Abstract
We purified an inhibitor of oriC plasmid replication and determined that it is a truncated form of ribosomal protein L2 evidently lacking 59 amino acid residues from the C-terminal region encoded by rplB. We show that this truncated form of L2 or mature L2 physically interacts with the N-terminal region of DnaA to inhibit initiation from oriC by apparently interfering with DnaA oligomer formation, and the subsequent assembly of the prepriming complex on an oriC plasmid. Both forms of L2 also inhibit the unwinding of oriC by DnaA. These in vitro results raise the possibility that one or both forms of L2 modulate DnaA function in vivo to regulate the frequency of initiation.
INTRODUCTION
In Escherichia coli, initiation of chromosomal DNA replication at the replication origin (oriC) leads to the assembly of a replisome at each replication fork [reviewed in (1)]. During initiation, DnaA protein recognizes 9-mer motifs termed DnaA boxes, other sequences named I- and τ-sites, and possibly the AT-rich region carrying three 13-mer motifs near the left boundary of oriC [reviewed in (2)]. DnaA complexed to ATP then unwinds the AT-rich region in a process that is stimulated by other proteins (see below). Following unwinding, a DnaA oligomer loads two DnaB helicase molecules complexed to DnaC on each separated strand of oriC to form the prepriming complex (3–7). Next upon interacting with DnaB, primase synthesizes primers that are extended by DNA polymerase III holoenzyme. The dynamic interactions of primase, DnaB helicase and the DNA polymerase advance each replication fork, which moves in opposite directions from oriC to duplicate the circular bacterial chromosome.
In vitro studies show that Integration Host Factor (IHF), HU and DiaA stimulate the DnaA-dependent unwinding of oriC (8–10). For HU and DiaA, they act by stabilizing the DnaA oligomer (9,10). These results correlate with in vivo studies, which showed that mutants of hupA encoding the α subunit of HU or of diaA initiate at abnormal times in the bacterial cell cycle (11–13). Similarly, mutation of the IHF binding site within oriC disrupts IHF binding and oriC function, and causes asynchronous initiation (14,15). As the anomalous timing of initiation is also observed with mutations in other loci to affect either the function of DnaA or oriC (16), these observations suggest that the stimulation of oriC unwinding by these proteins is physiologically relevant.
These findings raise the opposing possibility of factors that negatively regulate initiation at the step of oriC unwinding. In support, we recently found that Dps, which protects DNA from oxidative damage (17–19), interacts with DnaA to inhibit both the unwinding of oriC and DNA replication of an oriC-containing plasmid (20). Additional in vivo evidence suggests that Dps may act as a checkpoint during oxidative stress to reduce the frequency of initiation so that repair mechanisms can fix the oxidative damage in DNA before the genome is duplicated.
In E. coli and other free-living organisms, the initiation of DNA replication occurs at a specific time in the cell cycle, and is highly coordinated with cell growth. However, the mechanism of this coordination is not understood. Ribosome biogenesis also correlates with cell growth [reviewed in (21)]. The prevailing view is that the cellular concentration of ribosomes determines the rate of protein synthesis, which controls the rate of bacterial growth. These observations suggest that a factor that is required to assemble ribosomes may couple the initiation of DNA replication with cell growth.
During the purification of Dps for the work summarized above, we detected a factor that inhibited oriC plasmid replication. We purified it based on its inhibitory activity and identified it as ribosomal protein L2 by N-terminal sequence analysis and by immunoblotting with antibody specific for L2. However, electrospray mass spectrometry suggests that the protein isolated lacks 59 amino acid residues from the C-terminus. Because of the requirement of L2 for ribosome biogenesis, which is coupled to cell growth, we considered the possibility that it may inhibit DnaA function to affect the initiation process. If so, this essential protein, which is one of the most evolutionarily ancient among ribosomal proteins (22), may act to coordinate the initiation of DNA replication with cell growth. We show that this truncated form of L2 (hereafter called tL2) and also L2 physically interact with DnaA, and that the ribosomal protein inhibits the unwinding of oriC by DnaA and the assembly of the oriC prepriming complex.
MATERIALS AND METHODS
Reagents, proteins and DNAs
Commercial reagents and replication proteins have been described (9,20,23,24). L2 was a gift from Dr Knud Nierhaus at the Max-Planck-Institut für Molekulare Genetik. For experiments to assemble the prepriming complex, DnaA protein joined at its N-terminus to polyhistidine was used (25). This protein is essentially identical to wild-type DnaA based on the following evidence. We showed that a plasmid encoding wild-type dnaA joined at its N-terminal coding region to a DNA sequence for polyhistidine, or this plasmid lacking the sequence for polyhistidine equivalently maintained an oriC plasmid (pCM959-CmR) in a strain lacking the chromosomal dnaA gene [MS3898 (genotype: asnB32 relA1 spoT1 thi-1 ilv-192 zia::pKN500 (pKN500=mini-R1) ΔdnaA mad-2 (F-) recA1 (λimm434)]. The size of colonies was similar. We also showed that polyhistidine-tagged DnaA is essentially identical to wild-type DnaA in vitro in assays of oriC plasmid replication, duplication of M13 A-site ssDNA, ATP binding, ATPase activity, sequence-specific binding to a DNA fragment carrying oriC, Form 1* formation, unwinding of oriC as measured by sensitivity to P1 nuclease, prepriming complex assembly and interaction with DnaB measured by enzyme-linked immunosorbent assay (ELISA) or surface plasmon resonance [(4,25,26), Makowska-Grzyska and Kaguni, unpublished data]. Bovine carbonic anhydrase II and P1 nuclease were obtained from Sigma Chemical Co., St Louis, MO, USA. Hda carrying a polyhistidine tag (MGHHHHHHHHHHSSGHIQGRH) at its N-terminus, GrpE and Drosophila melanogaster topoisomerase I were laboratory stocks. Purified DnaAΔ129 lacking the N-terminal 129 residues (27) and DnaAΔ220-294 lacking residues 220–294 (27) are described in the cited references. Purified recombinant murine galectin-3 (28) and rabbit antiserum that specifically interacts with murine galectin-3 were gifts from Dr John Wang at Michigan State University. Rabbit antiserum that recognizes the C-terminal region (amino acids 370–467) of DnaA, affinity-purified rabbit polyclonal antibody for DnaB and L2 and the monoclonal antibody named M43 that recognizes DnaA (29) were prepared in our laboratory. M13oriC2LB5 supercoiled DNA contains the E. coli chromosomal origin inserted into the vector, M13ΔE101 (30). M13 A-site single-stranded DNA carries a DnaA box in a hairpin structure (31). pUC19 DNA was a laboratory stock.
Purification of an inhibitor of oriC plasmid replication
Replication of an oriC-containing plasmid requires that it is negatively supercoiled. Because topoisomerase I relaxes supercoiled DNA to inhibit oriC plasmid replication, we purified tL2 from a log phase culture of E. coli DM700 [Δ(topA-cysB)217 acrA11] (32) grown in Luria Bertani (LB) media at 37°C and harvested at a turbidity of 0.6 (595 nm). The cell pellet was resuspended in Buffer A (50 mM Tris–HCl, pH 8.0, 0.1 mM ethylenediamine-tetraacetic acid (EDTA), 5 mM dithiothreitol (DTT) and 10% glycerol) supplemented with 1.5 M NaCl, lysed with a French press, and the lysate supernatant was obtained by centrifugation for 30 min at 14 000 rpm in a Sorvall SS-34 rotor. All operations at this step and beyond were performed at 0–4°C. A solution of PEG 6000 was added to the supernatant with stirring to a final concentration of 8% (w/v). After 2 h of continuous stirring, the precipitate was collected by centrifugation as described above, resuspended in Buffer A containing 1.5 M NaCl and dialyzed against Buffer A. The dialyzed sample was then clarified by centrifugation as above but for 5 min, and applied to a heparin Sepharose (Amersham) column (72 ml) equilibrated in Buffer A. The column was washed extensively with Buffer A, and eluted with a 360 ml linear gradient of 0–1 M NaCl in Buffer A. Based on an assay of DNA replication of an oriC-containing plasmid, heparin Sepharose chromatography separated tL2 from two other inhibitory activities attributed to PepA and HU. Relative to the total inhibitory activity applied to the column, about 20% of this activity was due to tL2, which elutes between PepA and HU. Fractions containing tL2 were pooled and dialyzed overnight against Buffer B [25 mM N-(2-hydroxyethyl)-N′-(2-ethanesulfonic acid) (HEPES)–KOH, pH 8, 1 mM EDTA, 1 mM DTT and 10% (v/v) glycerol] supplemented with 50 mM NaCl. The insoluble material was collected by centrifugation at 18 000 rpm for 15 min in a Sorvall SS-34 rotor, and resuspended in a minimal volume of Buffer B containing 1 M NaCl. Compared with the supernatant that contained about 4% of the activity, this fraction contained about 14% relative to the activity in the sample before heparin Sepharose chromatography. After fractionation in a Superdex 75 HR 10/30 column (GE Healthcare) equilibrated in Buffer B supplemented with 400 mM NaCl, about 2% of inhibitory activity was recovered relative to the total activity in the fraction before heparin Sepharose chromatography. One unit of activity corresponds to that which causes 70% inhibition of in vitro DNA replication.
Electrospray ionization mass spectrometry
Analysis of purified tL2 isolated in Figure 1 was performed with a Waters Q-TOF Ultima APT spectrometer coupled to a Waters Alliance 2795 HPLC system. Samples were loaded onto a 5 μ pore size 10 × 1 mm Beta Basic CN column, followed by gradient elution starting with 0.1% formic acid in water and ending with acetonitrile. Data were collected in the positive ion mode with a capillary voltage of 3 kV, with a source temperature of 90°C and a cone voltage of 35 eV.
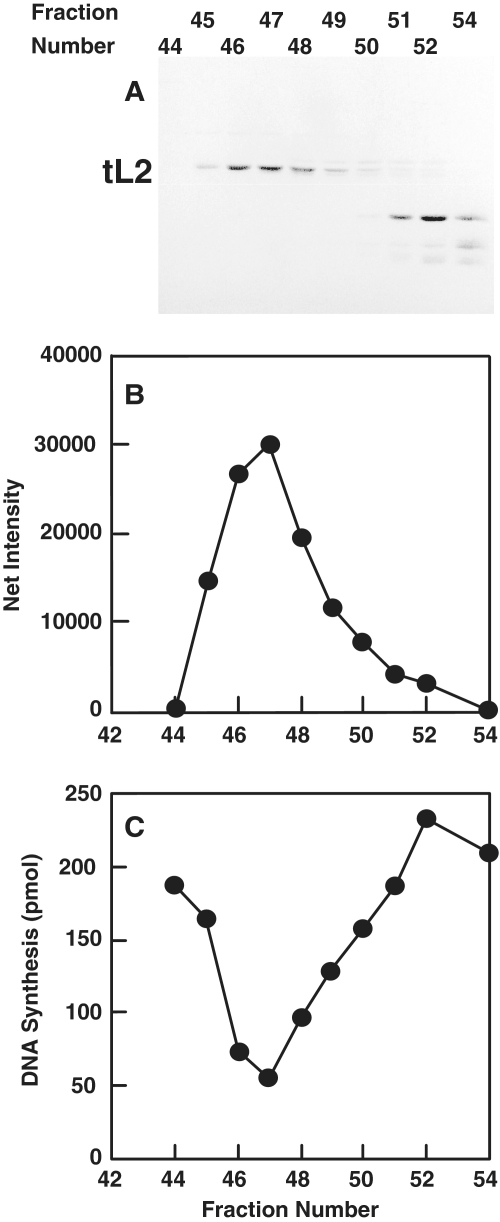
Figure 1.
Purification of a protein that inhibits DNA replication of an oriC-containing plasmid. (A) The indicated column fractions obtained by a Superdex 75 chromatography (GE Healthcare) were analyzed in a Coomassie blue-stained SDS–polyacrylamide gel (15% acrylamide). The elution volumes of monomeric DnaC (27.9 kDa) and other proteins were also determined for this column. (B) The amount of tL2 in (A) was quantified by densitometric analysis and expressed as net intensity, which is the background-subtracted pixel value for each band on the gel. (C) The extent of DNA synthesis was measured as described in ‘Materials and Methods’ section. DNA synthesis for an uninhibited reaction corresponds to about 320 pmol.
In vitro DNA replication of an oriC-containing plasmid
Each reaction mixture (25 µl) to measure DNA replication contained 40 mM HEPES–KOH, pH 7.6, 40 mM potassium glutamate, 10 mM magnesium acetate, 4 mM dithiothreitol, 0.1 mg/ml bovine serum albumin (BSA), 4% (w/v) sucrose, 2 mM ATP, 0.25 mM each of cytidine-5′-triphosphate (CTP), guanosine-5′-triphosphate (GTP) and uridine-5′-triphosphate (UTP), 100 µM each of dATP, dCTP, dGTP and [methyl-3H]-TTP (25–30 cpm/pmol), M13oriC2LB5 DNA (46 fmol; 200 ng) and the required purified replication proteins including DnaA (1 pmol; 50 ng) as described (33). The reactions were assembled at 0°C, and incubated at 30°C for 30 min to measure DNA synthesis after adding tL2 or L2. Total nucleotide incorporation of DNA (as pmol) was measured by liquid scintillation spectrometry after trichloroacetic acid precipitation onto glass fiber filters (Whatman GF/C).
DNA topology assays
To measure the sequestration of negative supercoils of a plasmid, reactions (23 µl) contained M13oriC2LB5 supercoiled DNA (46 fmol; 200 ng) in 25 mM HEPES–KOH, pH 7.6, 15% (v/v) glycerol, 1 mM DTT, 0.1 mM EDTA, 0.1 mM magnesium acetate, 2 mM ATP and the indicated amounts of either form of L2 or HU. After incubation at 30°C for 20 min with D. melanogaster topoisomerase I (9 ng), 5 µl of 40% sucrose, 10 mM EDTA, 3% sodium dodecyl sulfate (SDS) and 0.25% bromophenol blue was added to each reaction, followed by incubation at 86°C for 5 min. The samples were then electrophoresed at room temperature for about 12 h in a 0.8% agarose gel containing 90 mM Tris-borate buffer and 2 µg/ml chloroquine. To measure the production of Form 1* DNA, reactions (25 µl) were assembled essentially as described (6,9), and contained 40 mM HEPES–KOH, pH 7.6, 20 mM Tris–HCl, pH 7.5, 4% (v/v) sucrose, 4 mM DTT, 2 mM ATP, 10 mM magnesium acetate, 6 mM creatine phosphate, 0.1 µg/ml creatine kinase, M13oriC2LB5 DNA (200 ng, 46 fmol), HU (5 ng, α dimer), DnaA (0.8 pmol; 40 ng except as noted), DnaB (50 ng), DnaC (50 ng), single-strand DNA binding protein (SSB) (320 ng), DNA gyrase A subunit (360 ng), DNA gyrase B subunit (510 ng) and the indicated amounts of tL2 or L2. After incubation at 30°C for 25 min, the reactions were stopped by the addition of EDTA and SDS to final concentrations of 10 mM and 1% (w/v), respectively, followed by incubation at 65°C for 5 min and agarose gel electrophoresis. To measure the localized unwinding of oriC, assays (10 µl) contained 25 mM HEPES–KOH, pH 7.6, 12% (v/v) glycerol, 1 mM CaCl2, 0.2 mM EDTA, 5 mM ATP, 0.1 mg/ml BSA, M13oriC2LB5 (23 fmol; 100 ng), HU (5 ng, α dimer), DnaA (0.5 pmol; 25 ng unless noted) and the indicated amounts of tL2 as described (9,34,35). Incubation at 38°C was for 15 min, followed by the addition of 2 U of P1 nuclease, as noted, and incubation for 40 s. The reactions were stopped as described above for the Form 1* DNA assay followed by agarose gel electrophoresis.
ELISA
ELISAs were performed essentially as described (9,20). As indicated, tL2, L2 or HU was added to one set of wells of a microtiter plate in 100 μl of PBS buffer (10 mM Na2HPO4, 1.76 mM KH2PO4, 2.7 mM KCl and 0.137 M NaCl). After incubation for 1 h at 20°C, unbound protein was removed with three changes of 200 μl of PBS buffer supplemented with 0.05% (v/v) Tween-20 and 2% (w/v) non-fat milk; the last buffer change was incubated for 30 min. The indicated amounts of wild-type DnaA, galectin-3, and also the various mutant DnaAs at 50 ng each were then added in triplicate in 100 μl of PBS buffer to wells containing tL2, L2 or HU, followed by incubation for 1 h at 20°C. As controls, the indicated amounts of wild-type DnaA, murine galectin-3 (50 ng) and the mutant DnaAs (50 ng) were immobilized in triplicate to separate wells of the microtiter plate that lacked tL2, L2 or HU. After removing unbound protein as described above, the wells containing DnaA, the mutant DnaAs or galectin-3 were incubated overnight at 4°C with antisera diluted in PBS buffer that recognize either the C-terminal region of DnaA or galectin-3, respectively. After removing the unbound antibody as described above, immune complexes were detected colorimetrically at 490 nm with goat anti-rabbit antibody conjugated to horseradish peroxidase.
Isolation of the prepriming complex
The prepriming complex was assembled with 10-fold greater amounts of DnaA, DnaB, DnaC, HU and the oriC-containing plasmid than in a standard DNA replication reaction as described (4,6,9,26), but with 0.25 mM magnesium acetate, and either tL2 or L2 protein as indicated. Where indicated, tL2 (2.8 µg) or L2 (2 µg) was added prior to the isolation at 10-fold higher amounts than that of a standard DNA replication reaction. SSB was omitted at this stage. After incubation for 10 min at 37°C, the prepriming complex was isolated in the void volume of a Sepharose CL-4B (Pharmacia) gel filtration column equilibrated in 40 mM HEPES–KOH, pH 8.0, 40 mM potassium glutamate, 0.25 mM magnesium acetate, 4% (w/v) sucrose, 4 mM dithiothreitol, 100 µg/ml BSA and 5 mM ATP at room temperature. To measure DNA replication, the indicated void volume fractions (20 µl) were supplemented with SSB, primase, DNA polymerase III holoenzyme, DNA gyrase, magnesium acetate (10 mM final concentration) and other required components at standard amounts for DNA replication in a final volume of 25 µl followed by incubation at 30°C for 30 min. Portions of the void volume fractions were also electrophoresed in an agarose gel in parallel with known amounts of M13oriC2LB5 DNA, which was used to prepare a standard curve. After ethidium bromide staining, the amount of DNA in the isolated prepriming complex was quantified. The amounts of DnaA and DnaB in the isolated complex were determined by quantitative immunoblotting relative to the known amounts of the respective proteins, which were electrophoresed and transferred in parallel. M43 monoclonal antibody was used to detect DnaA protein (29), and affinity-purified polyclonal antibody was used for DnaB. The chemiluminescent signal (SuperSignal, Pierce) of horseradish peroxidase conjugated to the secondary antibody in immune complexes was quantified with a Kodak 4000R imaging system.
RESULTS
Isolation and identification of a factor that inhibits oriC plasmid replication in vitro
We discovered a factor that inhibited oriC plasmid replication in vitro, and purified it by standard liquid chromatographic methods based on this assay (see ‘Materials and Methods’ section). By gel permeation chromatography (Superdex 75, GE Healthcare), the inhibitory activity correlates with a polypeptide that elutes at the position of DnaC, a monomer of 28 kDa (Figure 1). Automated Edman degradation revealed that the N-terminal sequence (AVVKXKPTSPGRRHV where X represents an unknown amino acid) of the protein corresponds to that of ribosomal protein L2, but lacking the first methionine encoded by rplB.
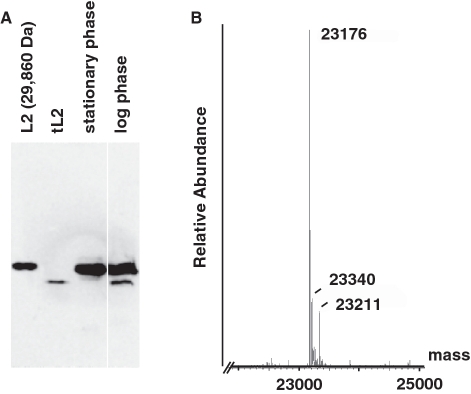
We confirmed the identity of the polypeptide by immunoblot analysis with an affinity-purified antibody that reacts specifically with L2 (Figure 2A). However, the polypeptide isolated has an electrophoretic mobility that is greater than L2 obtained as a gift from the Nierhaus laboratory. Its position is very similar to one of two polypeptides in a whole-cell lysate of a log phase culture of E. coli DM700 from which we purified this form of L2, but only the larger L2 was detected in a stationary phase culture. We also detected this truncated form of L2 (tL2) in log phase cultures of E. coli MG1655 and W3110 (data not shown), suggesting that tL2 is present in many other E. coli strains. Hence, tL2 does not apparently arise by proteolysis during purification. These findings raise the possibility that L2, like other ribosomal proteins summarized in the ‘Discussion’ section, has dual roles. We do not know if all log phase cells contain a basal level or if tL2 only appears in a subpopulation of cells in response to a signal (see ‘Discussion’ section).
Figure 2.
Immunoblot analysis reveals that the purified protein is a truncated form of L2, which exists in log phase cells. In (A), L2 (25 ng) from the Nierhaus laboratory, purified tL2 (15 ng) and whole-cell lysates from stationary (0.4 OD595nm) and log phase cells (0.3 OD595nm) were analyzed by immunoblotting with affinity-purified polyclonal antibody that specifically recognizes L2. Electrophoretic separation was in a 15% SDS–polyacrylamide gel. In (B), electrospray ionization mass spectrometry of purified tL2 isolated in Figure 1 was performed as described in ‘Materials and Methods’ section.
To estimate the size of the truncation, we analyzed tL2 by SDS–polyacrylamide gel electrophoresis (Supplementary Figure S1). Migrating more rapidly than the Nierhaus laboratory’s L2 that has a predicted mass of 29 860 Da, tL2 has a similar electrophoretic mobility as Hda (30 899 Da) and bovine carbonic anhydrase (28 982 Da). Because SDS–polyacrylamide gel electrophoresis only approximates the molecular weight of a protein, we determined the mass of tL2 by the more sensitive method of electrospray ionization mass spectrometry, which has an accuracy of about 0.02% or 4–5 Da for a protein of 23 kDa. The most abundant polypeptide has an observed mass of 23 176 Da (Figure 2B), which correlates with a mass of 23 171 Da for a polypeptide lacking the N-terminal methionine and 59 C-terminal residues of the intact form of L2. Despite our efforts to identify the mechanism that gives rise to tL2 (see ‘Discussion’ section), we have not determined if tL2 arises via a protease or by premature termination of translation, possibly via ribosome frameshifting. Under either mechanism, post-translational modification of the truncated polypeptide may play a role. Experiments to purify tL2, and to characterize its effect on DNA replication of the oriC-containing plasmid in comparison with L2 that are described herein were reproducible.
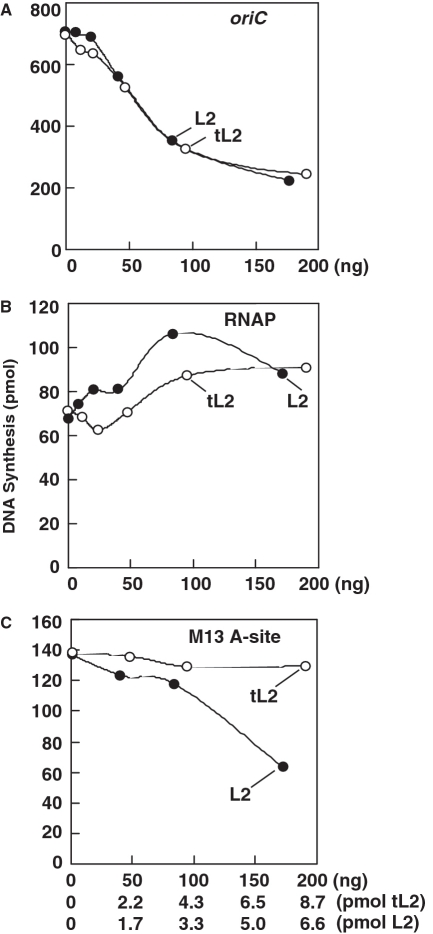
tL2 specifically inhibits oriC plasmid replication
The inhibitory effect of tL2 on DNA replication of the oriC-containing plasmid suggests that L2 is also inhibitory, which we confirmed (Figure 3A). We also compared both proteins in other DNA replication assays. In a replication system that includes RNA polymerase and any plasmid DNA, but does not require DnaA (36,37), both forms of L2 were modestly stimulatory (Figure 3B). In contrast, only L2 inhibited DNA replication of a single-stranded DNA carrying a DnaA box in a hairpin structure (Figure 3C). With this single-stranded DNA for which only a subset of DnaA functions is necessary (38), DnaA bound to the hairpin loads the DnaB–DnaC complex (38). After DnaC dissociates from DnaB, primase interacts with DnaB to form primers that are extended by DNA polymerase III holoenzyme to convert the single-stranded DNA to a duplex molecule. These results indicate that tL2 specifically inhibits oriC plasmid replication, whereas L2 additionally inhibits DNA replication of the single-stranded DNA. The following experiments investigate their mechanism of inhibition.
Figure 3.
tL2 specifically inhibits DNA replication of an oriC-containing plasmid. In (A), assays to measure DNA replication of an oriC plasmid (M13oriC2LB5 DNA; 46 fmol or 200 ng) were performed with DnaA (1 pmol; 50 ng), and the indicated amounts of tL2 or L2 as described in ‘Materials and Methods’ section. In (B), reactions were assembled as in (A) but lacked DnaA and instead were supplemented with RNA polymerase (0.56 µg), which supports DNA synthesis that is independent of DnaA and the oriC sequence. Reactions in (C) were assembled as in (A), but contained a single-stranded DNA carrying a DnaA box in a hairpin (M13 A-site DNA; 17 fmol or 80 ng) instead of the oriC-containing plasmid and lacked HU and DNA gyrase. Reactions were incubated at 30°C for 20 (A and B) or 10 min (C) to measure DNA synthesis.
tL2 does not inhibit oriC plasmid replication by sequestering negative supercoils
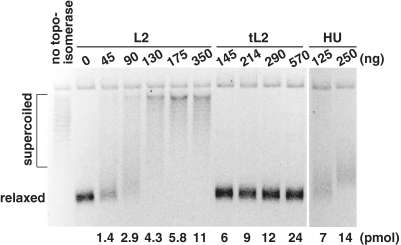
Because DNA binding proteins such as HU or FIS at an equal weight ratio to DNA inhibit oriC plasmid replication by sequestering its negative superhelicity (39–41), we considered whether this mechanism could explain the inhibitory effect of the ribosomal proteins. To test this possibility, we incubated the oriC plasmid with increasing amounts of either form of L2, and included a topoisomerase to stabilize any topological alteration followed by electrophoresis in an agarose gel containing chloroquine. Like HU, we found that L2 but not tL2 sequestered the negative supercoils of the oriC plasmid in proportion with the amount added (Figure 4), which could explain L2′s ability to inhibit DNA replication of the oriC plasmid. However, the results described below suggest that both forms of L2 interfere with initiation in vitro by an independent mechanism.
Figure 4.
Compared with L2, tL2 does not sequester the supercoils of a plasmid DNA. The change in DNA topology of a supercoiled plasmid was measured as described in ‘Materials and Methods’ section. The figure shows the reverse contrast image of DNAs with different superhelix densities that were visualized by ethidium bromide fluorescence.
Both tL2 and L2 interfere with the production of Form 1* DNA at the stage of DnaA-dependent unwinding of oriC
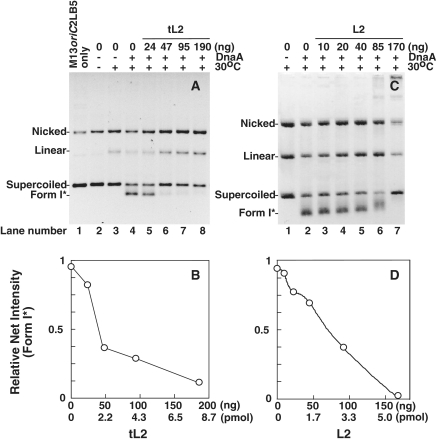
We examined tL2 and L2 in assays that measure specific events in the initiation process. One involves the formation of a highly negatively supercoiled DNA named Form 1* (42), which requires the localized unwinding of a region within oriC by DnaA (5,6,34). DnaB helicase from the DnaB–DnaC complex binds to the unwound region, and then unwinds the parental duplex DNA. The inclusion of DNA gyrase to remove the positive superhelicity that would otherwise accumulate in the duplex portion of the oriC plasmid leads to the appearance of Form 1* DNA. Detected by agarose gel electrophoresis (Figure 5), the production of Form 1* DNA was inhibited by tL2 and L2. At the higher levels of tL2 and all but the highest level of L2, the increased amounts of nicked and linear DNA suggest that a nuclease is responsible. However, other experiments failed to detect a nuclease in various preparations of purified tL2 or L2 (Figure 6; data not shown). Instead, we ascribe the increase of these DNAs to DNA gyrase because their appearance was dependent on both of its subunits (data not shown) that introduce breaks in DNA during strand passage that are revealed by treatment with SDS [see (43) and references therein]. The effect of tL2 or L2 on the level of nicked and linear DNA suggests that the ribosomal protein may stabilize DNA gyrase complexed to DNA at the step of strand cleavage, but we have not tested this idea.
Figure 5.
tL2 and L2 inhibit the formation of a highly negatively supercoiled DNA named Form 1*. Assays were performed as described in ‘Materials and Methods’ section. In (A) and (C), the reverse contrast image after ethidium bromide staining is shown. Where indicated, DnaA was added at 0.8 pmol (40 ng) and incubated at 30°C for 25 min. The leftmost lane in (A) contains purified M13oriC2LB5 DNA as a marker. In (B) and (D), the relative abundance of Form 1* DNA was determined by densitometric analysis of the stained agarose gel. The relative net intensity in (B) and (D) has been normalized to the pixel value for the amount of Form 1* determined in lane 4 of (A), or lane 2 of (C), respectively.
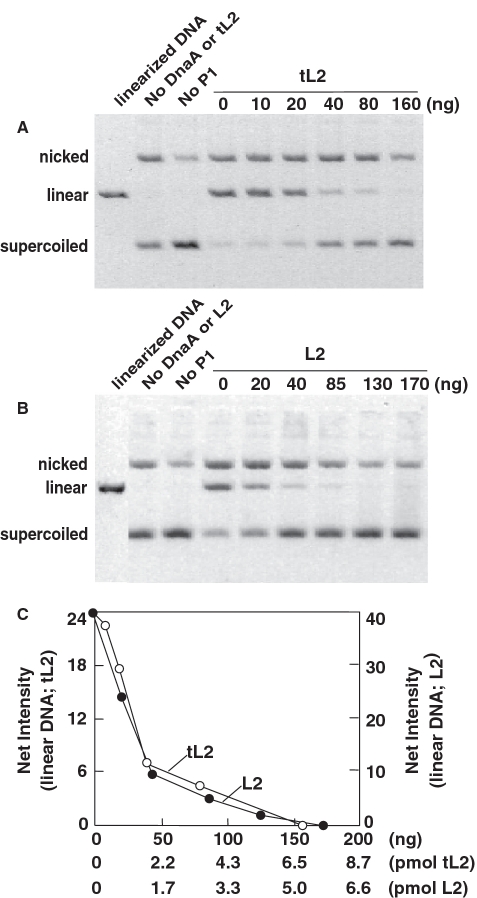
Figure 6.
tL2 and L2 inhibit the DnaA-dependent unwinding of oriC. The localized unwinding of oriC in a supercoiled plasmid was measured as described in ‘Materials and Methods’ section. In the leftmost lane of (A) and (B), which shows the reverse contrast image of ethidium bromide-stained agarose gels, M13oriC2LB5 DNA was linearized with HindIII endonuclease. In (C), densitometric analysis of the ethidium-bromide-stained gels was performed to quantify the relative abundance of the linear DNA. Net intensity (×10−3) is described in Figure 1.
Speculating that the ribosomal proteins may inhibit DnaA to interfere with the production of Form 1* DNA, we assessed their effect on the localized opening of oriC. As the single-stranded region within oriC is sensitive to cleavage by P1 nuclease, which linearizes the oriC plasmid, we quantified the amount of linear DNA after nuclease treatment and agarose gel electrophoresis. We confirmed by restriction mapping that P1 nuclease digested the plasmid within oriC (data not shown). Our results show that both proteins inhibit the DnaA-dependent unwinding of oriC (Figure 6). As the production of Form 1* DNA requires an oriC-containing plasmid that is supercoiled, which tL2 fails to sequester, the inhibition by tL2 in Figure 6 suggests a separate mechanism of inhibition.
ATP binding by DnaA and the hydrolysis of ATP bound to DnaA are unaffected by tL2 or L2
Because the ATP-bound form of DnaA is more active in the unwinding of oriC than DnaA complexed to ADP or the nucleotide-free form of DnaA (44), the ribosomal proteins may either interfere with the binding of ATP to DnaA or stimulate the hydrolysis of the bound ATP. Testing the former possibility, we found that tL2 and L2 at levels that inhibit oriC plasmid replication, Form 1* formation, and the unwinding of oriC had a negligible effect on ATP binding (Supplementary Figure S2). ATP hydrolysis by DnaA was also essentially unaffected. Neither tL2 nor L2 alone could bind ATP (data not shown).
tL2 and L2 bind non-specifically to DNA
The ribosomal protein may inhibit oriC plasmid replication by binding specifically to oriC to disrupt the binding of DnaA to one or more DnaA boxes and/or I-sites. To test this possibility, we performed gel mobility shift assays and compared a DNA fragment carrying oriC with a DNA encoding part of the ampicillin-resistance gene from pUC19 (Supplementary Figure S3). As a control, we showed that DnaA (2 and 4 ng) formed discrete complexes with the oriC-containing DNA fragment, reflecting the binding of DnaA to respective DnaA boxes of oriC (45). At the highest DnaA level, the oriC-containing DNA fragment was retained at the top of the gel. In contrast, DnaA bound negligibly to the other DNA (Supplementary Figure S3). At 12 and 17 ng with either DNA, tL2 produced heterogeneous complexes that migrated as a smear, suggesting non-specific DNA binding. At these levels of L2, the complexes were near the wells of the gel. Thus, neither protein appears to bind specifically to oriC to obstruct the binding of DnaA. Although their non-specific binding to DNA suggests that this activity may contribute to the inhibition of oriC plasmid replication, neither form of L2 inhibited non-specific DNA replication (Figure 3B), which presumably requires the recognition of promoters by RNA polymerase (36,37). In support, overproduced L2 does not inhibit but stimulates transcription from the rrnD P1 promoter in vivo (46), so L2 apparently does not occlude the binding of RNA polymerase. Hence, non-specific DNA binding may have a small or even marginal inhibitory effect on DNA replication of the oriC-containing plasmid.
tL2 and L2 interact with the N-terminal region of DnaA
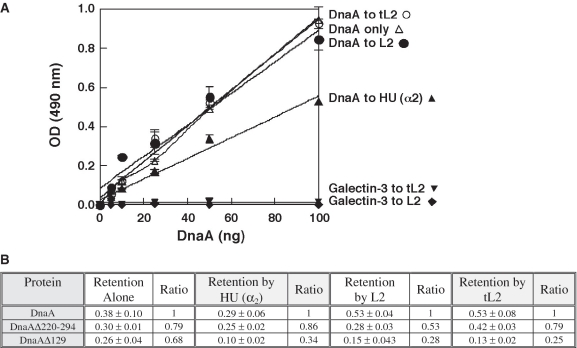
In other work, we discovered that a DnaA affinity column and not a BSA column specifically retained L2 from a soluble lysate (Vicente and Kaguni, data not shown). DnaA was also found to interact with L2 via a proteomics approach (47). To extend these observations, we showed in a solid phase binding assay that tL2 and L2 interacts with wild-type DnaA, or a mutant DnaA (DnaAΔ220-294) lacking residues 220–294, but that this interaction was reduced by about 4-fold with DnaAΔ129 lacking the N-terminal 129 amino acids (Figure 7). DnaAΔ129 was also defective in interaction with HU, confirming previous results (9). We know that the preparation of DnaAΔ129 is as active as DnaA+ in oriC unwinding, and in binding to ATP or to an oriC-containing DNA fragment (20,35), so the mutant DnaA is not grossly misfolded. As negative controls, we showed that tL2 or L2 does not bind to DnaB (data not shown) or murine galectin-3 (Figure 7A), which is involved in pre-mRNA splicing and is not expected to interact (48). These results suggest that both forms of L2 inhibit DNA replication of the oriC-containing plasmid by interacting with the N-terminal region of DnaA. As described above, it is possible that their non-specific DNA binding activity may also contribute to the inhibition.
Figure 7.
Both forms of L2 require the N-terminal region of DnaA to interact. In (A), the indicated amounts of DnaA or galectin 3 were added in triplicate to the wells containing immobilized tL2 (100 ng), L2 (100 ng) or HU (590 ng; α dimer). The vertical lines indicate the standard deviation. For comparison with the plot of DnaA retained by immobilized HU or tL2 or L2, the plot labeled ‘DnaA only’ represents the amount of DnaA (6, 12, 25, 50 and 100 ng) that was immobilized directly to the microtiter plate (see ‘Materials and Methods’ section). In (B), the column labeled ‘Retention Alone’ reflects the amount of DnaA or the respective mutant DnaA (each at 50 ng) that was immobilized when added directly to the microtiter plate. The columns labeled ‘Retention by HU (α2)’, ‘Retention by L2’ and ‘Retention by tL2’ compare the retention of DnaA or the different mutant DnaAs (each at 50 ng) by immobilized HU (α dimer), L2 or tL2, respectively. The columns labeled ‘Ratio’ represent the normalized absorbance relative to wild-type DnaA, which was set at 1.
tL2 or L2 interferes with prepriming complex assembly at oriC
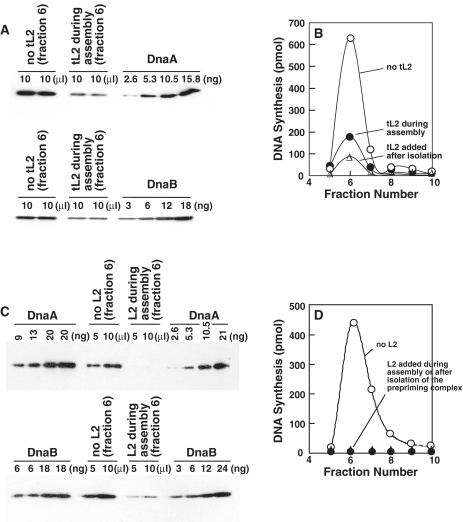
Recent work indicates that HU or DiaA stabilizes the DnaA oligomer by interacting with DnaA’s N-terminal region (9,10), which acts in self-oligomerization (4,26). We considered the converse possibility that both forms of L2 inhibit initiation by destabilizing DnaA oligomerized at oriC. To test this idea, we assembled the prepriming complex in the presence and absence of tL2 or L2 as described (6,26). After separating the complex from unbound proteins by gel filtration chromatography, we quantified the amounts of DnaA and DnaB in the complex by immunoblotting relative to a standard curve prepared with known amounts of each protein (Figure 8). We quantified DnaC in a separate study (7). Relative to the amount of supercoiled oriC plasmid in the complex (data not shown), both ribosomal proteins led to reduced amounts of DnaA and DnaB in the isolated complex. Hence, the interaction of tL2 or L2 with DnaA’s N-terminal region apparently destabilizes the DnaA oligomer to inhibit the loading of DnaB at oriC.
Figure 8.
tL2 and L2 destabilize DnaA bound to oriC. The prepriming complex was assembled as described in ‘Materials and Methods’ section. After isolation of the complex, the amounts of DnaA and DnaB in the complex assembled in the presence or absence of tL2 (A) or L2 (C) were measured by immunoblot analysis. In (B) and (D), the activity of the isolated prepriming complex was measured by the addition of required reagents and proteins needed for DNA synthesis to 20 µl of the indicated column fractions. After adjusting the concentration of magnesium acetate to 10 mM in a reaction volume of 25 µl, DNA synthesis was measured. Where indicated, tL2 (190 ng) or L2 (180 ng) was added to determine its effect on the isolated prepriming complex that was assembled in its absence.
We also measured the replication activity of the isolated prepriming complex by incubating it with other necessary components, and replication proteins (SSB, primase, DNA polymerase III holoenzyme and DNA gyrase) that act at the subsequent stage of DNA synthesis (Figure 8B and D). Compared with the activity of the isolated complex assembled in the absence of tL2, its inclusion during assembly was inhibitory. The addition of either to the isolated complex formed in the absence of the ribosomal protein also inhibited DNA replication, suggesting that they destabilize DnaA already bound to oriC. The results suggest that DnaA must remain oligomerized after DnaB has loaded at oriC for initiation to occur.
For these experiments, DnaA with an N-terminal polyhistidine sequence was used. As tL2 or L2 similarly inhibits the prepriming complex when assembled with either form of DnaA, the polyhistidine tag does not seem to interfere with the interaction between DnaA and tL2 or L2.
DISCUSSION
Dual functions for proteins needed for translation
Our observations strongly suggest that tL2 and L2 modulate the activity of DnaA in DNA replication of an oriC-containing plasmid in vitro. Of interest, a recent study described that L2 interacts with the alpha subunit of RNA polymerase to enhance transcription from the rrnD promoter (46). Other ribosomal proteins that have dual roles are S10 (NusE) [reviewed in (49)] and S4 (50) that act as antitermination factors in transcription, and S1 that is a subunit of Qβ replicase (51). Whereas in vivo evidence has not been obtained to validate the importance of the following biochemical studies, other examples are L14 that stimulates unwinding by Rep helicase (52), S9 that may interact with UmuC (53) and S3 that is identical to H protein and may be a component of the nucleoid (54). As E. coli grown in rich media contains substantial amounts of free ribosomal proteins (55), they are available to perform auxiliary functions. Although not a ribosomal protein, an isoform of IF2 named IF2-2 that lacks the N-terminal end of full-length IF2-1 was isolated by its requirement during bacteriophage Mu DNA replication in vitro (56). IF2-2 but not IF2-1 appears to remove MuA transposase from Mu DNA integrated at the target site. Transposase disassembly is necessary to permit the binding of PriA in the PriA-PriC pathway of Mu DNA replication.
How does tL2 arise?
The combined results from N-terminal sequence analysis and electrospray mass spectrometry suggest that the C-terminal residue of tL2 is arginine 214. Hence, a trypsin-like protease may be responsible for cleavage, but enzymes such as protease II, protease In or protease IV are not specifically expressed during log phase growth, which is when we detected tL2. Based on the crystallographic structure of the E. coli ribosome and biochemical studies (57–59), the part of L2 containing arginine 214 is accessible from the solvent with the distal C-terminal region interacting with the 23S rRNA. Thus, cleavage may occur when L2 is assembled in the ribosome, but the protease may act prior to assembly. Alternatively, tL2 may arise during translation by premature termination of rplB, which may also involve ribosomal frameshifting and post-translational modification by covalent addition of small molecules.
tL2 apparently does not coordinate the stringent response with inhibition of initiation of DNA replication
A number of stress conditions induce the stringent response [reviewed in (60,61)], which also inhibits the initiation of DNA replication (62,63). Because tL2 may inhibit initiation from oriC during the stringent response, we tested if such stress conditions led to a relative increase of tL2. Specifically, we added valine (0.5 mg/ml) to a log phase culture grown in supplemented M9 media to inhibit isoleucine biosynthesis, or transferred the culture into M9 media lacking glucose, phosphate or ammonium ion. We also exposed the culture to nitrogen gas to promote anaerobiosis, or changed the ionic strength of the media. With an antibody that specifically recognizes both L2 and tL2, we were unable to identify conditions that changed the relative level of tL2 to L2 to implicate tL2 in inhibition of initiation during the stringent response.
DnaA as a sensor
Like tL2 or L2, Dps inhibits the unwinding of oriC by DnaA and its activity in initiation (20). In contrast, HU and DiaA appear to stimulate initiation by stabilizing the DnaA multimer at oriC, which is necessary for helicase loading (4,24,35,64). Hence, one set of proteins has the opposing effect as the second set on DnaA function, enhancing or impairing DnaA to modulate initiation under certain conditions. As these proteins interact with the N-terminal region of DnaA (9,20,65), this region may act as a sensor in responding to these proteins. These observations evoke the β clamp, which interacts via a C-terminal domain with MutS, DNA ligase, Hda and several E. coli DNA polymerases (66). MutS and DNA ligase may exploit the β clamp bound to DNA to access and repair lesions in DNA. For Hda, studies show that Hda complexed with the β clamp stimulates the hydrolysis of ATP bound to DnaA to inhibit DnaA function in initiation [reviewed in (1)]. The β clamp also interacts with E. coli DNA polymerases that act in chromosomal DNA replication or translesion DNA synthesis (67). It is interesting to consider that dnaA and dnaN, which encodes the β clamp, are in the same operon, and interact with other proteins via a specific region during the different stages of DNA replication.
Supporting the biochemical evidence summarized above, physiological results suggest that Dps, HU and DiaA affect DnaA function to modulate the initiation process in vivo (11,12,20). For either form of L2, we attempted to demonstrate that its interaction with DnaA is physiologically important. First using a plasmid encoding the respective alleles under araBAD promoter control, we showed that an elevated level of tL2 or L2 reduced the frequency of colony formation of E. coli MC1061 by ≥102-fold compared with the non-induced control (Felczak and Kaguni, unpublished data). We also determined that an increased tL2 level lowered the frequency of colony formation by >4 × 104-fold or >4 × 103-fold in isogenic ΔrecA::kanR or ΔrecB::kanR strains, respectively. In contrast, the frequency of colony formation under comparable conditions was unaffected for these strains bearing the empty vector or a plasmid encoding isoleucine for Thr129, and cysteine for Arg175 of tL2. This plasmid was isolated from one of the spontaneously arising transformants that were viable in the presence of arabinose. We then examined the effect of an elevated tL2 level in a strain synchronized to initiate DNA replication. We did not test L2. Briefly, we shifted an exponentially growing dnaC2(Ts) mutant from 30°C to 42°C for 1 h. At 42°C, ongoing replication forks proceed to completion but new initiations are blocked (68). After returning the dnaC mutant to permissive temperature, we measured new initiations at various time intervals by the ratio of oriC to relE, a locus in the terminus region of the chromosome, by quantitative real-time PCR analysis. In recent work with Dps, we showed that an elevated level reduced the frequency of initiation after the temperature downshift (20). However, we were unable to show that an increased level of tL2 inhibits initiation in vivo, but the conditions may not have been appropriate. For example, the inhibition of initiation by tL2 may require a signal that causes neutralization of a factor that otherwise counterbalances the effect of tL2. As we do not know the nature of this signal or the factor, the negative evidence described above does not demonstrate that tL2 is unimportant in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health; Michigan Agricultural Experiment Station. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Knud Nierhaus for the gift of L2 protein and Dr John Wang for murine galectin-3 and rabbit antiserum that recognizes this protein. We also thank Drs Lijun Chen and A. Daniel Jones for the mass spectrometry analysis of tL2.
REFERENCES
- 1.Ozaki S, Katayama T. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid. 2009;62:71–82. doi: 10.1016/j.plasmid.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Leonard AC, Grimwade JE. Initiating chromosome replication in E. coli: it makes sense to recycle. Genes Dev. 2009;23:1145–1150. doi: 10.1101/gad.1809909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmons LA, Felczak M, Kaguni JM. DnaA protein of Escherichia coli: oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol. 2003;49:849–858. doi: 10.1046/j.1365-2958.2003.03603.x. [DOI] [PubMed] [Google Scholar]
- 4.Felczak MM, Simmons LA, Kaguni JM. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 2005;280:24627–24633. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
- 5.Fang L, Davey MJ, O'Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 6.Carr KM, Kaguni JM. Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J. Biol. Chem. 2001;276:44919–44925. doi: 10.1074/jbc.M107463200. [DOI] [PubMed] [Google Scholar]
- 7.Makowska-Grzyska M, Kaguni JM. Primase directs the release of DnaC from DnaB. Mol. Cell. 2010;37:90–101. doi: 10.1016/j.molcel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 9.Chodavarapu S, Felczak MM, Yaniv JR, Kaguni JM. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 2008;67:781–792. doi: 10.1111/j.1365-2958.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- 10.Keyamura K, Abe Y, Higashi M, Ueda T, Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 2009;284:25038–25050. doi: 10.1074/jbc.M109.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahloul A, Boubrik F, Rouviere-Yaniv J. Roles of Escherichia coli histone-like protein HU in DNA replication: HU-beta suppresses the thermosensitivity of dnaA46ts. Biochimie. 2001;83:219–229. doi: 10.1016/s0300-9084(01)01246-9. [DOI] [PubMed] [Google Scholar]
- 12.Keyamura K, Fujikawa N, Ishida T, Ozaki S, Su'etsugu M, Fujimitsu K, Kagawa W, Yokoyama S, Kurumizaka H, Katayama T. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weigel C, Messer W, Preiss S, Welzeck M, Morigen, Boye E. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 2001;40:498–507. doi: 10.1046/j.1365-2958.2001.02409.x. [DOI] [PubMed] [Google Scholar]
- 14.Polaczek P. Bending of the origin of replication of E. coli by binding of IHF at a specific site. New Biol. 1990;2:265–271. [PubMed] [Google Scholar]
- 15.Roth A, Urmoneit B, Messer W. Functions of histone-like proteins in the initiation of DNA replication at oriC of Escherichia coli. Biochimie. 1994;76:917–923. doi: 10.1016/0300-9084(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 16.Boye E, Lobner-Olesen A, Skarstad K. Timing of chromosomal replication in Escherichia coli. Biochim. Biophys. Acta. 1988;951:359–364. doi: 10.1016/0167-4781(88)90107-8. [DOI] [PubMed] [Google Scholar]
- 17.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 19.Ilari A, Stefanini S, Chiancone E, Tsernoglou D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct. Biol. 2000;7:38–43. doi: 10.1038/71236. [DOI] [PubMed] [Google Scholar]
- 20.Chodavarapu S, Gomez R, Vicente M, Kaguni JM. Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 2008;67:1331–1346. doi: 10.1111/j.1365-2958.2008.06127.x. [DOI] [PubMed] [Google Scholar]
- 21.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa A, Nakashima T, Taniguchi M, Hosaka H, Kimura M, Tanaka I. The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; a protein at the peptidyl transferase center of the ribosome. EMBO J. 1999;18:1459–1467. doi: 10.1093/emboj/18.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang DS, Kaguni JM. Interaction of dnaA46 protein with a stimulatory protein in replication from the Escherichia coli chromosomal origin. J. Biol. Chem. 1988;263:10633–10640. [PubMed] [Google Scholar]
- 24.Marszalek J, Kaguni JM. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- 25.Walker JR, Severson KA, Hermandson MJ, Blinkova A, Carr KM, Kaguni JM. Escherichia coli DnaA protein: specific biochemical defects of mutant DnaAs reduce initiation frequency to suppress a temperature-sensitive dnaX mutation. Biochimie. 2006;88:1–10. doi: 10.1016/j.biochi.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Felczak MM, Kaguni JM. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 2004;279:51156–51162. doi: 10.1074/jbc.M409695200. [DOI] [PubMed] [Google Scholar]
- 27.Sutton MD, Kaguni JM. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem. 1997;272:23017–23024. doi: 10.1074/jbc.272.37.23017. [DOI] [PubMed] [Google Scholar]
- 28.Agrwal N, Sun Q, Wang SY, Wang JL. Carbohydrate-binding protein 35. I. Properties of the recombinant polypeptide and the individuality of the domains. J. Biol. Chem. 1993;268:14932–14939. [PubMed] [Google Scholar]
- 29.Marszalek J, Zhang W, Hupp TR, Margulies C, Carr KM, Cherry S, Kaguni JM. Domains of DnaA protein involved in interaction with DnaB protein, and in unwinding the Escherichia coli chromosomal origin. J. Biol. Chem. 1996;271:18535–18542. doi: 10.1074/jbc.271.31.18535. [DOI] [PubMed] [Google Scholar]
- 30.Kaguni JM, Kornberg A. Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. Cell. 1984;38:183–190. doi: 10.1016/0092-8674(84)90539-7. [DOI] [PubMed] [Google Scholar]
- 31.Masai H, Arai KI. DnaA-dependent assembly of the ABC primosome at the A site, a single-stranded DNA hairpin containing a dnaA box. Eur. J. Biochem. 1995;230:384–395. doi: 10.1111/j.1432-1033.1995.tb20573.x. [DOI] [PubMed] [Google Scholar]
- 32.Sternglanz R, DiNardo S, Voelkel KA, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang JC. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc. Natl Acad. Sci. USA. 1981;78:2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marszalek J, Kaguni JM. Defective replication activity of a dominant-lethal dnaB gene product from Escherichia coli. J. Biol. Chem. 1992;267:19334–19340. [PubMed] [Google Scholar]
- 34.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 35.Sutton MD, Carr KM, Vicente M, Kaguni JM. E. coli DnaA protein: the N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa T, Baker TA, van der Ende A, Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: contributions of RNA polymerase and primase. Proc. Natl Acad. Sci. USA. 1985;82:3562–3566. doi: 10.1073/pnas.82.11.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker TA, Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 38.Carr KM, Kaguni JM. Escherichia coli DnaA protein loads a single DnaB helicase at a DnaA box hairpin. J. Biol. Chem. 2002;277:39815–39822. doi: 10.1074/jbc.M205031200. [DOI] [PubMed] [Google Scholar]
- 39.Rouviere-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 40.Skarstad K, Baker TA, Kornberg A. Strand separation required for initiation of replication at the chromosomal origin of E. coli is facilitated by a distant RNA–DNA hybrid. EMBO J. 1990;9:2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margulies C, Kaguni JM. The FIS protein fails to block the binding of DnaA protein to oriC, the Escherichia coli chromosomal origin. Nucleic Acids Res. 1998;26:5170–5175. doi: 10.1093/nar/26.22.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker TA, Sekimizu K, Funnell BE, Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986;45:53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- 43.Pfeiffer ES, Hiasa H. Determination of the primary target of a quinolone drug and the effect of quinolone resistance-conferring mutations by measuring quinolone sensitivity based on its mode of action. Antimicrob. Agents Chemother. 2007;51:3410–3412. doi: 10.1128/AAC.00362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 45.Margulies C, Kaguni JM. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J. Biol. Chem. 1996;271:17035–17040. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 46.Rippa V, Cirulli C, Di Palo B, Doti N, Amoresano A, Duilio A. The ribosomal protein L2 interacts with the RNA polymerase alpha subunit and acts as a transcription modulator in Escherichia coli. J. Bacteriol. 2010;192:1882–1889. doi: 10.1128/JB.01503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 48.Patterson RJ, Wang W, Wang JL. Understanding the biochemical activities of galectin-1 and galectin-3 in the nucleus. Glycoconj. J. 2004;19:499–506. doi: 10.1023/B:GLYC.0000014079.87862.c7. [DOI] [PubMed] [Google Scholar]
- 49.Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres M, Condon C, Balada JM, Squires C, Squires CL. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 2001;20:3811–3820. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blumenthal T, Carmichael GG. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 52.Yancey JE, Matson SW. The DNA unwinding reaction catalyzed by Rep protein is facilitated by an RHSP-DNA interaction. Nucleic Acids Res. 1991;19:3943–3951. doi: 10.1093/nar/19.14.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc. Natl Acad. Sci. USA. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruckner RC, Cox MM. The histone-like H protein of Escherichia coli is ribosomal protein S3. Nucleic Acids Res. 1989;17:3145–3161. doi: 10.1093/nar/17.8.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulbrich B, Nierhaus KH. Pools of ribosomal proteins in Escherichia coli. Studies on the exchange of proteins between pools and ribosomes. Eur. J. Biochem. 1975;57:49–54. doi: 10.1111/j.1432-1033.1975.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 56.North SH, Kirtland SE, Nakai H. Translation factor IF2 at the interface of transposition and replication by the PriA-PriC pathway. Mol. Microbiol. 2007;66:1566–1578. doi: 10.1111/j.1365-2958.2007.06022.x. [DOI] [PubMed] [Google Scholar]
- 57.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 58.Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, et al. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi T, Tahara M, Iwasaki K, Kouzuma Y, Kimura M. Requirement for C-terminal extension to the RNA binding domain for efficient RNA binding by ribosomal protein L2. Biosci. Biotechnol. Biochem. 2002;66:682–684. doi: 10.1271/bbb.66.682. [DOI] [PubMed] [Google Scholar]
- 60.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 61.Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 62.Levine A, Vannier F, Dehbi M, Henckes G, Seror SJ. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J. Mol. Biol. 1991;219:605–613. doi: 10.1016/0022-2836(91)90657-r. [DOI] [PubMed] [Google Scholar]
- 63.Schreiber G, Ron EZ, Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr. Microbiol. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- 64.Abe Y, Jo T, Matsuda Y, Matsunaga C, Katayama T, Ueda T. Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 2007;282:17816–17827. doi: 10.1074/jbc.M701841200. [DOI] [PubMed] [Google Scholar]
- 65.Ishida T, Akimitsu N, Kashioka T, Hatano M, Kubota T, Ogata Y, Sekimizu K, Katayama T. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 2004;279:45546–45555. doi: 10.1074/jbc.M402762200. [DOI] [PubMed] [Google Scholar]
- 66.Wijffels G, Dalrymple B, Kongsuwan K, Dixon NE. Conservation of eubacterial replicases. IUBMB Life. 2005;57:413–419. doi: 10.1080/15216540500138246. [DOI] [PubMed] [Google Scholar]
- 67.Lopez de Saro FJ, Georgescu RE, Goodman MF, O'Donnell M. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 2003;22:6408–6418. doi: 10.1093/emboj/cdg603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Withers HL, Bernander R. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 1998;180:1624–1631. doi: 10.1128/jb.180.7.1624-1631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.