LSM 510 Image Browser software.
Cell lysate preparation
Cells were harvested and washed twice in PBS. Whole cell extracts were prepared using WCE buffer (20 mM HEPES, pH 7.4, 0.2 M NaCl, 0.5% Triton X-100, 5% glycerol, 1 mM EDTA, 1 mM EGTA, 10 mM β-glycerophosphate, 2 mM Na3VO4, 1 mM NaF, 1 mM DTT, cocktail protease inhibitor). For preparation of nuclear extracts, cell pellets were resuspended in 1 ml of lysis buffer (10 mM Tris–HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40 and cocktail protease inhibitor) per 107 cells. After 10-min incubation on ice, nuclei were collected by centrifugation (500g, 5 min) and washed with lysis buffer devoid of NP-40. After centrifugation, the pellet was resuspended in 100 µl nuclei lysis buffer (10 mM Tris–HCl pH 7.4, 400 mM NaCl, 1 mM EDTA, 1 mM DTT and cocktail protease inhibitor), mixed thoroughly for 15 min at 4°C. The nuclei lysates were diluted 10-fold in WCE buffer and centrifuged (16 000g, 20 min, 4°C) to obtain the nuclear fraction. Lysates were boiled in 2× urea sample buffer dye (100 mM Tris–HCl, pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mM β-mercaptoethanol, 8 M urea), and then fractionated by SDS–PAGE.
Western blot analysis and immunoprecipitation
Western blot analysis was performed after electrophoretic separation of polypeptides by 7.5 or 12.5% SDS–PAGE and transfer to Immobilon-P/PVDF membrane (Millipore). Blots were probed with the indicated primary and appropriate secondary antibodies. Immuno-bands were subsequently detected by the enhanced chemiluminescence reaction (ECL) (PerkinElmer; Waltham, MA, USA). All immunoprecipitations were performed with equal amounts of cell extract protein (1 mg) incubated with the indicated antibodies (2.5 μg) at 4°C for 2 h with rotation. The immunocomplexes were captured with protein G-sepharose (30 μl) (Millipore) for 2 h at 4°C with rotation. The protein G-antigen-antibody complexes were washed four times with the WCE buffer, and boiled in 2× urea sample buffer dye for subsequent PAGE and immunoblotting analysis as described above. To examine the RNase sensitivity of the co-precipitation, 293T cell extracts were first treated with or without 0.5 mg/ml of RNase A at 37°C for 30 min prior to immunoprecipitation.
Chromatin immunopricipitation
Chromatin immunopricipitation (ChIP) experiments were done essentially as described before (35,36). Crosslinked, sonicated chromatin was precleared before being incubated with 2.5 µg of the indicated antibodies and rotated at 4°C overnight. Normal mouse and/or rabbit IgG (Millipore) was used for the mock immunoprecipitation. After extensive washes, immunocomplexes were treated with Proteinase K and decrosslinked. Bound DNA in the ChIP, as well as input DNA (1/10 fragmented chromatin), was extracted, purified, and subjected to PCR analysis using primers corresponding to the satellite regions (37) as shown in the Supplementary Table. After amplification, PCR products were run on a 1.5% agarose gel with ethidium bromide staining, analyzed under UV light, and presented in inversed intensity. Signals taken by snapshots were quantified by ImageJ software. In some experiments, bound DNA was subjected to quantitative real-time PCR analysis. For the sequential ChIP assay, immunocomplexes from the first round were recovered by an elution solution (0.1% SDS, 0.5 M NaCl and 25 mM DTT in TE buffer) and diluted 10× by binding buffer, prior to a second round of ChIP using the Dicer antibody.
Real-time PCR
DNA samples from ChIP preparations were quantified by real-time PCR using the Bio-Rad iQ5 Gradient Real Time PCR system. All reactions were performed using the 2× SYBR Green Master mix (Bio-Rad, USA), Primer sequences for amplifying the satellite regions are the same as above. Results were corrected for nonspecific binding to IgG/beads and presented as percentage of input DNA. Triplicate PCRs for each sample were carried out.
RNA isolation and reverse transcription-PCR
Total RNA from NIH-3T3 mouse fibroblasts was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Genomic DNA was removed by digestion with 2U of DNase I (Ambion; Foster City, CA, USA). cDNA were synthesized using random hexamers and Superscript II reverse transcriptase (Invitrogen), and amplified by PCR. The primers used in PCR analyses were based on previous reporter (9) and listed in the Supplementary Table. The PCR products were resolved on 1% agarose gels and stained with ethidium bromide.
Northern blot analysis
To enrich for the small RNA fraction with sizes of ≤200 nt, a mirVana RNA isolation kit (Ambion) was used according to the manufacturer’s instructions. For northern blot analysis, 50 μg of total RNA (Figure 3C) or 25 μg of the small RNA fraction (Figure 3D) was separated, respectively, on 8 or 12% polyacrylamide/8 M urea denaturing gels and transferred to GeneScreen Plus membranes (PerkinElmer). The oligonucleotides used to probe for satellite repeats (major: mixture of Maj23 and Maj12; minor: mixture of Min1, Min2 and Min3) and U6 snRNA were designed based on a previous report (16) and are listed in the Supplementary Table. Oligonucleotide probes were 5′-end-labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England Biolabs; Ipswich, MA, USA). Prehybridization of the filters was carried out in 6× SSC, 0.2% SDS, 5× Denhardt’s solution and 20 mg/ml sheared, denatured salmon sperm DNA. Hybridizations were performed in the same solution at 42°C, with addition of the denatured, labeled probes. After hybridization, the blots were subjected to stringent washes with 6× SSC, 0.2% SDS at 42°C twice for 10 min, and subsequently autoradiography.
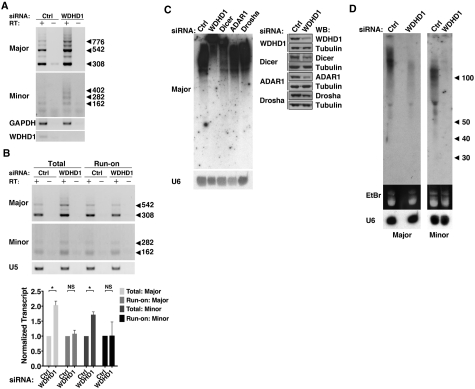
Figure 3.
Roles of WDHD1 in the expression of centromeric repeat non-coding RNA. (A) Expression levels of centromeric non-coding RNA spanning major and minor satellite repeats was examined by RT–PCR analysis of RNA isolated from control (ctrl) or WDHD1 knockdown NIH-3T3 cells. Total RNA samples were treated with DNase I prior to reverse transcription. ‘−’ denotes RT-minus reactions in which no reverse transcriptase was added. Expression levels of the housekeeping gene GAPDH, and WDHD1 are also shown. (B) Effect of WDHD1 knockdown on the transcription rates of minor and major satellite repeat region, as indicated. Nuclear run-on assays were performed to monitor newly transcribed centromeric RNA from nuclei of control (ctrl) and WDHD1 knockdown NIH-3T3 cells. U5 snRNA, which remained unchanged in both cell types, was used to demonstrate uniformity of input RNA. ‘–’ denotes RT-minus reactions in which no reverse transcriptase was added. Quantitative results are shown by bar graph below, and represent the mean ± SD of three independent experiments. For statistical significance of quantitative comparisons, calculations were done by (*P < 0.01; NS, P > 0.05). (C) NIH-3T3 cells were treated with siRNA targeting the indicated genes or control siRNA (ctrl). Degree of knockdown was monitored by western blot analysis as shown on the right. Equal amounts of total RNA isolated from these cells were resolved in a denaturing 8% acrylamide gel. The gel was subjected to northern blot analysis using probes specific for major satellite repeats. Blot was re-hybridized with a U6 snRNA-complementary probe to demonstrate equal loading (bottom on the left). (D) Expression of the homologous small RNA fragments. Total RNA from control (ctrl) or WDHD1 knockdown NIH-3T3 cells was size fractionated, and equivalents amounts of small RNA fraction (<200 nt) were resolved in a denaturing 12% acrylamide gel (EtBr staining shown in the middle). The gel was subjected to northern blot analysis using probes specific for major or minor satellite repeats, as indicated on the bottom. Blot was re-hybridized with a U6 snRNA-complementary probe to demonstrate equal loading (bottom).
RNA fluorescence in situ hybridization
RNA fluorescence in situ hybridization (FISH) was performed essentially as described previously (38), except with 3′-end Dig-labeled major satellite oligonucleotide probe in the hybridization step. Probe detection was carried out as follows. The slides were blocked with 3% BSA in 0.1 M Tris–HCl, pH 7.4, 0.15 M NaCl, 0.05% Tween-20 (TNT) at room temperature for 20 min and then incubated with sheep anti-Dig antibody (1:500, Roche) diluted with the blocking reagent for 3 h. Unbound antibodies were removed by washing six times in TNT. The slides were incubated with Alexa594-conjugated donkey anti-sheep IgG antibody (1:500, Invitrogen) for 2 h and then washed six times in TNT. After washing, the slides were coverslipped with Vectashield (Vector Laboratories) containing DAPI. Images were taken with a Zeiss LSM 510 microscope as described above. Sequence of the major satellite is: 5′-ATCACGGAAAATGAGAAATACACACTTTAGGACGTGAAATATGGCGAGGAAAACTGAAAAAGGTGGAAAATTTAGAAATGTCCACTGTAGGACGTGGAATATGGCAAGA-3′.
Nuclease accessibility assay
Nuclei of the control or knockdown NIH-3T3 cells were isolated using Nuclei PURE Prep Nuclei Isolation Kit (Sigma). To perform the Micrococcal nuclease (MNase) digestion, 2 × 105 nuclei were incubated in digestion buffer with 0, 0.6, 1.2, 2.4 and 4.8 U of MNase for 5 min at room temperature. For DNA purification, the samples were removed of protein by Proteinase K treatment and then extracted with phenol/chloroform/IAA (25:24:1). Upon ethanol precipitation, the DNA was resuspended and treated with RNase A. Digested DNA was eletrophoresed on a 1% agarose gel, transferred onto a Biodyne membrane (Pall Life Sciences, Ann Arbor, MI, USA) with the Pressure Blotter (Stratagene, La Jolla, CA, USA), and then crosslinked with UV light. To generate detecting probes for the Southern blot analysis, amplified DNA fragments corresponding the minor satellite (162 nt) or major satellite (308 nt) sequences were 5′-end labeled with [γ-32P] ATP using T4 polynucleotide kinase (NEB). Prehybridization of the membrances was carried out in 6× SSC, 5 % SDS, 10× Denhardt’s solution and 20 mg/ml denatured salmon sperm DNA at 42°C. Hybridizations were performed in the same solution at 42°C, with addition of the denatured, labeled probes. After hybridization, the membranes were washed at low stringency in 2× SSC, 0.1% SDS at room temperature twice for 10 min and then at high stringency in 0.1× SSC, 0.1% SDS at 42°C twice for 10 min, before being subjected to autoradiography.
Methylation-sensitive restriction analysis
Methylation-sensitive restriction analysis was carried out as previously described (39). Genomic DNA was isolated using QIAamp DNA Mini Kit (QIAGEN; Hilden Germany). To examine the global methylation at pericentromeric and centromeric regions, genomic DNA was digested overnight with Hpa II, a methyl-sensitive restriction enzyme (NEB) using 100 units of enzyme per 10 µg DNA. As control of digestion, another set of samples was digested with Msp I, a methyl-sensitive restriction enzyme that recognizes the same target sequence as Hpa II. An additional 20 units of enzyme was added the next morning to each sample to ensure complete digestion. For Southern blot analysis, 10 µg of purified DNA was separated on 1% agarose gel, transferred to Biodyne membrane, crosslinked with UV light and hybridized as described above. Primers for generating the mitochondria DNA probe are listed in the Supplementary Table.
Nuclear run-on assay
The nuclear run-on assay was carried out largely as described previously (40), with some modifications. We used Digitonin (40 µg/ml; Sigma) in place of NP-40 to permeabilize NIH-3T3 cells. Cells were washed twice with ice-cold 1× PBS and removed from the culture plate using a cell scraper in 1 ml of 1× PBS per 10-cm dish and collected by centrifugation (500g, 5 min). Cell pellets were resuspended in 1 ml of lysis buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 40 µg/ml Digitonin) per 107 cells. After 10-min incubation on ice, nuclei were then collected by centrifugation (500g, 5 min) and washed with lysis buffer devoid of Digitonin. To perform run-on reactions, aliquots of nuclei were mixed with 100 µl of 2× reaction buffer (20 mM Tris–HCl pH 8.0, 5 mM MgCl2, 200 mM KCl, 4 mM dithiothreitol, 1 mM each of ATP, CTP and GTP, 200 mM sucrose and 20% glycerol) and biotin-16-UTP (Roche) in a final volume of 200 µl at 29°C for 30 min. A total of 60 U of RNase-free DNaseI (Fermentas; Burlington, Ontario, Canada) and 6 µl 250 mM CaCl2 were added, and the reaction mixture was incubated for an additional 10 min at 37°C. The nuclear run-on RNA and total RNA were then digested with DNase I (Ambion) to further remove contaminating genomic DNA. Biotinylated RNA was purified by Dynabeads M-280 (Invitrogen), a magnetic bead covalently linked to streptavidin. Dynabeads resuspended in binding buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA and 2 M NaCl) were mixed to an equal volume of run-on RNA and subjected a 2-h incubation at room temperature. Beads were separated by the magnetic apparatus and washed once with 500 µl 2× SSC-15% formamide for 10 min and twice with 1 ml 2× SSC for 5 min each. Random hexamer-primed cDNA was synthesized using 10 µl biotinylated RNA and 500 ng total RNA, and subsequently subjected to semi-quantitative PCR to assay for major and minor satellite RNA transcription rate. PCR was performed as described above. PCR products were resolved by agarose gels and quantified with the ImageJ software. To ensure the efficiency of the reverse transcription, the intensities of PCR products were normalized to those of U5 snRNA (primers are listed in the Supplementary Table).
RNA pull-down assay
For in vitro synthesis of biotinylated transcripts corresponding to approximately one unit of the major and minor satellite repeats, templates were first generated by PCR reactions using chimeric oligonucleotide primers that encompass T7 RNA polymerase promoter sequence (Supplementary Table). Templates corresponding to partial 18S rRNA sequence that are of equivalent lengths to the minor and major satellite repeats (162 and 300 bp, respectively) were used as control. In order to synthesize biotinylated transcripts, AmpliScribe™ T7-Flash™ Biotin-RNA Transcription Kit (EPICENTRE; Madison, WI, USA) was then used according to the manufacturer’s instructions. NIH-3T3 nuclear extracts were prepared as described above. To remove endogenous WDHD1, immunodepletion was performed with 2.5 mg of total nuclei extracts. The supernatants were incubated with 2.5 µg primary antibody for 3 h with gentle agitation and subsequently with the addition of protein G-agarose beads (Millipore) for additional 1 h. The supernatants were subjected to a second round of depletion by the same procedure. Control depletions were performed using pre-immune rabbit IgG. All steps of the pull-down assay were performed at 4°C. Nuclei extracts were precleared with 12.5 μl streptavidin Sepharose (GE Healthcare; Piscataway, NJ, USA), in the presence of SUPERase·In (0.05 U/ml) (Ambion) and yeast tRNA (25 μg/ml) (Sigma), for 1 h with rotation. After centrifugation, 2 μg of in vitro transcribed biotinylated RNA was added to the supernatant and the mixture was further incubated for 1 h. The protein-biotinylated RNA complexes were recovered by addition of 30 μl streptavidin Sepharose (1 h incubation with rotation), and the bound complexes were washed four times with WCE buffer and subsequently analyzed by 7.5% SDS–PAGE and western blot.
RNA immunoprecipitation
RNA immunoprecipitation was performed essentially as described for ChIP except with some modifications. In brief, cells were fixed in 1% formaldehyde for 10 min at room temperature, washed twice with ice-cold 1× PBS, and then collected from the culture plate. Nuclei were isolated based on the above procedure and subsequently resuspended in 100 µl nuclei lysis buffer (10 mM Tris–HCl pH 7.4, 400 mM NaCl, 1 mM EDTA, 1 mM DTT and proteinase inhibitor cocktails) containing RNase inhibitor (125 U/100 µl of SuperRNAsin; Ambion). The nuclear lysates were diluted 10-fold in WCE buffer, and centrifuged (12 000g, 15 min, 4°C) to obtain the nuclear fraction. Immunoprecipitation was performed by adding the WDHD1 or Dicer antibody to the precleared nuclear extracts and incubating at 4°C overnight. Magnetic protein-G beads (Invitrogen) were added to each IP sample and rotated for 1 h at 4°C. The beads were pelleted and washed twice with 0.5 ml of WCE buffer and another two times with WCE buffer adjusted to 300 mM NaCl, and subsequently resuspended in 100 μl of de-crosslink buffer containing 10 mM Tris–HCl pH 7.4, 400 mM NaCl and incubated at 65°C for 2 h. Then, 10 U of DNase I (Fermentas) and 10× reaction buffer was added and incubated at 37°C for 15 min to remove all contaminating DNA. The elute fraction was treated with Proteinase K, extracted with phenol/chloroform, and precipitated by ethanol. The immunoprecipitated RNA was dot-blotted onto a nitrocellulose membrane as described previously (41) and hybridized it with oligonucleotide probes corresponding to satellite repeats as described above in the ‘northern blot’ section.
RESULTS
Distinct focal localization of WDHD1 during mid-to-late S phase
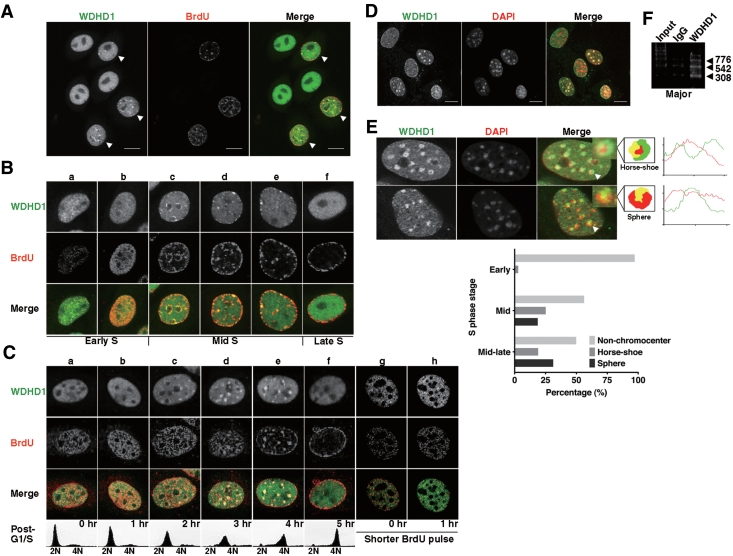
To begin investigating the biological function of WDHD1, we first performed indirect immunofluorescence analysis to examine its cellular localization in cycling HeLa cells. Consistent with previous observations, WDHD1 localized to the nucleus (Figure 1A, left panel). Interestingly, in a sub-population of cells, WDHD1 displayed a unique, punctate staining (indicated by arrowheads). These cells corresponded to those actively undergoing DNA replication, as indicated by BrdU incorporation (Figure 1A, middle panel). Furthermore, confocal images revealed a partial overlap between WDHD1 and BrdU staining signals (Figure 1A, right panel, arrowheads). A detailed breakdown of the S-phase cells (Figure 1B), based on the distribution of DNA replication sites, further pinpointed the distinct punctate appearance temporally at the mid-to-late stage of S phase (Figure 1B, c–e).
Figure 1.
Dynamic patterns of WDHD1 subnuclear localization during S phase. (A) HeLa cells were grown on coverslips, pulse-labeled with BrdU (for marking cells in S phase) before being fixed, and subjected to immunostaining. Left, WDHD1; middle, BrdU; right, merged image (WDHD1, green; BrdU, red). Individual and merged images were captured by laser scanning confocal microscope and single sections are shown (scale bar is 10 µm). Arrowheads mark cells with punctate staining patterns of WDHD1. (B) Distinct focal localization of WDHD1 (green) and its partial colocalization (see merge) with BrdU (red) in S-phase cells. Immunostaining and laser confocal microscopy were performed as in (A). Cells in the indicated sub-stages of S phase (early, mid, or late) were distinguished on the basis of the BrdU incorporation patterns (a–f). (C) Cell-cycle localization of WDHD1 during S phase by immunofluorescence microscopy. NIH-3T3 cells were synchronized at G1/S junction and released into S (a–f). Before being fixed at the indicated post-release time points, cells were pulse-labeled with BrdU (15 min) for visualizing DNA replication site. S phase progression was also analyzed based on DNA content by flow cytometry (shown on the bottom). Images of early S-phase cells subjected to a shorter pulse of BrdU labeling (8 min) are shown for a more defined representation of early replication site (g and h, ‘shorter BrdU pulse’). (D) NIH-3T3 cells were briefly treated to remove the soluble pool of proteins, fixed, and subsequently stained to reveal WDHD1 and pericentric heterochromatin (DAPI). Confocal microscopy was done as above (scale bar is 10 µm). (E) Distinct patterns of WDHD1 localization at centromeric foci. NIH-3T3 cells in S phase were collected as in (C), and subjected to indirect immunofluorescence analysis. Three types of WDHD1 subnuclear localization pattern can be readily distinguished, based on staining signals relative to the DAPI foci: non-chromocenter, horse-shoe and sphere. Representative immunostaining images for the latter two types are shown. The merged images (‘merge’) and the schematic cartoon figure illustrate the relative positions of WDHD1 (‘WDHD1’, green) and the centromeric heterochromatin in nucleus (‘DAPI’, blue, but pseudo-color red in the merged images). The insets represent enlarged images denoted by arrows. The histograms on the right represents the local intensity distribution (diagonal white lines through the images) of WDHD1 in green and DAPI in red. Graph on the bottom shows quantification of cells with the indicated distinct focal patterns of WDHD1 staining at centromeres at 2, 3 and 4 h after release into S phase. One hundred percent represents total number of cells (n = 200) and percentages given are averages of three independent experiments. (F) WDHD1 occupancy of centromeric repeat regions. ChIP assays were performed on crosslinked chromatin from NIH-3T3 cells using antibodies specific for WDHD1 or control rabbit antibodies (IgG). Products of final PCR analysis using primers specific to major satellite repeat DNA sequence are resolved in agarose gel.
We further characterized the dynamics of WDHD1 localization in mouse NIH-3T3 cells selectively during S phase, after synchronization at G1/S junction and release into S phase (Figure 1C). In the early S-phase cells, we observed a small punctate distribution of WDHD1 characteristic of euchromatin structure and a partial, albeit limited, overlap between these signals with the BrdU incorporation sites (Figure 1C, a and b, g and h). More importantly, we were able to observe a similarly cell cycle-dependent focal staining pattern for WDHD1, time window of which was also refined to the mid-to-late stage of S phase (Figure 1C, c–e), as in HeLa cells. Interestingly, the S phase-associated focal appearance was much more distinct in this cell line, and colocalized to a large extent with the DAPI spots (Figure 1D and E). Such distinctive nuclear staining pattern by DAPI corresponds to the densely packed chromatin structure contiguous with the chromosomal centromeres, called pericentric heterochromatin. Thus, we next wanted to determine if the punctate pattern of WDHD1 indeed reflects heterochromatin association. Upon removal of soluble WDHD1 pool by triton extraction, immunofluorescence analysis revealed that the focal distribution of WDHD1 and its overlap with DAPI signals remained (Figure 1D), consistent with the inaccessible nature of the WDHD1-associated structure. Notably, mid-to-late S-phase culture exhibited an enrichment for cells with two discrete types of focal staining (Figure 1E, confocal images on the top and quantitative results on the bottom). One is reminiscent of the ‘horse-shoe’ replication foci that is also shared by PCNA and CAF-1. The other represents a more ‘sphere’ distribution characteristic of structural proteins such as HP1 (42). Additionally, as a more direct means to probe its possible centromere association, we performed ChIP assay and verified WDHD1’s binding to the major satellite and minor satellite repeat regions (Figure 1F and data not shown). Based on these results, we conclude that WDHD1 is enriched transiently in nuclear spots on centromeric chromatin during the second half of S phase, which may coincide with molecular events at this heterochromain domain, such as DNA replication.
Centromeric association of WDHD1 is RNA-independent but requires ongoing RNA transcription
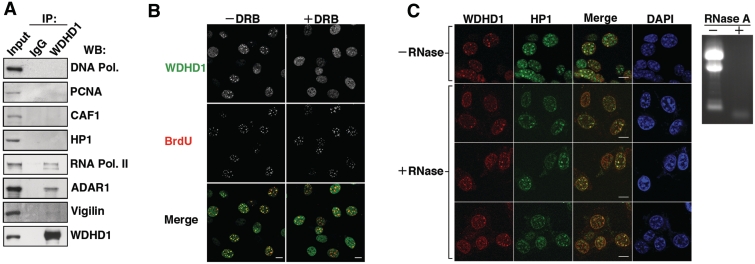
Having established the spatiotemporal patterns of WDHD1 nuclear localization, we next aimed to further explore the molecular basis of such attribute. Towards this end, we first performed selective profiling of WDHD1’s associated proteins in the NIH-3T3 cell nucleus (Figure 2A). Immunoprecipitation assays revealed that, in contrast to the previous findings, our WDHD1 specific antibodies did not co-precipitate DNA polymerase δ. Architectural components of pericentric replication foci, PCNA, CAF-1 and HP1α, also were not present in the WDHD1 immunoprecipitates. Conversely, we were able to detect association of RNA Pol II as well as known post-transcriptional regulators of the CT/PCT, such as ADAR and Vigilin, with WDHD1. These findings collectively raised the possibility that the distinct overlap of WDHD1 with PCT replication foci may actually be coupled with transcriptional and/or post-transcriptional events taking place in the vicinity. In line with such notion, we observed that inhibition of Pol II transcription by DRB treatment in a mid-late S phase population of cells resulted in the disappearance of WDHD1 staining at pericentric heterochromatin domains in the vast majority of cells (Figure 2B), thus suggesting that such localization requires ongoing RNA transcription. However, this change in staining did not result from a loss of RNA, as RNase-treated cells retained significant WDHD1 focal signals (Figure 2C). Incidentally, colocalization was evident between the RNase-resistant WDHD1 foci and signals corresponding to the RNase-insensitive, replication-specific HP1α pool (42) (Figure 2C), again affirming the spatiotemporal mode of WDHD1 pericentromere recruitment. Given that transcription of the satellite repeats underlies the establishment of replicated CT/PCT heterochromatin, our results may support a link between WDHD1 and the regulation of satellite repeats-encoded RNA expression in cells.
Figure 2.
Characterization of the molecular determinants underlying WDHD1′s centromeric localization. (A) WDHD1 was isolated from NIH-3T3 cell lysates by immunoprecipitation (IP) using α-WDHD1 antibody. Western blot analysis was subsequently performed with antibodies against selected components of the DNA replication and RNA transcription complexes, as indicated. IgG refers to control rabbit or mouse Ab IP using whole cell lysates derived from the same cells. Input equals 1/10 of IP. (B and C) Indirect immunofluorescence analysis was performed to observe localization of endogenous WDHD1 (red) and HP1α (green) in the inhibitor-treated NIH-3T3 cells. (B) Before immunostaining, cells arrested at the G1/S junction were released in the presence of 100 µM DRB for 2 h to inhibit Pol II activity (+DRB) or in its absence (−DRB) and subsequently pulse-labeled with BrdU. Nuclear DNA was stained by DAPI (blue). Images were captured by laser scanning confocal microscopy and single sections are shown (scale bar is 10 µm). (C) NIH-3T3 cells grown on coverslips were first permeablized, and subsequently mock- (−) or RNase A- (+) treated before fixation. Immunostaining were performed with antibodies specific for WDHD1 (red) and HP1α (green). Nuclear DNA was stained by DAPI (blue). Confocal microscopy was done as above (scale bar is 10 µm). Removal of RNA was monitored by gel electrophoresis of total RNA extracted from the cells, as shown by the EtBr staining image (right).
Abrogation of WDHD1 expression alters the post-transcriptional processing of the centromere-encoded RNA transcripts
We further investigated if WDHD1 could contribute to the transcriptional regulation of the repetitive sequences within the CT/PCT regions. For that purpose, we analyzed the expression levels of major- and minor-satellite transcripts in control and WDHD1-depleted NIH-3T3 cells, using semi-quantitative reverse transcription (RT)–PCR on total RNA. The results revealed a substantial increase in the transcript levels of both the major and minor satellite repeats in the WDHD1 knockdown cells compared to control cells (Figure 3A), suggesting a repressive role for WDHD1 in the expression of centromeric RNA. Given the altered transcript abundance observed following WDHD1 knockdown and WDHD1’s association with RNA Pol II (Figure 2A), we next sought to characterize the involvement of WDHD1 in regulating the transcription efficiency of the CT and PCT repeats. We conducted nuclear run-on assay, in which intact nuclei isolated from control and WDHD1-RNAi cells were pulsed with biotin-16-UTP. Upon run-on reaction, newly synthesized biotin-labeled RNA transcripts and total RNA were subjected to semi-quantitative RT-PCR assay to measure changes in the transcription activity of major and minor repeats. As shown in Figure 3B, while WDHD1 depletion led to a consistent increase in the steady-state levels of major and minor satellite transcripts in the overall RNA pools, we did not detect significant differences in the newly synthesized fraction. Therefore, these data exclude the possibility that WDHD1 exerts its role in centromeric RNA expression through transcriptional regulation.
Earlier reports have demonstrated that, in fission yeast and higher eukaryotic cells, CT/PCT-encoded transcripts exhibit heterogeneous size distribution and are processed by a Dicer-dependent RNAi-like mechanism into small RNA molecules that are critical for centromere silencing and integrity (43). Our current findings thus led us to speculate that accumulation of satellite RNA observed in WDHD1-depleted cells might stem from abnormal processing of these transcripts. To test this hypothesis, total RNA was isolated from cells treated with siRNA targeting WDHD1, Dicer or post-transcriptional regulators Drosha and ADAR1, and resolved in a denaturing acrylamide gel. Northern blot analysis using major satellite-specific oligonucleotide probes was then performed and, in agreement with the previous observations, displayed diverse sizes of RNA species in the control cells (Figure 3C). Furthermore, in line with its established catalytic role in cleavage of centromeric RNA transcripts, Dicer ablation caused an enrichment of uncharacteristically larger major satellite RNA. Interestingly, RNAi depletion of WDHD1 altered the transcript sizes in a similar fashion, thus signifying a defective post-transcriptional processing. This finding is also consistent with the above RT-PCR analysis, which demonstrated that transcripts from these heterochromatic regions were more abundant in the total RNA pool of knockdown cells than in that of the control (Figure 3A). In contrast, knockdown of Drosha or ADAR1 expression did not trigger discernible changes in RNA size distribution, therefore excluding their involvement in such process. To further ascertain the role of WDHD1 in the generation of small RNAs homologous to the satellite repeats, we next analyzed small RNA species (<200 nt) isolated from cells. Based on results of the northern blot analysis, expression of the small RNA molecules derived from the major and minor satellite repeats were reduced to a great extent in the absence of WDHD1 (Figure 3D). Knockdown of Dicer also triggered a similar small RNA down-regulation (Supplementary Figure S1). Finally, RNA-FISH analysis revealed normal nuclear punctate distribution of the satellite transcripts in the absence of WDHD1, indicating that this altered transcript processing did not arise from transcript mislocalization (Supplementary Figure S2). Taken together, our data implicate WDHD1 in the regulation of CT/PCT-encoded small RNAs expression, a function likely mediated at the post-transcriptional step.
WDHD1 associates with the centromere-encoded RNA transcripts
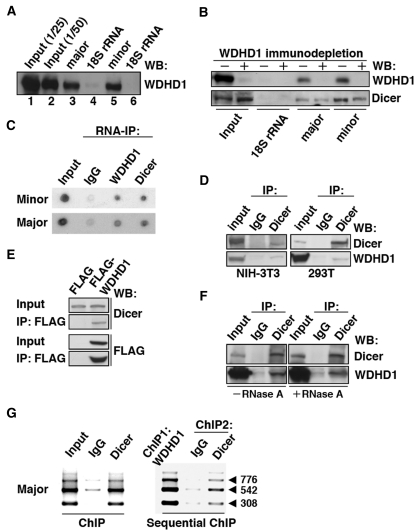
Next, to unequivocally elucidate whether WDHD1 is directly involved in the processing of centromeric transcripts, we sought to determine if WDHD1-centromeric RNAs complexes exist. To demonstrate such association, we first performed RNA pull-down assays using streptavidin-coated beads and in vitro transcribed, biotinylated minor and major satellite RNAs, and probed for the presence of endogenous WDHD1 in the precipitated material. The immunoblotting results showed that WDHD1 in nuclear extracts was efficiently retained on the major and minor satellite RNA (respectively, lanes 3 and 5 of Figure 4A). As a control, no association was observed between WDHD1 and in vitro 18S rRNA transcripts (lanes 4 and 6). Furthermore, we also detected a specific pull-down of Dicer by the major satellite RNA (Figure 4B), consistent with its previously reported role. Next, to characterize the functional relevance of the observed WDHD1-RNA association, we assessed RNA binding of Dicer in the absence of WDHD1. To this end, cell extracts for the pull-down assay were first subjected to immunodepletion by the control or a WDHD1-specific antibody. After pull-down reactions, precipitates were then probed by immunoblotting. Interestingly, our results revealed that the amount of recovered Dicer was diminished after WDHD1 depletion (Figure 4B), implying that a stable association of Dicer with centromeric RNA may require WDHD1.
Figure 4.
WDHD1 associates with the centromere-encoded RNA transcripts. (A and B) RNA pull-down assay was carried out using biotinylated RNA transcripts. (A) In vitro transcribed RNAs corresponding to approximately one repeat of the centromeric major (lane 3) and minor (lane 5) satellite sequences were used as the baits. Precipitated proteins were visualized by western blotting using anti-WDHD1 antibody. Input represents direct loading that equals to 1/25 or 1/50 of the lysate protein used in the pull-down (lanes 1 and 2). Transcripts corresponding to 300 nt (lane 4) and 162 nt (lane 6) of the 18S rRNA sequence serve as controls for the major (lane 3) and minor (lane 5) satellite transcripts, respectively. (B) Dicer association with centromeric RNA sequences attenuated upon WDHD1 depletion. Cell lysates were subjected to immunodepletion using control IgG or WDHD1-specific antibodies, followed by RNA pull-down assay as in (A). (C) RNA-immunoprecipitation assay was performed using antibodies against the indicated proteins (WDHD1 and Dicer). RNA was extracted from the immunoprecipitates and subjected to dot blot analysis using probes corresponding to the sequences of the minor (upper) or major (lower) satellite repeats. (D–F) Association of WDHD1 with Dicer. (D) Anti-Dicer immunoprecipitates (IP) using extracts (input) prepared from NIH-3T3 (left) or 293T (right) cells were probed with antibodies against WDHD1 (top) and Dicer (bottom). (E) 293T cells were transfected with constructs encoding FLAG or FLAG-tagged WDHD1. Immunoprecipitation (IP) of the cell lysates was done using FLAG antibodies, and subsequently analyzed by immunoblotting with antibodies against FLAG (top) and Dicer (bottom). Extent of overexpression and subcellular localization of the FLAG-WDHD1 protein are shown in the Supplementary Information (Supplementary Figure S3). (F) 293T cell lysates were treated with (+) or without (−) RNase A prior to anti-Dicer immunoprecipiation as above. (G) Dicer occupancy of centromeric repeat regions. ChIP assays were performed on crosslinked chromatin from NIH-3T3 cells using antibodies specific for Dicer or control rabbit antibodies (IgG) (left panel). Sequential ChIP analysis was done to assess co-occupancy of the centromere by both WDHD1 and Dicer (right panel). Products of final PCR analysis using primers specific to major satellite repeat DNA sequence are resolved in agarose gel.
To further validate the association of WDHD1 with the CT and PCT transcripts in cells, we conducted anti-WDHD1 and anti-Dicer immunoprecipitation assays, followed by dot-blot analysis of co-precipitated endogenous RNA. Using probes corresponding to major or minor satellite sequences, we uncovered specific co-precipitation of both minor and major satellite-encoded RNA transcripts with WDHD1 and Dicer (Figure 4C). Consistent with a functional interaction, endogenous (Figure 4D) and transiently overexpressed (Figure 4E and Supplementary Figure S3) WDHD1 were also found in association with Dicer. Intriguingly, despite the association of these two proteins with centromeric transcripts, their complex formation was independent of RNA (Figure 4F).
Although Dicer is generally regarded as a cytoplasmic enzyme, its nuclear localization has been reported previously (44–46) and was demonstrated as well by our immunofluorescence analysis (Supplementary Figure S4). The WDHD1-Dicer association therefore likely occurs in the nucleus compartment, a notion further corroborated by the ChIP findings that Dicer also associated with the centromeric chromatin regions (Figure 4G, left panel) and, more importantly, exhibited co-occupancy with WDHD1 (Figure 4G, right panel). Together, these data provide strong evidence for a post-transcriptional function of WDHD1 in centromeric RNA expression.
WDHD1 is important for maintaining centromeric epigenetic markers
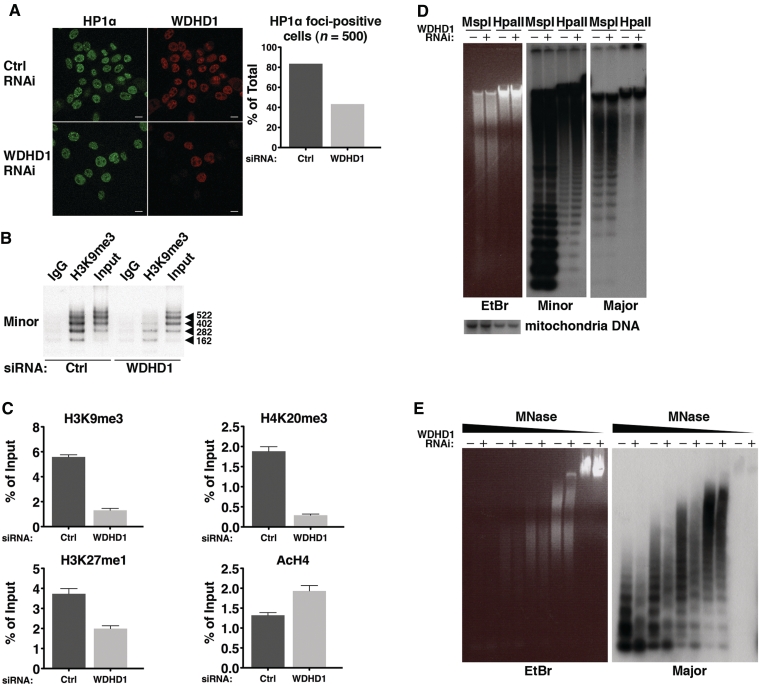
Because small RNA molecules transcribed from the CT/PCT regions are important for the initiation and maintenance of repressive chromatin modifications and structure, we next aimed to dissect the role of WDHD1 in regulating these physical attributes. We first examined the status of HP1α on heterochromatin following WDHD1 knockdown in mouse NIH-3T3 cells. Indirect immunofluorescence analysis revealed a significant loss of HP1α nuclear foci in the knockdown versus control cells (Figure 5A), suggesting a delocalization of this heterochromatin structural protein. Next, ChIP assays followed by PCR using primers spanning the minor (Figure 5B) or major (Figure 5C) satellite regions were performed to compare the levels of several resident histone modifications between control and WDHD1 RNAi cells. As shown in Figure 5B and C, these regions underwent a decline in the repression-associated marks (H3K9me3, H4K20me3 and H3K27me1) in response to WDHD1 knockdown. Significant reduction in the association of H3K9me3 with pericentric heterochromatin was also illustrated by the immunostaining analysis (Supplementary Figure S5A). On the contrary, there was a coincident increase in the level of acetylated H4, an activating mark. We further confirmed that such altered centromeric heterochromatin association was not due to changes in the overall abundance of these epigenetic marks (Supplementary Figure S5B).
Figure 5.
Loss of heterochromatic features at the centromere in the absence of WDHD1. (A) Representative indirect immunofluorescence of HP1α in NIH-3T3 cells transfected with control (ctrl) or WDHD1 siRNA. Loss of HP1α was also measured by calculating the percentage of cells stained positively for HP1α foci in each cell type (n = 500). Percentages given are averages of three independent experiments. (B and C) Chromatin fragments were prepared from control (ctrl) or WDHD1 knockdown cells. ChIP was carried out with control (IgG), H3K9me3, H4K20me3, or H3K27me antibody, as denoted. Semi-quantitative determination of the bound minor satellite DNA in the anti-H3K9me3 ChIP is depicted by the gel figure in (B). Input chromatin was also loaded and equals to 1/10 of IP. Quantitative determination of the bound major satellite DNA, as assessed by quantitative real-time PCR, is shown by bar graphs in (C). Results are expressed as percentages of input chromatin, normalized to control IgG samples, and represent the mean ± SD of at least three independent experiments. (D) DNA methylation analysis of genomic DNA derived from control (−) and knockdown (+) cells. Equivalent amounts of DNA were digested with MspI and HpaII, and subsequently purified and gel separated (EtBr staining on the left). Southern blot analysis was next carried out with hybridizing probes corresponding to minor (middle) or major (right) satellite sequences. Blot was re-hybridized with a mitochondria DNA probe to demonstrate equal loading (bottom). A shorter-exposed version of the blot is shown in the Supplementary Figure S6A. (E) Chromatin structure on the centromeric region was examined in the control (−) and WDHD1 knockdown (+) cells based on the Micrococcal nuclease (MNase)-Southern blot analysis. Isolated intact nuclei were treated with 4.8, 2.4, 1.2, 0.6 and 0 U of MNase. DNA was subsequently purified and separated on an agarose gel (EtBr staining shown in the left panel). In the right panel, the gel was blotted and probed with a radiolabeled probe specific for major satellite repeats.
Moreover, as the centromeric regions are rich in methylated DNA residues (9), we also investigated whether the DNA methylation status of satellite tandem repeats were altered in the absence of WDHD1. Equivalent amounts of DNA from the control and knockdown cells were digested either with the methylation-sensitive enzyme HpaII, or its methylation-insensitive isoschizomer MspI. Southern blot analysis subsequently demonstrated that the CT and PCT regions of genomic DNA from WDHD1-RNAi cells were more easily digested by HpaII (Figure 5D and it shorter-exposed version in Supplementary Figure S6A), suggesting that it was significantly less methylated than DNA isolated from the control. In contrast, comparable digestion by MspI (Figure 5D) as well as equivalent signals corresponding to the mitochondrial DNA (Figure 5D, bottom) verified equal loading of DNA in the digestion reactions and in agarose gel electrophoresis, respectively. We further excluded the possibility that such DNA methylation alteration may stem from disproportionate numbers of replicating cells in the culture by showing that both the control and WDHD1 knockdown cells exhibited similar levels of DNA replication (Supplementary Figure S6B). Therefore, abrogation of WDHD1 specifically led to a decrease in the levels of DNA methylation at the CT and PCT.
Heterochromatin structural alteration in the absence of WDHD1
Since the above findings strongly imply that silencing of WDHD1 might compromise the heterochromatin structure surrounding the CT/PCT regions, we next set out to examine the role of WDHD1 on the associated chromatin architecture. In this regard, we employed a nuclease accessibility assay using Micrococcal nuclease (MNase). Equivalent amounts of nuclei isolated from cells harboring control and knockdown siRNA were digested with increasing amounts of MNase, which cuts chromatin at the inter-nucleosomal linker. DNA were then purified from these nuclei and detected by Southern blot analysis using minor and major satellite-specific probes. Using this assay, we detected at equal loading a larger extent of chromatin DNA digestion in the knockdown cells (Figure 5E), indicative of a more decompacted chromatin structure assembled at these sequences when WDHD1 was abrogated. This finding of chromatin structure relaxation, together with those for the reduction of the repressive epigenetic marks, is consistent with a reversal of heterochromatin maintenance in the centromeric regions and further emphasizes the importance of WDHD1 in this regulatory process.
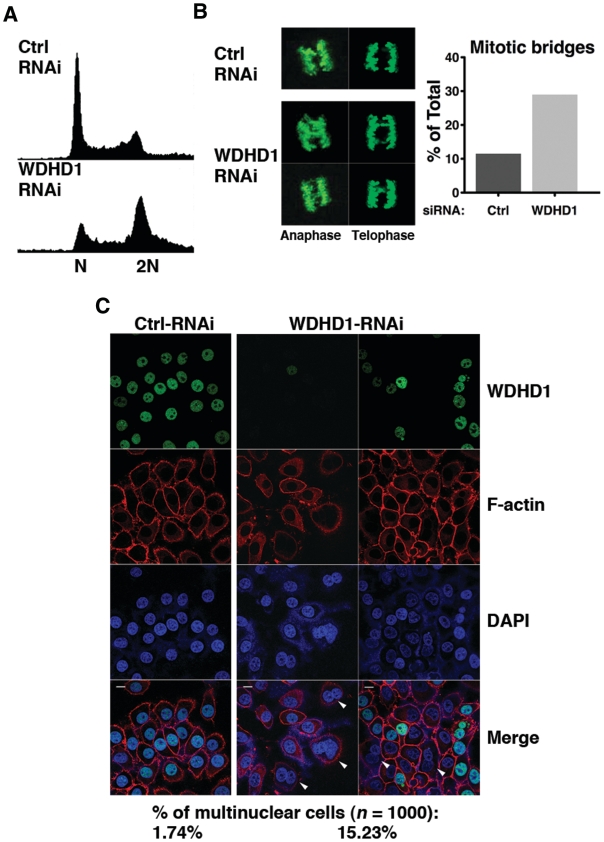
Down-regulation of WDHD1 leads to cell cycle progression deficiencies associated with defective centromere
Next, to address the physiological significance of WDHD1-mediated regulation of centromere integrity, we wanted to assess the effects of WDHD1 knockdown upon the progression and fidelity of the cell cycle, particularly in the mitotic phase. Flow cytometry analysis demonstrated an accumulation of cells with a DNA content of 4 N (representative of G2/M phase) in the WDHD1 knockdown culture (Figure 6A), implying possibly a delay in G2 and/or mitosis progression. Immunofluorescence staining with phospho-H3 antibody, which demarcates condensed mitotic chromosomes, further revealed that silencing of WDHD1 resulted in pronounced increase in the occurrence of mitotic chromosome bridges (11.5–29%, Figure 6B), indicative of defects in chromosome segregation. This phenotype was also accompanied by abnormal nuclear morphology (marked by arrowheads in Figure 6C), characterized by significantly higher incidence of cells with multiple nuclei (15.2% in the knockdown cells compared to 1.7% in the control). We conclude that WDHD1 down-regulation, giving rise to dramatic loss of the centromeric small RNA and disruption of the associated heterochromatin structure, likely impacts on the centromere integrity and kinetochore assembly, and consequently faithful chromosome segregation during mitosis.
Figure 6.
Down-regulation of WDHD1 leads to cell cycle progression deficiencies attributable to defective centromere. (A) Cell cycle profiles of the control (ctrl) and WDHD1 knockdown cells. Mock or WDHD1-knockdown 293T cells were subjected to FACS for measurement of DNA content. (B) Mitotic fidelity was assessed by scoring mitotic cells with chromosome bridges. Condensed mitotic chromosomes were visualized by phospho-histone H3 immunostaining. Representative immunofluorescence images are depicted on the left. Both anaphase and telophase nuclei are shown, as indicated. Panel on the right shows the percentages of mitotic cells with chromosome bridges in control (ctrl) versus knockdown cells (n = 250). The results represent averages of three independent experiments. (C) Nuclear morphology was altered in WDHD1-deficient cells. Control (ctrl) and knockdown HeLa cells were immunostained for WDHD1, and counterstained for F-actin (for demarcating cell membrane) and DNA (DAPI). Confocal microscopy was done as described before (scale bar is 10 µm). Percentages of cells with more than one nuclei staining, examples of which are indicated by arrowheads, are shown on the bottom (n = 1000) and represent averages of three independent experiments.
DISCUSSION
Due to the importance of the centromere to the maintenance and propagation of eukaryotic genetic materials, its structural integrity is thought to be under intricate control by intercalated mechanisms. To the previously discovered effectors in these regulatory networks, we have added a new important one. In our present report, a functional link of mammalian WDHD1 protein to the centromeric silencing process was demonstrated. Our findings also unequivocally revealed some aspects of the molecular mechanism underlying such involvement, which entails post-transcriptional processing of CT- and PCT-encoded RNA transcript. This mode of action, which resembles the transcriptional gene silencing pathway previously documented in the fission yeast, may also engage the Dicer machinery. To our knowledge, this is the first report of a hitherto unrecognized link of an HMG-domain containing protein to heterochromatin maintenance.
Consistent with its significance in maintaining the histone code and structural attributes of the centromeric heterochromatin (Figure 5), abrogation of WDHD1 expression distinctly compromised mitotic fitness of the cell cycle (Figure 6). The observed phenotypes are in accordance with those previously reported for other centromere-associated epigenetic or structural factors. For instance, deficiency in KDM2A and SUV39h1 (9,10,47) or drug inhibition (48), which abrogates proper expression of defining histone marks, was found to affect centromere/kinetochore organization and sister chromatid segregation. Intriguingly, one of the cell cycle abnormalities that distinctively arose in WDHD1 knockdown HeLa cells was the accumulation of multinuclear cells (Figure 6C). Such aberrant nuclear morphology is conceived as a hallmark of mitotic catastrophe, a cellular response to chromosome missegregation and mitotic DNA damage (49). The occurrence of this nuclear aberration, including binucleation and multimicronucleation, attests to the critical role of WDHD1 in maintaining centromere structure, which underlies proper segregation of duplicated chromosomes. Furthermore, consistent with a role in the proliferation-associated centromeric silencing process, WDHD1 was recently shown to exhibit a high expression in the testis tissue (50). However, upon further examination of protein expression data in the public databases (the Human Protein Atlas), no significant enrichment of WDHD1 was found that particularly correlated with the proliferation state or tissue type (Supplementary Tables S2 and S3). Comparative profiling of EST sequences in normal and tumor tissues (the Cancer Genome Anatomy Project) also did not reveal significant differences in WDHD1 transcript representation (data not shown). Such wide distribution of WDHD1 is therefore in line with its potential relevance in fundamental cellular processes, such as centromere regulation.
A role in centromere integrity and chromosome segregation fidelity has previously been ascribed to the yeast homologues of WDHD1 (27,29–31). Several reports have preliminarily implied that such role may be mediated through a DNA replication-dependent mechanism (20–23). In this capacity, yWDHD1 may act as an accessory factor for DNA polymerase or interacts with replication factors GINS and/or Tipin to facilitate heterochromatin DNA synthesis. It is thus a formal possibility that WDHD1 travels with the replication machinery as it acts on the centromere. Indeed, there was a partial overlap in the immunostaining signals of WDHD1 and BrdU, indicative of a possible spatial association (Figure 1A). However, our current findings provide several lines of evidence that strongly argues instead for a post-transcriptional role of WDHD1 in the regulation of centromere identity. First, despite colocalization with the BrdU incorporation sites, the temporal change in spatial patterns displayed by WDHD1 (horse-shoe versus sphere) during mid-to-late S phase is in contrast to the persistent localization of replication machinery on the periphery of the pericentric heterochromatin foci (51). In addition, such focal association occurs subsequently to the accumulation of CT- and PCT-encoded transcripts, which was reported to be at the G1/S junction (52). Second, the focal immunostaining patterns of WDHD1 are sensitive to RNA Pol II inhibitor (Figure 2A), suggesting that such localization is coordinated with RNA synthesis. Third, the alteration in the processing of CT- and PCT-encoded transcripts as a consequence of WDHD1 knockdown (Figure 3) phenocopies defects previously observed in the Dicer−/− MEF cells. This, together with the observation of the WDHD1-centromeric RNA association (Figure 4), clearly implies that this RNA species may be the site of WDHD1’s post-transcriptional action. Finally, results from our co-IP experiments did not reveal association between WDHD1 and DNA replication machinery (Figure 2A). In addition, no delay in S phase progression was evident in the WDHD1 knockdown cells (Figure 6A), thus suggesting that WDHD1 may not be directly linked to DNA replication regulation at the centromere.
How WDHD1 is recruited to the centromeric regions at distinct stage of the S phase remains an unresolved issue. Due to the presence of the carboxyl-terminal HMG domain, it is likely that WDHD1 recognizes specific sequence or structure characteristic of the centromere DNA (53). Interestingly however, while yeast homolog Mcl1 lacks such distinguishing feature of the mammalian protein, it can still localize and regulate the centromeric region. In spite of such discrepancy, no concrete evidence is yet present to exclude the possibility that HMG may still play a role in stabilizing WDHD1’s centromeric binding. WDHD1’s centromeric occupancy may also depend on other factors, such as epigenetic and/or structural proteins, or particular histone modification(s), that may serve as adaptor for such distinct localization. Finally, as the focal immunostaining patterns of WDHD1 is transcription-dependent (Figure 2B), and by virtue of WDHD1’s interaction with RNA pol II (Figure 2A), WDHD1 may very likely rely on RNA pol II and associated factors to become connected with the centromeric heterochromatin regions during S phase. Understanding the mechanism of its centromere recruitment may shed new light on the functional role of WDHD1 in the regulation of centromeric silencing process.
FUNDING
National Science Council of Taiwan (NSC99-2321-B-002-001 to S.-C.L., NSC97-2320-B-182-027-MY3 to B.C-M.T., NSC98-2312-B-182-001-MY3 to H.L.); Chang Gung Memorial Hospital (CMRPD170302 to B.C-M.T.); National Health Research Institute of Taiwan (NHRI-EX99-9923SC to B.C-M.T.); Ministry of Education, Taiwan (to Chang Gung University). Funding for open access charge: Chang Gung Memorial Hospital, Taiwan.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Thomas Jenuwein for the H3K9me3 antibody. We are grateful to Simon Silver, Scott Schuyler and members of the B.C-M.T. lab for critical reading of the article and important discussions.
REFERENCES
- 1.Amor DJ, Kalitsis P, Sumer H, Choo KH. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 2004;14:359–368. doi: 10.1016/j.tcb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Warburton PE, Cooke HJ. Hamster chromosomes containing amplified human alpha-satellite DNA show delayed sister chromatid separation in the absence of de novo kinetochore formation. Chromosoma. 1997;106:149–159. doi: 10.1007/s004120050234. [DOI] [PubMed] [Google Scholar]
- 3.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 4.Eymery A, Horard B, El Atifi-Borel M, Fourel G, Berger F, Vitte AL, Van den Broeck A, Brambilla E, Fournier A, Callanan M, et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res. 2009;37:6340–6354. doi: 10.1093/nar/gkp639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 6.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128:647–650. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 9.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 10.Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA, Pagano M. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle. 2008;7:3539–3547. doi: 10.4161/cc.7.22.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papait R, Pistore C, Grazini U, Babbio F, Cogliati S, Pecoraro D, Brino L, Morand AL, Dechampesme AM, Spada F, et al. The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol. Biol. Cell. 2008;19:3554–3563. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol. Cell. 2007;18:1098–1106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 14.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 15.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez HR, Kavi HH, Xie W, Birchler JA. Heterochromatin: on the ADAR radar? Curr. Biol. 2005;15:R132–R134. doi: 10.1016/j.cub.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr. Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 19.Kohler A, Schmidt-Zachmann MS, Franke WW. AND-1, a natural chimeric DNA-binding protein, combines an HMG-box with regulatory WD-repeats. J. Cell Sci. 1997;110(Pt 9):1051–1062. doi: 10.1242/jcs.110.9.1051. [DOI] [PubMed] [Google Scholar]
- 20.Bermudez VP, Farina A, Tappin I, Hurwitz J. Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 2010;285:9493–9505. doi: 10.1074/jbc.M109.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Errico A, Cosentino C, Rivera T, Losada A, Schwob E, Hunt T, Costanzo V. Tipin/Tim1/And1 protein complex promotes Pol alpha chromatin binding and sister chromatid cohesion. EMBO J. 2009;28:3681–3692. doi: 10.1038/emboj.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshizawa-Sugata N, Masai H. Roles of human AND-1 in chromosome transactions in S phase. J. Biol. Chem. 2009;284:20718–20728. doi: 10.1074/jbc.M806711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Formosa T, Nittis T. Dna2 mutants reveal interactions with Dna polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams DR, McIntosh JR. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot. Cell. 2002;1:758–773. doi: 10.1128/EC.1.5.758-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams DR, McIntosh JR. Mcl1p is a polymerase alpha replication accessory factor important for S-phase DNA damage survival. Eukaryot. Cell. 2005;4:166–177. doi: 10.1128/EC.4.1.166-177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamnun YM, Katayama S, Toda T. Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem. Biophys. Res. Commun. 2006;350:125–130. doi: 10.1016/j.bbrc.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 29.Natsume T, Tsutsui Y, Sutani T, Dunleavy EM, Pidoux AL, Iwasaki H, Shirahige K, Allshire RC, Yamao F. A DNA polymerase alpha accessory protein, Mcl1, is required for propagation of centromere structures in fission yeast. PLoS One. 2008;3:e2221. doi: 10.1371/journal.pone.0002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petronczki M, Chwalla B, Siomos MF, Yokobayashi S, Helmhart W, Deutschbauer AM, Davis RW, Watanabe Y, Nasmyth K. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 2004;117:3547–3559. doi: 10.1242/jcs.01231. [DOI] [PubMed] [Google Scholar]
- 31.Xu H, Boone C, Brown GW. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics. 2007;176:1417–1429. doi: 10.1534/genetics.107.072876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan BC, Lee SC. Nek9, a novel FACT-associated protein, modulates interphase progression. J. Biol. Chem. 2004;279:9321–9330. doi: 10.1074/jbc.M311477200. [DOI] [PubMed] [Google Scholar]
- 33.Wright JH, Munar E, Jameson DR, Andreassen PR, Margolis RL, Seger R, Krebs EG. Mitogen-activated protein kinase kinase activity is required for the G(2)/M transition of the cell cycle in mammalian fibroblasts. Proc. Natl Acad. Sci. USA. 1999;96:11335–11340. doi: 10.1073/pnas.96.20.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, Russell J, Lee SC, Zomerdijk JC. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Tan BC, Tseng KH, Chuang CP, Yeh CW, Chen KD, Lee SC, Yung BY. Nucleophosmin acts as a novel AP2alpha-binding transcriptional corepressor during cell differentiation. EMBO Rep. 2007;8:394–400. doi: 10.1038/sj.embor.7400909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Corput MP, Grosveld FG. Fluorescence in situ hybridization analysis of transcript dynamics in cells. Methods. 2001;25:111–118. doi: 10.1006/meth.2001.1220. [DOI] [PubMed] [Google Scholar]
- 39.Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 40.Patrone G, Puppo F, Cusano R, Scaranari M, Ceccherini I, Puliti A, Ravazzolo R. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. Biotechniques. 2000;29:1012–1014. doi: 10.2144/00295st02. 1016–1017. [DOI] [PubMed] [Google Scholar]
- 41.Kafatos FC, Jones CW, Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979;7:1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eymery A, Callanan M, Vourc’h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int. J. Dev. Biol. 2009;53:259–268. doi: 10.1387/ijdb.082673ae. [DOI] [PubMed] [Google Scholar]
- 44.Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Developmental Cell. 2010;18:102–113. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 45.Giles KE, Ghirlando R, Felsenfeld G. Maintenance of a constitutive heterochromatin domain in vertebrates by a Dicer-dependent mechanism. Nature Cell Biol. 2010;12:94–99. doi: 10.1038/ncb2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PloS one. 2010;5:e12175. doi: 10.1371/journal.pone.0012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 48.Heit R, Rattner JB, Chan GK, Hendzel MJ. G2 histone methylation is required for the proper segregation of chromosomes. J. Cell Sci. 2009;122:2957–2968. doi: 10.1242/jcs.045351. [DOI] [PubMed] [Google Scholar]
- 49.Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 50.Sato N, Koinuma J, Fujita M, Hosokawa M, Ito T, Tsuchiya E, Kondo S, Nakamura Y, Daigo Y. Activation of WD repeat and high-mobility group box DNA binding protein 1 in pulmonary and esophageal carcinogenesis. Clin. Cancer Res. 2010;16:226–239. doi: 10.1158/1078-0432.CCR-09-1405. [DOI] [PubMed] [Google Scholar]
- 51.Dimitrova DS, Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- 52.Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 2007;179:411–421. doi: 10.1083/jcb.200706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonstrup AT, Thomsen T, Wang Y, Knudsen BR, Koch J, Andersen AH. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIalpha. Nucleic Acids Res. 2008;36:6165–6174. doi: 10.1093/nar/gkn640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.