Abstract
In pre-mRNA splicing, a conserved AG/G at the 3′-splice site is recognized by U2AF35. A disease-causing mutation abrogating the G nucleotide at the first position of an exon (E+1) causes exon skipping in GH1, FECH and EYA1, but not in LPL or HEXA. Knockdown of U2AF35 enhanced exon skipping in GH1 and FECH. RNA-EMSA revealed that wild-type FECH requires U2AF35 but wild-type LPL does not. A series of artificial mutations in the polypyrimidine tracts of GH1, FECH, EYA1, LPL and HEXA disclosed that a stretch of at least 10–15 pyrimidines is required to ensure normal splicing in the presence of a mutation at E+1. Analysis of nine other disease-causing mutations at E+1 detected five splicing mutations. Our studies suggest that a mutation at the AG-dependent 3′-splice site that requires U2AF35 for spliceosome assembly causes exon skipping, whereas one at the AG-independent 3′-splice site that does not require U2AF35 gives rise to normal splicing. The AG-dependence of the 3′-splice site that we analyzed in disease-causing mutations at E+1 potentially helps identify yet unrecognized splicing mutations at E+1.
INTRODUCTION
In higher eukaryotes, generation of functional mRNA is dependent on the removal of introns from pre-mRNA by splicing (1). The splicing process occurs in the spliceosome, the major components of which include five small nuclear RNAs and their associated proteins (U1, U2, U4, U5 and U6 snRNPs) in addition to a large number of non-snRNP proteins (2). In the first step of assembly of the spliceosome, U1 snRNP, SF1, U2AF65 and U2AF35 bind to the splicing cis-elements at the 5′ splice site (ss), the branch point sequence (BPS), the polypyrimidine tract (PPT) and the acceptor site, respectively, to form complex E (3).
Yeast has a well conserved BPS of UACUAAC (4), whereas we recently reported that human carries a highly degenerate BPS of yUnAy, where ‘y’ and ‘n’ represent pyrimidines and any nucleotides, respectively (5). Degeneracy of the human BPS supports a notion that the human BPS is likely to be recognized along with the downstream PPT where U2AF65 binds and possibly with the invariant AG dinucleotide at the 3′ ss where U2AF35 binds (6,7). U2AF65 and U2AF35 also make a heterodimer (8). In PPT, uridines are preferred over cytidines (9,10). In addition, PPT with 11 continuous uridines is highly competent and the position of such PPT is not critical (10). On the other hand, PPTs with only five or six uridines are required to be located close the 3′ AG for efficient splicing. In addition, phosphorylated DEK binds to and cooperates with U2AF35 for proper recognition of the 3′ ss (11).
In the next step of the spliceosome assembly, the bound U2AF65 and U2AF35 facilitate substitution of SF1 for U2snRNP at the branch point to form complex A. Introns carrying a long stretch of PPT do not require U2AF35 for this substitution, which is called ‘AG-independent 3′ ss’ (12–15). On the other hand, introns with a short or degenerate PPT require both U2AF65 and U2AF35 for this substitution, which is called ‘AG-dependent 3′ ss’. Thereafter, the U4/U6.U5 tri-snRNP is integrated into the spliceosome to form complex B and the initial assembly of the spliceosome is completed.
The invariant AG dinucleotides are frequently reported targets of mutations causing human diseases, and the most frequent consequence is skipping of one or more exons (16). In addition, even mutations in highly degenerate BPS (5) and PPT (17) give rise to aberrant splicing causing genetic diseases (18). Disease-causing mutations also affect the first nucleotide of an exon (E+1), but their effects on pre-mRNA splicing have been rarely scrutinized. As far as we know, only three such mutations in FECH (19), GH1 (20) and EYA1 (21) have been reported to cause aberrant splicing. Similarly, two such mutations in LPL (22) and HEXA (23) have been reported to have no effect on splicing. In this communication, we dissected molecular bases that differentiate splicing-disrupting and splicing-competent mutations, and found that AG-dependent ss is vulnerable to a mutation at E+1, whereas AG-independent ss is tolerant.
MATERIALS AND METHODS
Minigene constructs and mutagenesis
Human genes of our interest were PCR-amplified from HEK293 cells using the KOD plus DNA polymerase (Toyobo). We introduced restriction enzyme-recognition sites at the 5′-end of the forward and reverse primers. We inserted the amplicon into the pcDNA3.1(+) mammalian expression vector (Invitrogen). We introduced patients’ or artificial mutations with the QuikChange site-directed mutagenesis kit (Stratagene). We confirmed the absence of unexpected artifacts with the CEQ8000 genetic analyzer (Beckman Coulter).
Cell culture and transfection procedures
HEK293 cells were maintained in the Dulbecco’s minimum essential medium (DMEM, Sigma-Aldrich) with 10% fetal bovine serum (FBS, Sigma-Aldrich). At ∼50% confluency (∼5 × 105 cells) in a 12-well plate, 1 ml of fresh Opti-MEM I (Invitrogen) was substituted for DMEM, and 500 ng of a minigene with 1.5 µl of the FuGENE6 transfection reagent (Roche Diagnostics) were then added. After 4 h, 2 ml of DMEM with 10% FBS was overlaid, and the cells were incubated overnight. The transfection medium was replaced with 2 ml of fresh DMEM with 10% FBS. RNA was extracted at 48 h after initiation of transfection.
RNA extraction and RT–PCR
Total RNA from HEK293 was extracted by Trizol reagent (Invitrogen) according to the manufacturer’s protocols. The quantity and quality of RNA was determined by spectrophotometry (NanoDrop Techonologies). Twenty percent of the isolated RNA was used as a template for cDNA synthesis with the Oligo(dT) 12–18 Primer (Invitrogen) and the ReverTra Ace (Toyobo). Ten percent of the synthesized cDNA was used as a template for RT–PCR amplification with T7 primer (5′-TAATACGACTCACTATAGGG-3′) and gene-specific primers for minigenes in pcDNA3.1(+). Image J software (National Institutes of Health) was used to quantify intensities of fragments. We employed JMP (SAS Institute) for statistical analysis.
RNA interference to knockdown U2AF35
We synthesized siRNA of 5′-GGCUGUGAUUGACUUGAAUdTdT-3′ (GenBank accession number NM_006758, nucleotides 459–479), which is against the shared sequence of U2AF35a and U2AF35b (15). We employed Lipofectamine 2000 (Invitrogen) to cotransfect plasmids and siRNAs according to the manufacturer’s protocols. Briefly, the transfection reagent included 300 ng of the plasmid, 50 pmol of siRNA, and 2 µl of lipofectamine 2000 in 100 µl of Opti-MEM I. The cells were harvested by western blotting for 48 h after transfection. The primary antibodies were goat polyclonal antibody for U2AF35 (Santa Cruz Biotechnology), and mouse monoclonal antibodies for U2AF65 (Santa Cruz Biotechnology) and PTB (Zymed Laboratories). The secondary antibodies were HRP-conjugated mouse anti-goat (Santa Cruz Biotechnology) or sheep anti-mouse (GE healthcare) antibodies. The immunoreactive proteins were detected by enhanced chemiluminescence (ECL, Amersham Biosciences).
For the siRNA rescue assay, we cloned the human U2AF35 cDNA (Open Biosystems) into the HindIII and EcoRI restriction sites of the p3XFLAG-CMV-14 vector (Sigma-Aldrich). We introduced four silent mutations into the siRNA target region using the QuikChange site-directed mutagenesis kit with a primer, 5′-GAAAAGGCTGTAATCGATTTAAATAACCGTTGGTT-3′, where artificial mutations are underlined (24).
RNA probe synthesis
We synthesized [α-32P]-CTP-labeled RNA using the Riboprobe in vitro transcription system (Promega) from a PCR-amplified fragment according to the manufacturer’s instructions. We used the same forward primer for all the probes with the sequence of 5′-TAATACGACTCACTATAGGGAGACAGG-3′, where the italicized is T7 promoter and the underlined is for annealing to the reverse primer. The four reverse primers were: wild-type FECH, 5′-TGGACCAACCTATGCGAAAGATAGACGAATGCGTAAGCCTGTCTC-3′; mutant FECH, 5′-TGGACCAAACTATGCGAAAGATAGACGAATGCGTAAGCCTGTCTC-3′; wild-type LPL, 5′-TGGATCGAGGCCTTAAAAGGGAAAAAAGCAGGAACACCCTGTCTC-3′; and mutant LPL, 5′-TGGATCGAGGACTTAAAAGGGAAAAAAGCAGGAACACCCTGTCTC-3′, where the underlined is for annealing to the forward primer.
Expression and purification of recombinant proteins
The human U2AF35 and U2AF65 cDNAs were obtained from Open Biosystems. U2AF35 and U2AF65 cDNAs were subcloned into the BamHI and EcoRI restriction sites of the pFastBac HTb vector. The recombinant baculoviruses were expressed using the Bac-to-Bac Baculovirus Expression System (Invitrogen) according to the manufacturer’s instructions. Infected Sf9 cells were harvested after 48 h and resuspended in the lysis buffer containing 50 mM sodium phosphate, 10 mM imidazole, 300 mM NaCl, 1% Triton X-100, 2 mM β-mercaptoethanol, the Complete Protease Inhibitor Cocktail (Roche Applied Science) and 5 U endonuclease in pH 7.0. His-tagged U2AF35 and U2AF65 proteins were purified using the TALON metal affinity resins (Clontech) under the denatured and native conditions, respectively. Purified U2AF35 was refolded by extended dialysis in dialysis buffer (50 mM sodium phosphate, 300 mM NaCl, 150 mM imidazole, pH 7.0). We determined the protein concentrations using the Pierce 660 nm Protein Assay Reagent (Thermo Scientific).
RNA-electrophoretic mobility shift assay
The radioactively labeled RNA (1 × 105 cpm) was incubated at room temperature with varying concentrations of recombinant proteins, 16 µg of yeast tRNA, and 1.6 U of RNasin (Toyobo) in a final volume of 20 µl of the binding buffer (20 mM HEPES pH 7.8, 50 mM KCl, 3 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM EDTA and 5% glycerol). After 20 min, the RNA–protein complexes were separated on 5% non-denaturing polyacrylamide gels in 1 × TBE buffer at 4°C. The gels were dried and complex formation was visualized using the Typhoon 8600 Imager (GE Healthcare).
In silico analysis of the human genome and ESE-motifs
We analyzed human genome annotations (NCBI Build 37.1, hg19) by writing Perl programs, and executing them either on the PrimePower HPC2500/Solaris 9 supercomputer (Fujitsu) or on the cygwin UNIX emulator running on a Windows computer. To search for ESE-motifs, we used the ESE Finder (http://rulai.cshl.org/ESE/) (25,26), the RESUCE-ESE server (http://genes.mit.edu/burgelab/rescue-ese/) (27), the FAS-ESS server (http://genes.mit.edu/fas-ess/) (28), the PESX server (http://cubweb.biology.columbia.edu/pesx/) (29,30), and the ESRsearch server (http://ast.bioinfo.tau.ac.il/) (31).
RESULTS
Recapitulation of normal and aberrant splicing in minigenes
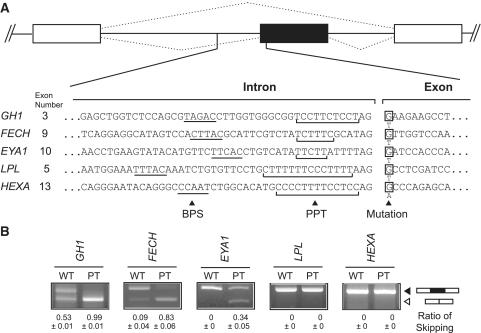
We first constructed minigenes of GH1, FECH, EYA1, LPL and HEXA, and introduced a previously reported disease-causing mutation at E+1 (Figure 1A). These minigenes successfully recapitulated normal and aberrant splicings: mutations in GH1, FECH and EYA1 caused exon skipping, whereas those in LPL or HEXA did not (Figure 1B).
Figure 1.
Recapitulation of normal and aberrant splicing of five genes. (A) Nucleotide sequences at the intron/exon junctions of five analyzed genes. Putative BPS is underlined. PPT is shown by a bracket. Mutant nucleotides are indicated at E+1. (B) RT–PCR of minigenes expressed in HEK293 cells carrying the wild-type (WT) or patient’s (PT) nucleotide. The mutations cause exon skipping in GH1, FECH and EYA1, but not in LPL and HEXA. Mean and SD of three independent experiments of the densitometric ratios of the exon-skipped product is shown at the bottom.
Down-regulation of U2AF35 increased exon skipping in wild-type GH1 and FECH, but not in wild-type EYA1, LPL and HEXA
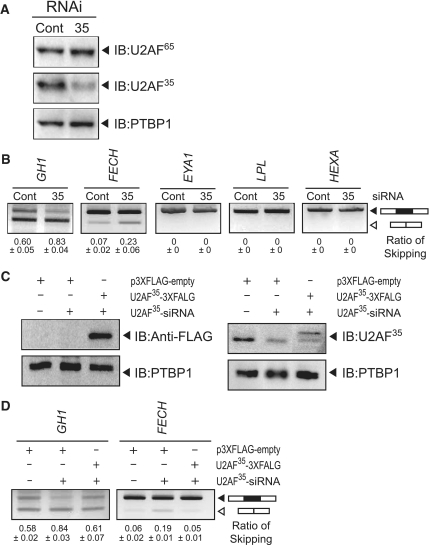
We predicted that a mutation at E+1 should disrupt binding of U2AF35. We thus hypothesized that GH1, FECH and EYA1 require binding of U2AF35 for the assembly of spliceosome, whereas LPL and HEXA do not require it. To prove this hypothesis, we first knocked down U2AF35 and analyzed its effect on the wild-type minigenes. We achieved an efficient down-regulation of U2AF35 in HEK293 cells (Figure 2A). We also confirmed that the U2AF35-siRNA had no effect on the expression level of U2AF65. As expected, the down-regulation of U2AF35 increased exon skipping of GH1 and FECH (Figure 2B) but not to the levels of the mutant constructs (Figure 1B). Again, as expected, we observed no effect on LPL and HEXA. Unexpectedly, however, EYA1 demonstrated no response to the down-regulation of U2AF35. Less efficient effects of U2AF35-siRNA on GH1, FECH and EYA1 (Figure 2B) compared to the mutant constructs (Figure 1B) were likely because the mutation abolished binding of U2AF35 in all the cells, whereas substantial numbers of cells failed to incorporate U2AF35-siRNA and gave rise to normally spliced products.
Figure 2.
Effects of down-regulation of U2AF35 on pre-mRNA splicing. (A) Western blots demonstrating that U2AF35-siRNA efficiently knocks down U2AF35 but not U2AF65 or PTBP1. (B) Down-regulation of U2AF35 facilitates exon skipping in wild-type GH1 and FECH, but not in wild-type EYA1, LPL and HEXA. (C) Introduction of an siRNA-resistant p3XFLAG-U2AF35 encoding 3× FLAG fused with U2AF35 is visualized by immunoblots against FLAG and U2AF35. (D) Exon skipping facilitated by U2AF35-siRNA is partially rescued by introduction of the siRNA-resistant p3XFLAG-U2AF35.
We additionally introduced the siRNA-resistant p3XFLAG-U2AF35 to ensure that the effect of siRNA-U2AF35 was not due to off-target effects (Figure 2C). As expected, coexpression of p3XFLAG-U2AF35 partially rescued the splicing defects in GH1 and FECH (Figure 2D).
U2AF35 is required for binding of U2AF65 to PPT in FECH but not in LPL
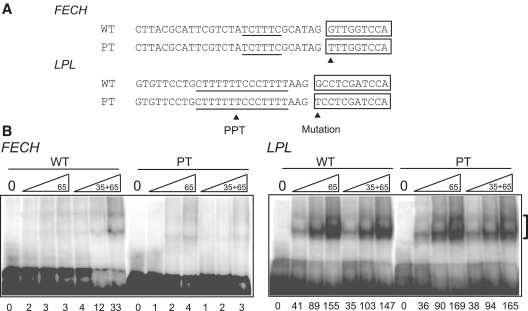
To further prove that U2AF35 is required for pre-mRNA splicing, we employed an electrophoretic mobility shift assay (EMSA) using wild-type and mutant RNA substrates of FECH and LPL (Figure 3A). His-tagged U2AF35 and U2AF65 were expressed using bacluovirus and were purified under denatured and native conditions, respectively. Denatured U2AF35 was refolded before RNA-EMSA. As expected, U2AF65 failed to bind to the wild-type FECH in the absence of U2AF35, and addition of U2AF35 gained its binding. For the mutant FECH, neither U2AF65 alone nor addition of both U2AFs showed binding of U2AFs. On the other hand, the wild-type LPL did not require U2AF35 to bind to U2AF65. Addition of U2AF35 did not substantially increased binding of U2AF65. These bindings were not affected by the mutation at E+1 of LPL (Figure 3B).
Figure 3.
RNA-EMSA. (A) Sequences of wild-type (WT) and mutant (PT) RNA probes of FECH and LPL employed for RNA-EMSA. (B) RNA-EMSA of wild-type and mutant FECH and LPL with increasing amounts of U2AF65 with or without U2AF35. His-tagged U2AF65 and U2AF35 are expressed in Sf9 cells and are purified. Wild-type FECH requires U2AF35 to bind to U2AF65, whereas wild-type LPL does not require U2AF35. A mutation at E+1 abrogates binding of U2AF65 in FECH but not in LPL. Concentrations of U2AF35 are 5, 10 and 20 ng/µl; and those of U2AF65 are 10, 20 and 40 ng/µl. Numbers at the bottom indicate intensities of the retarded fragments in arbitrary units.
These results indicate that the mutation in FECH compromises a binding affinity for U2AF35, which in turn abrogates binding of U2AF65 and results in aberrant splicing. On the other hand, wild-type LPL does not need to bind to U2AF35 and the mutation at E+1 has no effect on the assembly of spliceosome.
PPT determines the splicing consequences of the mutations
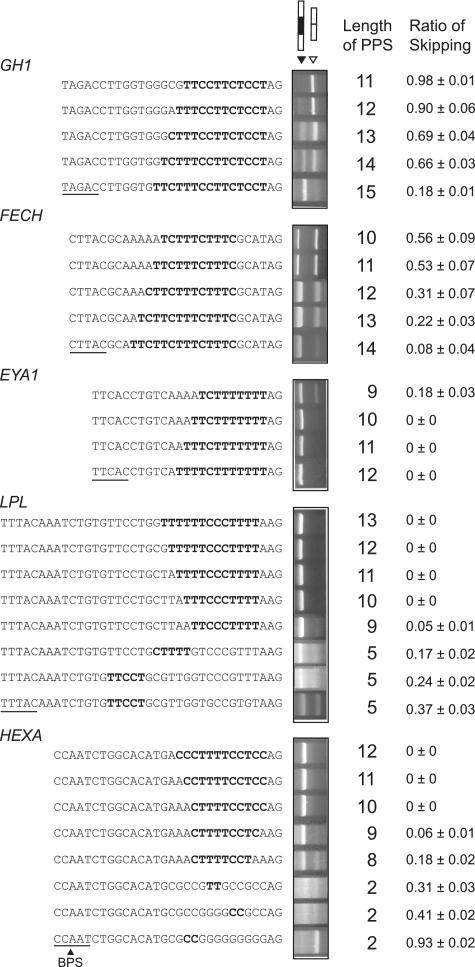
In an effort to delineate effects of the PPT sequences on the splicing consequence of a mutation at E+1, we introduced a series of mutations into the PPT in the presence of the mutation at E+1. Extensions of the polypyrimidine stretch ameliorated aberrant splicing in GH1, FECH and EYA1. Conversely, truncations or disruptions of the polypyrimidine stretch caused exon skipping in LPL and HEXA (Figure 4).
Figure 4.
RT–PCR of HEK293 cells transfected with minigenes carrying artificially extended or disrupted PPT’s. All the constructs harbor a mutation at E+1. The top construct of each gene represents the patient’s sequence. Only the nucleotide sequences of the 3′-end of an intron are indicated. The longest stretches of the polypyrimidines are shown in bold. Underlines indicate putative BPS’s. The rightmost column shows the mean and SD of three independent experiments of the densitometric ratios of the exon-skipped product.
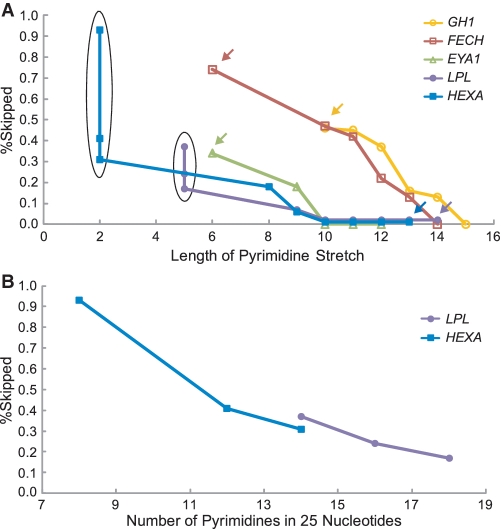
Length of the polypyrimidine stretch best predicts the splicing consequences
We next sought for parameters that differentiate normal and aberrant splicings in these minigenes. Analysis of parameters that potentially dictate the strength of the PPT indicated that the length of pyrimidine stretch, the number of pyrimidines in 25 or 50 nt at the 3′-end of an intron correlated with the ratio of exon skipping with correlation coefficients of more than 0.6 (Supplementary Table S1). The number of pyrimidines in 25 or 50 nt at the 3′-end of an intron, however, failed to predict splicing consequences of nine other constructs shown in Figure 6, and is likely to be overfitted parameters unique to the 35 constructs in Figure 4. Coolidge and colleagues report that (GU)11 in PPT is partly functional, but we did not observe alternative purine and pyrimidine residues in our PPTs and did not quantify effects of alternative nucleotides (10). We thus took the length of pyrimidine stretch as a best parameter to dictate the strength of the PPT (Figure 5A). The native GH1, FECH and EYA1 carry a stretch of 6–10 pyrimidines, whereas the native LPL and HEXA harbor a stretch of 14 and 13 pyrimidines, respectively (arrows in Figure 5A). For highly degenerate PPTs in the artificial constructs, the total number of pyrimidines in a stretch of 25 nt at the 3′-end of an intron well predicts the ratio of exon skipping (Figure 5B). These analyses revealed that the length of the polypyrimidine stretch should be at least 10–15 nt to ensure normal splicing even in the presence of a mutation at E+1.
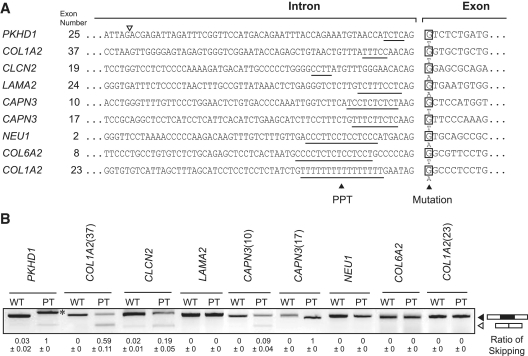
Figure 6.
RT–PCR analysis of nine disease-causing mutations at E+1. (A) Sequences at the intron/exon junctions of nine pairs of wild-type and mutant constructs. The longest polypyrimidine stretches are underlined. (B) RT–PCR of minigenes transfected into HEK293 cells. Five mutant constructs are aberrantly spliced, whereas the remaining four mutants are normally spliced. Numbers in the parentheses indicate exon numbers. In PKHD1, a cryptic 3′-splice site (open arrowhead in panel A) at 55 nt upstream of the native site is activated (asterisk). Mean and SD of three independent experiments of the densitometric ratios of the exon-skipped product is shown at the bottom.
Figure 5.
Ratios of exon skipping are plotted against the lengths of the polypyrimidine stretch (A) and the numbers of pyrimidines in 25 nt at the 3′-end of an intron (B). The ordinate (percent skipped) represents the ratios of exon skipping compared to that of the wild-type construct. The data are obtained from RT–PCR shown in Figure 4. Arrows indicate the original constructs carrying the patient’s sequence, and the others are artificial constructs. Six constructs indicated by ovals in (A) are plotted in (B).
Identification of effects on pre-mRNA splicing of nine disease-associated mutations at the first nucleotide of an exon
We next examined other mutations at E+1 in which splicing consequences have not been previously analyzed. We first identified 224 mutations that abrogate the first ‘G’ nucleotide of an exon in the Human Gene Mutation Database at http://www.hgmd.cf.ac.uk/ (data not shown). Among these, we arbitrarily chose nine mutations causing neuromuscular and musculoskeletal disorders (Figure 6A).
We constructed nine pairs of wild-type and mutant minigenes, and introduced them into HEK293 cells. We observed aberrant splicing in PKHD1, COL1A2 (exon 37), CLCN2, CAPN3 (exons 10 and 17), but not in LAMA2, NEU1, COL6A2 and COL1A2 (exon 23) (Figure 6B). The lengths of the polypyrimidine stretch of the five aberrantly spliced constructs ranged from 4 to 10 nt, whereas those of the four normally spliced constructs ranged from 9 to 16 nt. These results are in concordance with a notion that the short polypyrimidine stretches are predisposed to aberrant splicing due to a mutation at E+1, whereas long polypyrimidine stretches are tolerant to such mutations. Among the 224 mutations affecting ‘G’ at E+1, only three mutations have been reported to cause aberrant splicing. We here analyzed nine mutations and identified five more such mutations. It is thus likely that most splicing mutations at E+1 still remain unrecognized to date.
Analysis of the 3′-splice sites of the human genome
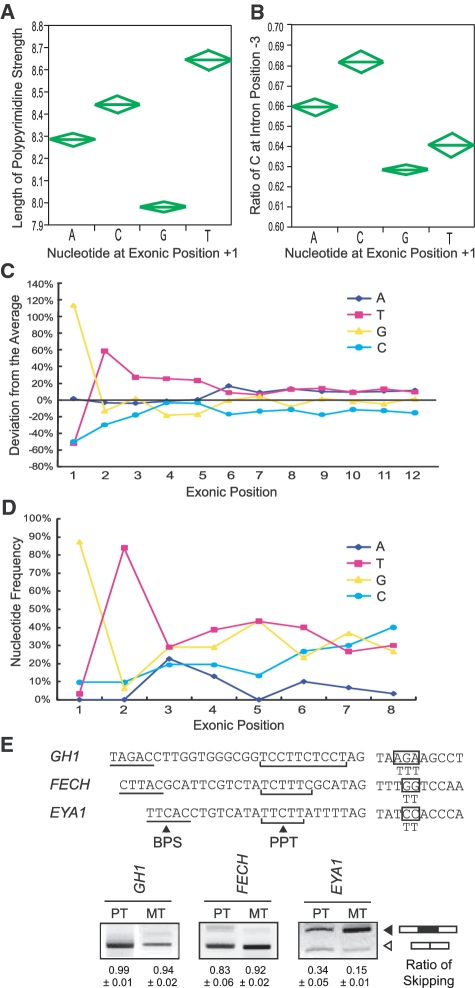
We next analyzed PPTs of 176 809 introns of the entire human genome. The length of the pyrimidine stretch was shorter when E+1 was the conserved ‘G’ (Figure 7A). This also supports a notion that AG-dependent 3′ ss harboring G at E+1 has a short polypyrimidine stretch (12). In addition, the ratio of ‘C’ at intronic position −3 was lower when E+1 was the conserved ‘G’ (Figure 7B), which suggests that G at E+1 makes C at −3 dispensable for binding to U2AF35, although this is not directly relevant to the length of the PPT.
Figure 7.
(A) Polypyrimidine stretch and the first nucleotide of an exon in the human genome. The longest stretch of uninterrupted pyrimidines among 25 nt at the 3′-ends of an intron is counted for 176 809 introns of the human genome. Diamonds represent means and 95% confidence intervals. One-way ANOVA and Fisher’s-multiple range test revealed statistical significance of P < 0.0001. (B) Ratios of ‘C’ at position −3 in relation to the first nucleotide of an exon are analyzed for 176 809 introns of the human genome. Diamonds represent means and 95% confidence intervals. One-way ANOVA and Fisher’s-multiple range test revealed statistical significance of P < 0.0001. (C) Preferentially observed nucleotides at the 5′-end of an exon in human. Only wobbling nucleotides are counted in the human genome. (D) Nucleotide frequencies at exonic positions +1 to +8 according to the SELEX data of U2AF35 by Wu and colleagues (12). (E) Effects of ‘TTT’ at exonic positions +3 to +5 in GH1, FECH and EYA1 carrying the patient’s mutation at E+1. Artificially substituted exonic nucleotides are indicated by boxes. Mean and SD of three independent experiments of the densitometric ratios of the exon-skipped product is shown at the bottom.
Being prompted by a previous report that U2AF35 binds up to the 10th nucleotide of an exon (12), we examined nucleotide frequencies at exonic positions +1 to +12. We counted only wobbling nucleotides based on the human genome annotation NCBI Build 37.1 (hg19). As expected, ‘GT’ dinucleotide was frequently observed at exonic positions +1 and +2. We also observed preference for a ‘T’ nucleotide at positions +3 to +5 (Figure 7C). Alignment of SELEX results of U2AF35 by Wu and colleagues (12) similarly demonstrate overrepresentation of ‘T’ nucleotides at positions +3 to +6 (Figure 7D). We thus analyzed effects of ‘TTT’ at positions +3 to +5 using the GH1, FECH and EYA1 minigenes carrying the patient’s mutations. We found that introduction of ‘TTT’ at exonic position +3 to +5 had no effect in GH1 and FECH, but slightly enhanced exon recognition in EYA1 (Figure 7E).
DISCUSSION
We previously reported that the SD-score algorithm efficiently predicts splicing consequences of a mutation affecting the 5′ ss (32). We next identified that the human BPS consensus is simply yUnAy (5), and hoped to predict if a given mutation affecting the BPS causes aberrant splicing or not. The high degeneracy of the BPS consensus, however, prevented us from constructing an efficient algorithm. In this communication, we worked on mutations at E+1. As far as we know, only three such mutations have been reported to cause aberrant splicing, and only two such mutations have been reported not to affect splicing. Knockdown and RNA-EMSA of U2AF35, as well as analyses of artificial PPT mutations and nine disease-causing mutations at E+1 revealed that AG-dependence of 3′ ss determines the splicing consequences. In the presence of a mutation at E+1, a stretch of 15 or more pyrimidines ensures normal splicing, whereas a stretch of 10 or less pyrimidines are predisposed to aberrant splicing.
AG-dependent 3′ ss requires both U2AF65 and U2AF35 to bring U2snRNP to the branch point, whereas AG-independent 3′ ss has a long stretch of pyrimidines that can bind to U2AF65 without U2AF35 (13,15). U2AF35 potentially provides an additional RNA–protein interacting force and an additional SR protein-binding surfaces (33). An artificial G-to-C mutation at E+1 downstream of a stretch of five pyrimidines in the mouse IgM gene abrogates binding of U2AF35 and causes defective splicing (14). Similarly, in INSR exon 11 carrying an ‘A’ nucleotide at E+1, a stretch of 14 pyrimidines but not of 10 pyrimidines is properly spliced (34). Additionally, a stretch of eight pyrimidines upstream of the last exon with ‘C’ at E+1 of EIF3S7 is dependent on U2AF35, whereas a stretch of 14 pyrimidines upstream of the last exon with ‘A’ at E+1 of CUEDC1 is independent (15). Our observations and previous reports all point to a notion that effects on pre-mRNA splicing should be scrutinized for a mutation at E+1 if the preceding intron carries a short stretch of 10 or less pyrimidines. Indeed, in our analysis of nine disease-causing mutations, five of six mutants with 10 or less contiguous pyrimidines were aberrantly spliced (Figure 6), but no splicing analysis has been documented for any of them.
We first report overrepresentation of ‘T’ nucleotides at exonic positions +3 to +5 in the human genome, as well as in in vitro U2AF35-binding sites. Enhancement of exon recognition in EYA1 by introduction of ‘TTT’ at positions +3 to +5 also underscores a notion that ‘TTT’ at +3 to +5 is likely to enhance binding of U2AF35. Effects of ‘TTT’, however, were not observed in GH1 and FECH. As the patient’s mutation in GH1 and FECH resulted in almost complete skipping of an exon, whereas that in EYA1 gave rise to both exon-skipped and included products. The degrees of aberration of exon recognition may account for the ‘TTT’-responsiveness. Alternatively, although no ESE motif was detected in the ‘TTT’-introduced EYA1 by five different ESE search tools, an unrecognized ESE might have ameliorated exon skipping in EYA1. Further analysis is required to elucidate effects of overrepresentation of ‘T’ at positions +3 to +5.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aids from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Ministry of Health, Labor and Welfare of Japan. Funding for open access charge: Innovative Cell Biology by Innovative Technology granted by the Japan Science and Technology Agency (JST).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 3.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Gen. Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 4.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 5.Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36:2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 7.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 8.Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 9.Mullen MP, Smith CW, Patton JG, Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991;5:642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- 10.Coolidge CJ, Seely RJ, Patton JG. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–896. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 12.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 13.Guth S, Martinez C, Gaur RK, Valcarcel J. Evidence for substrate-specific requirement of the splicing factor U2AF(35) and for its function after polypyrimidine tract recognition by U2AF(65) Mol. Cell. Biol. 1999;19:8263–8271. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guth S, Tange TO, Kellenberger E, Valcarcel J. Dual function for U2AF(35) in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 2001;21:7673–7681. doi: 10.1128/MCB.21.22.7673-7681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacheco TR, Coelho MB, Desterro JM, Mollet I, Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell. Biol. 2006;26:8183–8190. doi: 10.1128/MCB.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vorechovsky I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefevre SH, Chauveinc L, Stoppa-Lyonnet D, Michon J, Lumbroso L, Berthet P, Frappaz D, Dutrillaux B, Chevillard S, Malfoy B. A T to C mutation in the polypyrimidine tract of the exon 9 splicing site of the RB1 gene responsible for low penetrance hereditary retinoblastoma. J. Med. Genet. 2002;39:E21. doi: 10.1136/jmg.39.5.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 19.Wang X-H, Poh-Fitzpatrick M, Chen T, Malavade K, Carriero D, Piomelli S. Systematic screening for RNA with skipped exons - splicing mutations of the ferrochelatase gene. Biochim. Biophys. Acta. 1995;1271:358–362. doi: 10.1016/0925-4439(95)00059-d. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi I, Takahashi T, Komatsu M, Sato T, Takada G. An exonic mutation of the GH-1 gene causing familial isolated growth hormone deficiency type II. Clin. Genet. 2002;61:222–225. doi: 10.1034/j.1399-0004.2002.610310.x. [DOI] [PubMed] [Google Scholar]
- 21.Okada K, Inoue A, Okada M, Murata Y, Kakuta S, Jigami T, Kubo S, Shiraishi H, Eguchi K, Motomura M, et al. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science. 2006;312:1802–1805. doi: 10.1126/science.1127142. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda Y, Takagi A, Nakata Y, Sera Y, Hyoudou S, Hamamoto K, Nishi Y, Yamamoto A. Novel compound heterozygous mutations for lipoprotein lipase deficiency. A G-to-T transversion at the first position of exon 5 causing G154V missense mutation and a 5′ splice site mutation of intron 8. J. Lipid Res. 2001;42:1072–1081. [PubMed] [Google Scholar]
- 23.Petroulakis E, Cao Z, Clarke JT, Mahuran DJ, Lee G, Triggs-Raine B. W474C amino acid substitution affects early processing of the alpha-subunit of beta-hexosaminidase A and is associated with subacute G(M2) gangliosidosis. Hum. Mutat. 1998;11:432–442. doi: 10.1002/(SICI)1098-1004(1998)11:6<432::AID-HUMU3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 24.Kralovicova J, Vorechovsky I. Allele-specific recognition of the 3′ splice site of INS intron 1. Hum. Genet. 2010;128:383–400. doi: 10.1007/s00439-010-0860-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 27.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences–the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Sahashi K, Masuda A, Matsuura T, Shinmi J, Zhang Z, Takeshima Y, Matsuo M, Sobue G, Ohno K. In vitro and in silico analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5′ splice sites. Nucleic Acids Res. 2007;35:5995–6003. doi: 10.1093/nar/gkm647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 34.Kosaki A, Nelson J, Webster NJ. Identification of intron and exon sequences involved in alternative splicing of insulin receptor pre-mRNA. J. Biol. Chem. 1998;273:10331–10337. doi: 10.1074/jbc.273.17.10331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.