Abstract
SyMAP (Synteny Mapping and Analysis Program) was originally developed to compute synteny blocks between a sequenced genome and a FPC map, and has been extended to support pairs of sequenced genomes. SyMAP uses MUMmer to compute the raw hits between the two genomes, which are then clustered and filtered using the optional gene annotation. The filtered hits are input to the synteny algorithm, which was designed to discover duplicated regions and form larger-scale synteny blocks, where intervening micro-rearrangements are allowed. SyMAP provides extensive interactive Java displays at all levels of resolution along with simultaneous displays of multiple aligned pairs. The synteny blocks from multiple chromosomes may be displayed in a high-level dot plot or three-dimensional view, and the user may then drill down to see the details of a region, including the alignments of the hits to the gene annotation. These capabilities are illustrated by showing their application to the study of genome duplication, differential gene loss and transitive homology between sorghum, maize and rice. The software may be used from a website or standalone for the best performance. A project manager is provided to organize and automate the analysis of multi-genome groups. The software is freely distributed at http://www.agcol.arizona.edu/software/symap.
INTRODUCTION
Comparative genomics is becoming increasingly important as more genomes are being sequenced. Comparative genomics software can be broadly classified as focusing on genome alignment or synteny computation. The algorithms vary as to whether they are optimized for large genomes, small genomes, regions or genes, along with the number of sequences that can be compared simultaneously. The available software packages also have variations on input, graphical displays and data management capabilities. Consequently, there is not one software solution to address all problems and the best package to use depends on the genomes compared and questions being asked. SyMAP (1) is a distributable software package that was originally developed to compute synteny blocks between a sequenced genome and a Fingerprint Contigs (FPC) map (2,3), and has been extended to support pairs of large sequenced genomes, with special attention to plant genomes. Hence, this background section emphasizes distributable synteny software packages for large sequenced genomes, i.e. software that only provides the graphical display (4–7) or is purely web-based (8–17) will not be included in this background discussion.
There is a range of terminology used for discussing conserved regions of genomes and a range of variations on the problem of detecting them. When comparing two genomes, an algorithm may detect homologous genes with conserved order, for which the following terms have been used: microcolinearity [ADHoRe (18)]; colinearity [DAGchainer (19) and ColinearScan (20)]; locally collinear blocks [Mauve (21)]; and conserved synteny [CYNTENATOR (22)]. When small microrearrangments are allowed, the following terms have been used: synteny blocks [GRIMM-Synteny (23), DiagHunter (24) and SyMAP (1)]; segmental homologs [FISH (25)]; and orthologous segments [OSfinder (26)]. The term ‘synteny block’ is used for both problems in OrthoCluster (27,28), where Ng et al. (28) defined ‘perfect synteny block’ as a region of perfectly conserved gene order and stranded-ness, and ‘imperfect synteny block’ as a region of conserved genes without regard to order or stranded-ness. Imperfect synteny is advantageous when there may be microrearrangements and also provides tolerance for misassembled sequences. SyMAP v3.4 computes imperfect synteny for both sequence-to-sequence and FPC-to-sequence comparisons.
Except for Mauve (21) and SyMAP, the above-referenced synteny programs all require preprocessing to compute the genes and homologous pairs, and file formatting to meet the input requirements. Mauve takes as input genome sequences, and performs its own multiple alignment followed by synteny computation; however, it does not identify large-scale synteny blocks, but rather smaller-scale ‘locally collinear blocks’, reflecting its original orientation toward bacterial genomes. SyMAP takes as input two sequenced genomes (one of which may be in unordered sequence contigs) and executes MUMmer (29) to compute the raw hits shared between the genomes, which allows it to include regions in syntenic blocks that were not detected by gene-finding programs. It also takes as input an optional file of gene annotations, which is used in the synteny computations and graphical display.
Synteny software packages often contain scripts to generate visualization [e.g. (18,26)], but only Mauve and SyMAP have fully interactive displays (both built using Java). The display of Mauve is well suited for multiple alignments of small genomes, but is somewhat limited for large genomes, as there is no unified way to handle multiple chromosomes except by concatenating them together, and there is only one view style supported. A salient aim of SyMAP is to allow ease of manually exploring the relations between multiple chromosomes and genomes, where the computed synteny blocks elucidate interesting regions. Toward this end, SyMAP v3.4 provides a range of Java views for genome or chromosome, which allow the user to zoom into regions of interest; additionally, it provides web-based block and circle views.
Several packages, such as i-ADHoRe 2.0 (30,31), OrthoCluster (27), CYNTENATOR (22), OSfinder (26), MCscan (32) and Mauve (21) can compute synteny between multiple genomes simultaneously. Additionally, i-ADHoRe and MCscan explicitly detect transitive homology [i.e. if segment A is homologous to B and C, then B and C are homologous (33)]. SyMAP is pair-oriented, so it does not explicitly compute multi-genome synteny or transitive homology; however, as will be shown in the ‘Results’ section, the graphics allow multiple genomes, chromosomes or regions to be viewed together, which allows easy investigation of duplicated regions and transitive homology.
To summarize the SyMAP v3.4 features, it is a software package aimed at detecting imperfect synteny in large, repetitive, duplicated genomes such as found in plant species, where the input is genome sequence files and optional annotation files. The user does not have to perform preprocessing on the data, nor adjust parameters, nor employ additional packages to visualize the data. SyMAP incorporates the MUMmer program (29) to compute the raw hits, which are clustered and filtered to form anchors, where a region may be in multiple anchors in order to detect duplications; the filtered anchors are input into the synteny algorithm, which is unchanged from version 1.0 (1). A set of genomes may be viewed together as a group, where each pair has been compared, and each genome may also be compared with itself. The Java graphics provides a full range of views from multiple genomes down to individual genes. The graphics can run standalone or from the web, where the web display also allows querying for specific genes, annotations or regions. Since the management of many genomes becomes easily disorganized, SyMAP v3.4 provides a project manager to organize the genomes and computations. Finally, as in the first version (1), SyMAP v3.4 can also align an FPC map to a sequenced genome. Hence, SyMAP v3.4 is a turnkey system that provides all software necessary to manage, compute, query and display synteny between multiple sequenced genomes. The software is freely available from www.agcol.arizona.edu/software/symap and a showcase of plant genomes is available from www.symapdb.org.
MATERIALS AND METHODS
The following describes the features added to SyMAP since its original publication (1), where the emphasis has been on synteny computation and display for multiple sequenced genomes (as opposed to the FPC maps supported in the first version). For each genome to be compared, the input is multiple sequence files and an optional GFF formatted annotation file (www.gmod.org/wiki/GFF3).
Anchor loading and synteny analysis
The original SyMAP synteny algorithm (1) works without change for the comparison of sequenced genomes, since its input is the anchor coordinates from the two genomes being compared. For FPC-to-sequence, BLAT (34) is used to compute the anchors; for sequence-to-sequence synteny, MUMmer (29) is used to compute individual ‘raw hits’, which SyMAP clusters into anchors. MUMmer has two modes of operation, NUCmer and PROmer, where the first finds nucleotide matches and the second finds amino acid matches. SyMAP uses NUCmer for same-genome comparisons, and PROmer when the genomes are different.
As shown in Table 1, the number of raw hits can be quite large due to repetitive sequences (i.e. sequences which participate in many hits). SyMAP reduces the number with several stages of clustering and filtering. First, the existing set of annotated genes (AGs) for each genome is augmented with un-annotated clusters (UCs), which are created from the MUMmer hits that do not overlap gene annotation on that genome. The purpose of retaining these hits is that some of them may be genes that were missed by annotation, or they may be non-genic conserved sequences. The query and target side of the hits are considered separately as the matches for each genome, where the matches are clustered into UCs using the average gene length; if there is no gene annotation, a default length of 1 kb is used. The second stage of processing is to group the MUMmer hits into ‘anchors’ using the AGs and UCs. Note that, by design, each MUMmer hit is contained in either an AG or a UC for each genome; therefore, each resulting anchor connects an AG or UC on one genome to an AG or UC on the second genome, and represents one or more original MUMmer hits connecting those regions. The score of the anchor is the sum of the lengths of each of its component hits.
Table 1.
Sorghum–maize clustering and anchor filtering
| Categorya | MUMmer raw hits | Filtered raw hitsb (%) | Clustered anchorsc | Filtered anchorsd | Synteny anchorse |
|---|---|---|---|---|---|
| AG-to-AG | 280 156 | 55 | 116 421 | 35 153 | 27 404 |
| AG-to-UC | 61 550 | 22 | 45 213 | 6720 | 4095 |
| UC-to-UC | 20 102 | 55 | 15 193 | 7396 | 6447 |
| Total | 361 808 | 49 | 176 827 | 49 269 | 37 946 |
aAnnotated genes (AGs) and un-annotated clusters (UCs).
bPercentage of raw hits contributing to the filtered anchors.
cAnchors formed by clustering all raw hits using the AGs and UCs.
dAnchors after applying the reciprocal top-2 filter.
eAnchors found to be part of synteny blocks.
The anchors are then passed through a reciprocal ‘top-N’ filter, where N = 2 by default. This is a modified version of ‘reciprocal best hit’ filtering, in which an anchor must be one of the top N anchor scores for both its query and target side. Traditional reciprocal-best-hit filtering corresponds to N = 1, but a value of a least 2 aids in detecting duplications [also, N = 1 rejects many true orthologs (35)]. The issue of duplications is an important question for most genomes, but especially important for plant genomes, as they exhibit more whole genome and segmental duplication compared to vertebrates (8,36).
The filtered anchors are then input into the synteny algorithm, which uses dynamic programming to compute chains of anchors, where small intervening inversions or rearrangements are allowed, hence detecting imperfect synteny. Though the details are given in ref. 1, we emphasize one point concerning SyMAP’s handling of gaps between anchors in a synteny block. It is generally not optimal to have the user explicitly set a ‘gap parameter’ for both genomes, since different blocks may require different gap parameters. For example, ancient duplications generally have a much lower density of anchors than those from the most recent divergence; also, some regions contain higher densities of transposons than others. The SyMAP algorithm mitigates this problem by exploring a large range of gap parameters for each block, and using the ones that produce the largest block, subject to several measures of quality.
Project management
The project manager (Supplementary Figure S1) is a Java application that automates the storage and analysis of multiple species, including running MUMmer or BLAT, loading anchors, analyzing synteny and launching the Java graphical interfaces. The original SyMAP synteny algorithm (1) was written in Perl, but has since been rewritten in Java, so it works seamlessly with the project manager. In order to make it easy for potential users to try SyMAP, the downloadable package contains all necessary programs (including MySQL) along with demo files.
Interactive graphics
The SyMAP v3.4 Java views include dot plots for multiple whole genomes and multiple chromosomes, a three-dimensional (3D) display that provides a global view of multiple aligned chromosomes, and a 2D display that allows zooming down into regions of interest; these will be illustrated in the ‘Results’ section. The Java views are available both from the standalone desktop application and from the web as a Java web applet. As illustrated in Supplementary Figure S2, the web system also includes the following CGI-based views: (A) a view that is similar to the Circos circular two-genome display (7), (B) a block-to-chromosome two-genome display, (C) an annotation and location search page and (D) a summary page of statistics with a table of blocks. All views (except the circle view) link to the Java 2D display, which allows drilling down into the details of the synteny.
All Java displays allow filtering on different attributes and manipulations of the displays, as described in the online documentation. An important manipulation is the ability to flip a region in the 2D view, as once zoomed into a region, it is easier to view details of the alignment if the majority of the synteny lines are not crossing. A second important filter is the ability to show all filtered anchors, regardless if they are part of a synteny block.
The GFF-format has a field for ‘type’, where SyMAP recognizes the annotation types gene, exon (or CDS), gap and centromere, which are displayed as features in the 2D view. The GFF ‘attribute’ field allows for arbitrary ‘keyword=value’ pairs in order to store other information, and SyMAP displays this information in the zoomed-in 2D view. If the attribute field contains a URL, a link to the external site will be provided. The web-based annotation search page allows searching on any of the values in the attribute field.
RESULTS
In the following, results from comparing the sequenced genomes of rice (37,38), sorghum (39) and maize (40) using SyMAP are discussed; this is not intended to be a robust coverage of the evolutionary events for these plants, but only as a way to illustrate the processing and types of information that can be extracted using the SyMAP interface.
Input and computation
The grasses share a duplication event ∼70 Mya followed by divergence ∼50 Mya (41). The ancestral sorghum genome and two progenitors of the maize genome diverged ∼11.9 Mya, and hybridization of the two maize progenitors occurred ∼4.8 Mya (42). The maize genome expanded in size due to retrotransposons during the last ∼3 million years (43), resulting in a large repetitive genome. The rice, sorghum and maize genomes are 400 MB, 760 MB and 2300 MB, respectively. The maize RefGen_v2 sequence and annotation were downloaded from maizesequence.org, rice (release 6.1) from rice.plantbiology.msu.edu, and sorghum bicolor v1.0 from www.phytozome.net/sorghum.
All synteny computations were performed without any parameter adjustment. The computations were run using eight threads on a 64-bit platform having dual 6-core AMD Opteron 8431 2.4 GHz CPUs, and 48G RAM. The processing time for each of the three inter-species comparisons was under 2.5 h, with the vast majority of time being used for MUMmer (the rest of the processing took under 2 min). Self-comparisons were also performed, with the unmasked rice taking 7 h. The maize and sorghum genomes were masked, as the difference in the times for MUMmer self-comparisons for masked versus unmasked is significant, e.g. the sorghum self-comparison without masking took 60 h as compared to 32 min for the masked sequence. For inter-species comparisons the time difference is much smaller, e.g. the three comparisons cited above take less than an hour longer when maize and sorghum are not masked.
Table 1 shows the results of anchor filtering and synteny analysis for the comparison of sorghum and maize. Of the ∼362 k raw hits found by PROmer, 280 k (77%) had both ends in an AG (i.e. annotated gene), 62 k (17%) had one end in an AG and the other in an UC (i.e. un-annotated cluster) and 20 k (5.5%) had both ends in an UC. After processing the hits into anchors and applying the synteny-finding algorithm to the anchors, the categorization of synteny anchors was 27 k AG-to-AG, 4 k AG-to-UC and 6 k UC-to-UC. That is, there were 10 k synteny anchors that would not have been found if only the annotation was used.
Graphical results
The following notations will be used: <species> chromosome <number> will be abbreviated <species>-<number>, e.g. rice chromosome 1 is rice-1. When referring to a set of chromosomes for a species, it will be abbreviated <species>-<list of numbers>, e.g. rice chromosome 1 and rice chromosome 2 is rice-1,2. The comparison of a pair of chromosomes will be in parentheses (<species>-<number>, <species>-<number>), e.g. (rice-1, maize-3) is the comparison between rice chromosome 1 and maize chromosome 3.
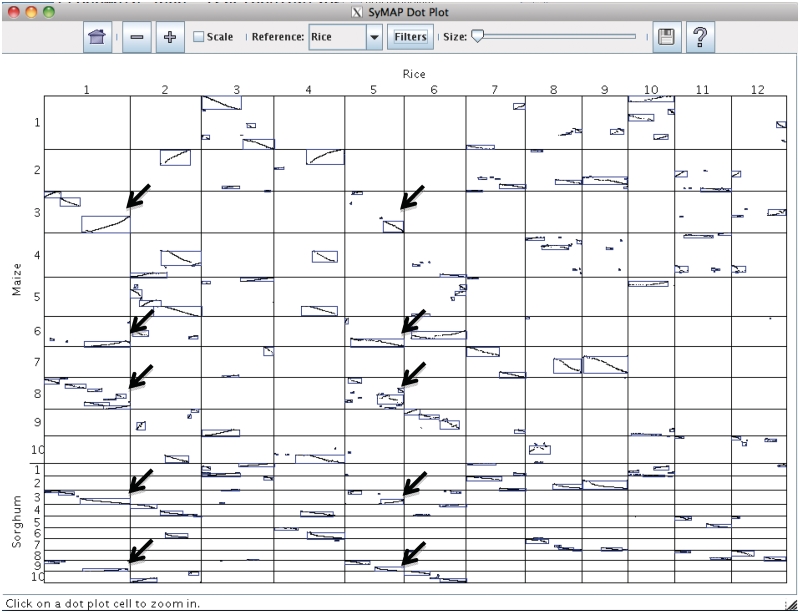
Figure 1 shows the whole-genome dot plot of the three genomes, where the 12 rice chromosomes are the reference (listed across the top) and are aligned to the 10 maize and 10 sorghum chromosomes (listed on the left side). The synteny blocks are enclosed in blue boxes with anchors shown as dots. As discussed in Wei et al. (44), a rice chromosome may share synteny blocks with up to four maize chromosomes, where two are ‘primary’ (from the recent maize duplication) and two are ‘secondary’ (from the ancient grass duplication). The dot plot provides visual evidence, for example, scanning down the vertical cells under rice-1, it shares synteny blocks with maize-3,6,8 and sorghum-3,9. Maize-3,8 have stronger synteny with rice-1 (more anchors) compared to maize-6, which provides evidence that maize-3,8 are the primary syntenic chromosomes; there is only one obvious secondary syntenic chromosome, which is maize-6. Scanning horizontally from these five chromosomes, they all share synteny blocks with rice-5, where maize-6 and sorghum-9 have stronger synteny than maize-3,8 and sorghum-3.
Figure 1.
SyMAP multiple genome dot plot. The reference chromosomes are listed across the top. Synteny blocks are outlined in blue and the dots are anchors. Each cell represents a comparison between the two respective chromosomes, e.g. cell (1,3) represents the comparison between rice-1 and maize-3. Anchor chains that slope from lower left to upper right represent inversions. The Filters button brings up a menu of filters that can be applied; for this example, the filter was set to show only the anchors that are part of a synteny block. The black arrows have been added to point to the upper right corner of the duplications discussed in the text.
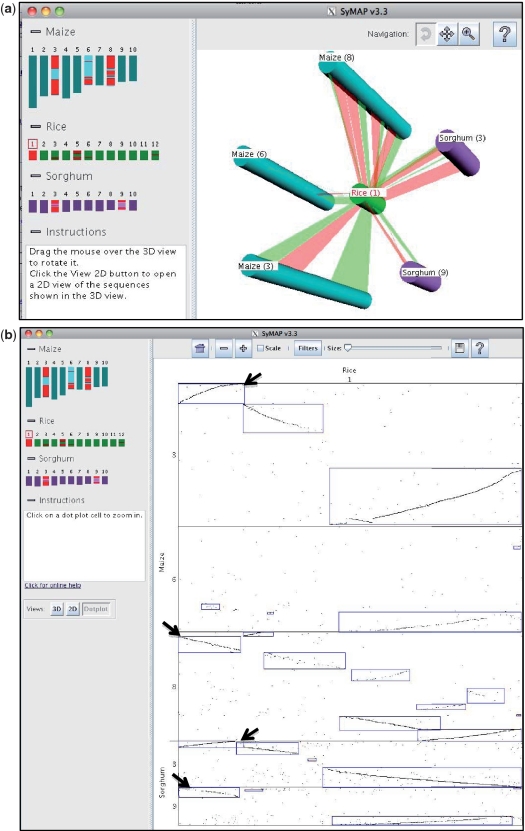
To further investigate this set of chromosomes, the ‘Chromosome 3D and 2D’ option was selected from the project manager; this leads to 3D, 2D and dot-plot display options for multiple chromosomes. On the left side of Figure 2a, rice-1 was selected as the reference chromosome, which automatically highlights maize-3,6,8 and sorghum-3,9. All five of these chromosomes were then selected, which aligned them in the 3D figure, as shown on the right side of Figure 2a. By selecting the ‘Dotplot’ button, the dot-plot view was displayed for the same set of chromosomes (Figure 2b). Anchor chains that slope from lower left to upper right are inversions, for example, the segment in the upper corner of (rice-1, maize-3). Scanning vertically from this block, it can be seen that maize-6 does not have the segment and maize-8 has the segment but it is not inverted; sorghum-3 has the segment inverted and sorghum-8 has the un-inverted segment.
Figure 2.
SyMAP multiple chromosome 3D view and dot plot. (a) On the left panel, the reference chromosome is selected by clicking its number, which highlights the regions of all other chromosomes that have shared synteny blocks. Selecting the body of chromosomes on the left adds them to the 3D view. Selecting them a second time removes them from the 3D display. The 3D display on the right can be rotated, zoomed and moved in order to inspect the syntenic relations. Red ribbons represent un-inverted synteny blocks, green ribbons are inverted (although, as SyMAP detects imperfect synteny, each may contain small regions of the opposite type). (b) Selecting the ‘Dotplot’ button brings up the dot plot for the same set of chromosomes as shown in the 3D display. The black arrows have been added to point to the inverted and un-inverted blocks discussed in the text.
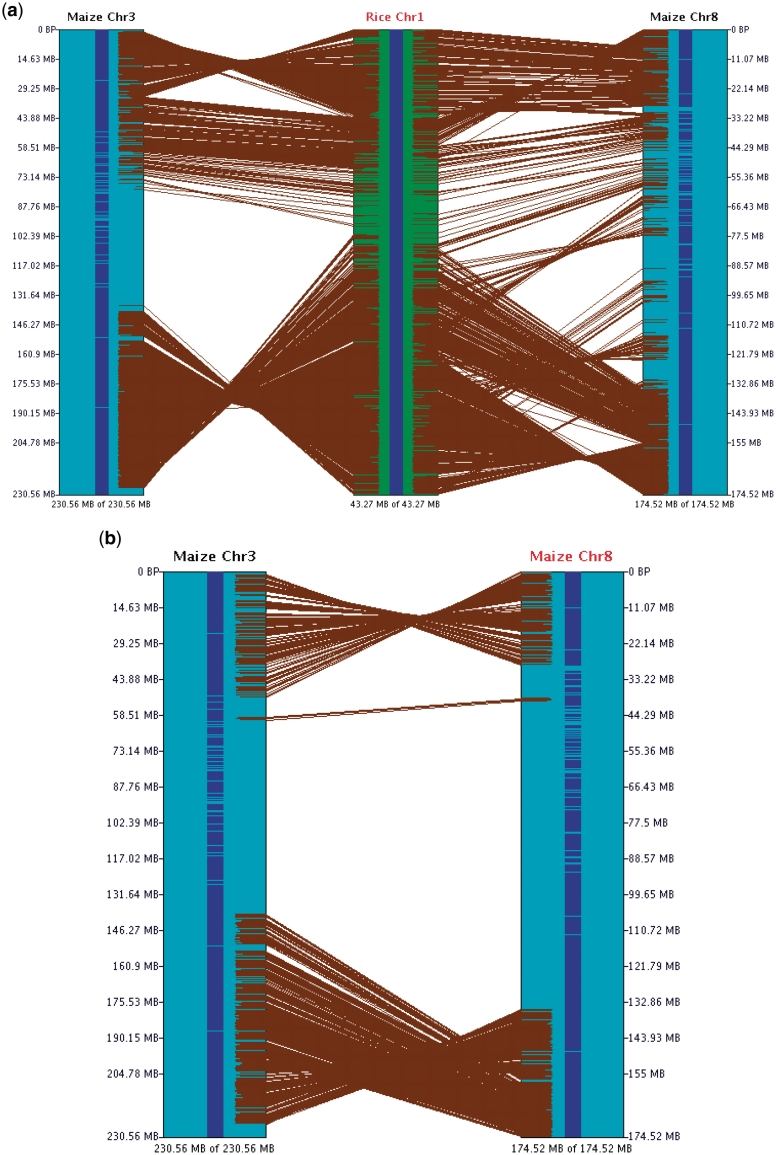
For a detailed view of maize-3 and maize-8 aligned to rice-1, all chromosomes were deselected except these two, then the 2D button was selected, which results in the view shown in Figure 3a. This view provides the most detail, where the large inversions and translocations are easy to see, and small changes can be viewed when zooming into a region. There is a large difference in the sizes of the chromosomes, where maize-3 is 230 Mb, rice-1 is 43 Mb and maize-8 is 174 Mb. This difference in size is only obvious by looking at the size at the bottom of each chromosome; however, the displayed regions may be drawn to scale using the SyMAP scale button (Supplementary Figure S3).
Figure 3.
SyMAP chromosome 2D view. (a) Three chromosomes in 2D. (b) Two chromosomes in 2D. These two displays show the effect of transitive homology, where there is more homology via rice-1 than directly between maize-3 and maize-8. Each brown line represents an anchor. The size of the displayed region is shown at the lower end of the chromosome.
After duplication and subsequent diploidiziation, gene loss and translocation can obscure the homology in a direct comparison, but it can often be revealed by comparing both segments to a third that shares the same ancestral segment [i.e. transitive homology (33)]. Evidence of this can be viewed in Figure 3b compared to Figure 3a. Both maize-3 and maize-8 have syntenic regions to the middle of rice-1, where they do not have corresponding syntenic regions to each other (some level of residual synteny may exist, but it is too weak for the algorithm to detect).
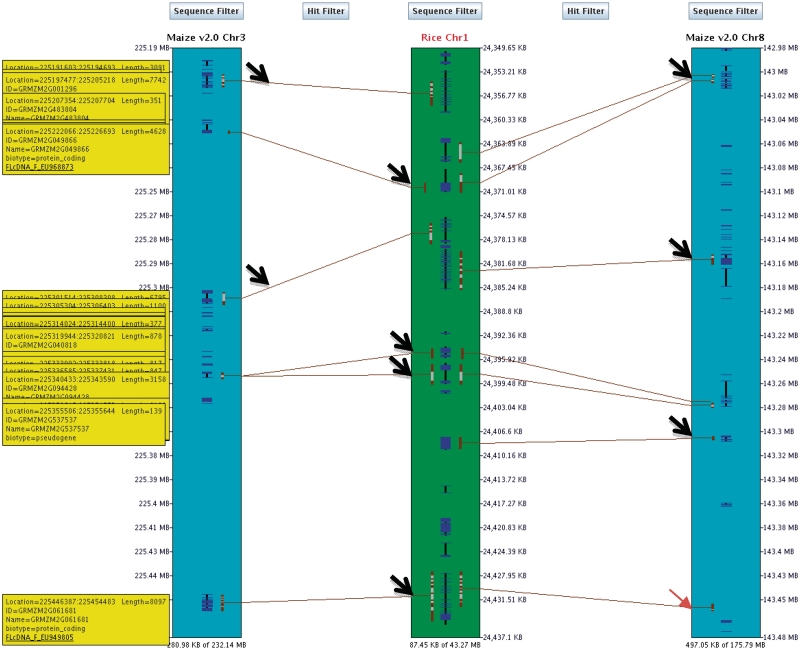
After duplications, one copy of the duplicated gene is often lost (45), which is very apparent when zooming in to various regions of Figure 3a, where one such region is shown in Figure 4. Two genes only occur between rice-1 and maize-3, four genes occur on all three chromosomes, and three genes only occur between rice-1 and maize-8 (shown in Figure 4 as the three columns of arrows, respectively). All anchor points but one are within annotations, where the small red arrow indicates the omission on maize-8.
Figure 4.
SyMAP region 2D view. Any region can be zoomed by dragging the mouse over the region in any of the chromosomes. The genes are represented in blue in the middle of each chromosome, where the thicker parts are exons. The brown bars represent the MUMmer anchors, where the light brown portions link clustered anchors. The yellow bars with text on the left are from the attributes field in the GFF file for maize, where the underlined text is an URL. The sizes of the regions are shown at the bottom of rice-1 (87 kb), maize-3 (281 kb) and maize-8 (497 kb). The three columns of arrows indicate sets of shared genes, as discussed in the text.
To view the information in the attributes field of the GFF file, it is necessary to be zoomed in close to see individual genes, and then turn on the ‘Show annotation descriptions’ option. Figure 4 shows this on the left most track, although further zooming would still be needed to separate the annotations clearly. For maize, we cross-referenced the annotations to the full-length cDNAs (46) and included the link to the FLcDNA site in the GFF file so that the user can find more information about the FLcDNA. If a researcher wants to know if there are any syntenic regions for their gene of interest, they can use the annotation search page (see Supplementary Figure S2C).
DISCUSSION
For synteny packages that depend on annotation, preprocessing is typically required before the synteny computation can be applied. First, the annotated gene set needs to be self-aligned, then a script needs to be written to parse the alignment output and the gene coordinates to create the appropriate input for the program. For SyMAP, the sequence and GFF files can directly be used as input without any preprocessing. In other words, the MUMmer execution within SyMAP is time-consuming (compared to using gene annotation only), but there is generally little human time necessary. There is also no post-processing necessary with SyMAP, as the results are already in graphical format (text file output is also provided). Another important benefit of using the genome sequence as input is that it allows for the possibility of detecting conserved regions that are non-genic or missed by gene prediction programs. As shown in the ‘Results’ section for maize–sorghum, this allowed for identification of potentially 10 542 synteny anchors for which one end was not annotated.
One limitation of the MUMmer-based approach is that MUMmer does not detect off-diagonal anchors for a chromosome aligned to itself (unless the –maxmatch option is used, which produces prohibitively large output). This means that SyMAP can compute synteny for different chromosomes from the same genome, but not within the same chromosome (i.e. duplicated or translocated regions within a single chromosome cannot be seen). For a future release, we will seek to support a genome alignment program that resolves this problem, while retaining acceptable performance.
Versatile graphical displays, such as provided by SyMAP, are important for effectively using the results of syntenic computations. Java is an excellent language to use for this application, as there are a range of graphical libraries to support the visualization, it provides the dynamic interactive displays of a full featured programming language (in contrast to Perl/CGI interfaces), and it can run standalone or from the web. The only drawback we have found is that the support for Java 3D in applets (i.e. in the web display) can have problems, especially on current Mac platforms, which have pre-installed out-of-date Java 3D libraries. The standalone version is easiest to use for intense exploration of synteny as Internet latency is avoided.
SUPPLEMENTARY DATA
Supplementary Data available at NAR Online.
FUNDING
Funding for open access charge: The USDA National Institute of Food and Agriculture (grant no. 2008-35300-04439).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: A system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16:1159–1168. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson W, Soderlund C. Integrating sequence with FPC fingerprint maps. Nucleic Acids Res. 2009;37:e36. doi: 10.1093/nar/gkp034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinform. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youens-Clark K, Faga B, Yap IV, Stein L, Ware D. CMap 1.01: a comparative mapping application for the Internet. Bioinformatics. 2009;25:3040–3042. doi: 10.1093/bioinformatics/btp458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann S, Lu D, Phillips J, Vision TJ. Phytome: a platform for plant comparative genomics. Nucleic Acids Res. 2006;34:D724–D730. doi: 10.1093/nar/gkj045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha AU, Meller J. Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinform. 2007;8:82. doi: 10.1186/1471-2105-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courcelle E, Beausse Y, Letort S, Stahl O, Fremez R, Ngom-Bru C, Gouzy J, Faraut T. Narcisse: a mirror view of conserved syntenies. Nucleic Acids Res. 2008;36:D485–D490. doi: 10.1093/nar/gkm805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 14.Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19:1497–1505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandepoele K, Saeys Y, Simillion C, Raes J, Van De Peer Y. The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. 2002;12:1792–1801. doi: 10.1101/gr.400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Shi X, Li Z, Zhu Q, Kong L, Tang W, Ge S, Luo J. Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinform. 2006;7:447. doi: 10.1186/1471-2105-7-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodelsperger C, Dieterich C. CYNTENATOR: progressive gene order alignment of 17 vertebrate genomes. PLoS One. 2010;5:e8861. doi: 10.1371/journal.pone.0008861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pevzner P, Tesler G. Genome rearrangements in mammalian evolution: lessons from human and mouse genomes. Genome Res. 2003;13:37–45. doi: 10.1101/gr.757503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannon SB, Kozik A, Chan B, Michelmore R, Young ND. DiagHunter and GenoPix2D: programs for genomic comparisons, large-scale homology discovery and visualization. Genome Biol. 2003;4:R68. doi: 10.1186/gb-2003-4-10-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calabrese PP, Chakravarty S, Vision TJ. Fast identification and statistical evaluation of segmental homologies in comparative maps. Bioinformatics. 2003;19(Suppl. 1):i74–i80. doi: 10.1093/bioinformatics/btg1008. [DOI] [PubMed] [Google Scholar]
- 26.Hachiya T, Osana Y, Popendorf K, Sakakibara Y. Accurate identification of orthologous segments among multiple genomes. Bioinformatics. 2009;25:853–860. doi: 10.1093/bioinformatics/btp070. [DOI] [PubMed] [Google Scholar]
- 27.Vergara IA, Chen N. Using OrthoCluster for the detection of synteny blocks among multiple genomes. Curr. Protoc. Bioinform. 2009;27 doi: 10.1002/0471250953.bi0610s27. 6.10.1–6.10.18. [DOI] [PubMed] [Google Scholar]
- 28.Ng MP, Vergara IA, Frech C, Chen Q, Zeng X, Pei J, Chen N. OrthoClusterDB: an online platform for synteny blocks. BMC Bioinform. 2009;10:192. doi: 10.1186/1471-2105-10-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simillion C, Vandepoele K, Saeys Y, Van de Peer Y. Building genomic profiles for uncovering segmental homology in the twilight zone. Genome Res. 2004;14:1095–1106. doi: 10.1101/gr.2179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simillion C, Janssens K, Sterck L, Van de Peer Y. i-ADHoRe 2.0: an improved tool to detect degenerated genomic homology using genomic profiles. Bioinformatics. 2008;24:127–128. doi: 10.1093/bioinformatics/btm449. [DOI] [PubMed] [Google Scholar]
- 32.Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van de Peer Y. Computational approaches to unveiling ancient genome duplications. Nat. Rev. Genet. 2004;5:752–763. doi: 10.1038/nrg1449. [DOI] [PubMed] [Google Scholar]
- 34.Kent WJ. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 36.Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Genet. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 37.IRGSP. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 38.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 40.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 41.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14:1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 44.Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai J, Ma J, Swigonova Z, Ramakrishna W, Linton E, Llaca V, Tanyolac B, Park YJ, Jeong OY, Bennetzen JL, et al. Gene loss and movement in the maize genome. Genome Res. 2004;14:1924–1931. doi: 10.1101/gr.2701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, Currie J, Angelova A, Collura K, Wissotski M, Ashley E, et al. Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 2009;5:e1000740. doi: 10.1371/journal.pgen.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.