SUMMARY
We previously reported that LuxS in Streptococcus mutans is involved in stress tolerance and biofilm formation. In this study, flowcells and confocal laser scanning microscopy were used to further examine the effects of LuxS-deficiency on biofilm formation. Similar to the wild-type strain (UA159), a strain deficient in LuxS (TW26D) bound efficiently to the flowcells and formed microcolonies 4 h after inoculation. Unlike UA159, which accumulated and formed compact, evenly distributed biofilms after 28 h, TW26D showed only loose, sporadic, thin biofilms. DNA microarray analysis revealed alterations in transcription of more than 60 genes in TW26D biofilms by at least 1.5-fold (P < 0.001). Among the upregulated genes were those for sugar-specific enzymes II of the phosphotransferase (PTS) system and the atp operon, which codes for the proton-pumping F-ATPase. Of the downregulated genes, several encode proteins with putative functions in DNA repair. Mutation of selected genes caused severe defects in the ability of the mutants to tolerate low pH and oxidative stress. These results provide additional proof that LuxS-deficiency causes global alterations in the expression of genes central to biofilm formation and virulence of S. mutans, including those involved in energy metabolism, DNA repair and stress tolerance.
Keywords: biofilms, dental caries, DNA array analysis, quorum sensing, Streptococcus mutans
INTRODUCTION
Streptococcus mutans, the primary etiological agent of human dental caries, exists almost exclusively in biofilms on the tooth surface in a high-density, high-diversity ecosystem known as dental plaque (Burne, 1998). Estimates indicate that nearly 750 different bacterial species can reside in dental plaque (Jenkinson & Lamont, 2005). Although relatively stable once established, the composition and spatial organization of these multi-species communities can be profoundly influenced by the source and availability of nutrients and pH (Burne, 1998; Quivey et al., 2000). Intra-and inter-species communication is also believed to play a crucial role in development of plaque, and ultimately in development of diseases, such as dental caries and periodontitis (Kolenbrander et al., 2002; Jenkinson & Lamont, 2005; Kuramitsu et al., 2007).
One mode of microbial cell–cell communication, commonly referred to as ‘quorum sensing’, involves production, secretion and responses to small signal molecules known as autoinducers (AI). Quorum sensing is widely used by both gram-positive and gram-negative bacteria and is believed to provide quorum-sensing populations with advantages in accessing complex nutrients, coping with fluctuating environmental conditions and enhancing defenses against other microorganisms or eukaryotic host defense mechanisms (Waters & Bassler, 2005; Ng & Bassler, 2009). Recently, a large number of eubacteria have been found to produce AI-2, which are furanones produced as by-products of the catalytic reaction of the LuxS enzyme. The AI-2 molecules have been shown to serve as signals that confer information about cell numbers, metabolic activity and other properties of cells in a population (Surette & Bassler, 1998; Chen et al., 2002; Ohtani et al., 2002; McNab et al., 2003; Wen & Burne, 2004; Merritt et al., 2005; Vendeville et al., 2005; Rickard et al., 2006). Data continue to accumulate that implicate LuxS (AI-2)-mediated signaling in the global regulation of physiological functions and virulence (reviewed by Henke & Bassler, 2004).
Evidence is also accumulating that inter-species LuxS-mediated signaling plays an important role in the development and spatial organization of oral biofilms, and so can impact the pathogenic potential of dental plaque (Fong et al., 2001; McNab et al., 2003; Yoshida et al., 2005; Rickard et al., 2006; Z.T. Wen and R.A. Burne, unpublished data). LuxS appears to be essential for the development of Porphyromonas gingivalis–Streptococcus gordonii mixed-species biofilms (McNab et al., 2003). Formation of mutualistic biofilms of Actinomyces naeslundii and Streptococcus oralis was also shown to be dependent on AI-2 of S. oralis (Rickard et al., 2006). These effects on biofilm development appear to be related to changes in gene expression stimulated by AI-2. For example, AI-2 produced by Aggregatibacter actinomycetemcomitans is able to complement a P. gingivalis luxS mutant to restore normal expression of the uvrB and hasF genes in the latter organism (Fong et al., 2001).
S. mutans possesses a functional homologue of LuxS. Recent studies revealed that LuxS in S. mutans affects acid-and oxidative-stress tolerance, bacteriocin production and biofilm formation (Merritt et al., 2003, 2005; Wen & Burne, 2004; Yoshida et al., 2005), traits that are thought to be crucial to the establishment, persistence and cariogenicity of this human pathogen. Recent studies using proteomics and DNA microarray analysis have provided evidence that LuxS-deficiency in S. mutans caused global alterations in the expression of genes (Sztajer et al., 2008; Z.T. Wen and R.A. Burne, unpublished data). In this study, flowcells and confocal laser scanning microscopy (CLSM) were used to further characterize the impact of LuxS-deficiency on the development and architecture of biofilms by S. mutans. DNA microarrays were also used to identify the genes involved in LuxS-regulated biofilm formation.
MATERIALS AND METHODS
Bacterial strains, growth conditions and construction of isogenic mutants
The S. mutans UA159 and its derivatives were maintained on brain–heart infusion (BHI; Difco Laboratories, Detroit, MI). When needed, erythromycin (10 μg ml−1) and kanamycin (1 mg ml−1) were added to the growth medium. All solid media were prepared similarly, but agar (Difco Laboratories) was added at a concentration of 1.5% (weight/volume). For biofilm formation, S. mutans strains were grown in a semi-defined biofilm medium (Loo et al., 2000) with glucose (20 mM, final concentration) (BMG), sucrose (10 mM) (BMS), or glucose (18 mM) plus sucrose (2 mM) (BMGS). All strains deficient for the selected genes were generated using polymerase chain reaction (PCR)–Ligation–Mutation strategy as described previously (Lau et al., 2002; Wen & Burne, 2004), and the deficiency was further verified by PCR and DNA sequence analysis. Unless otherwise stated specifically, all cultures were grown aerobically at 37°C in a 5% CO2 atmosphere under static conditions.
Biofilm formation, confocal laser scanning microscopy and image analysis
Biofilm assays on 96-well plates (Corning Inc., Corning, NY) were carried out by following the procedures described previously (Wen & Burne, 2002). Briefly, S. mutans strains were grown on BMG and BMS at 37°C in a 5% CO2 atmosphere under static conditions. After 24 h, biofilms developed on the polystyrene surface were stained using 0.1% crystal violet and measured quantitatively using a spectrophotometer. For confocal analysis, S. mutans strains were grown in BMGS in flowcells (4 × 40 × 1 mm, width × length × depth) (Stovall Life Science, Inc., Greensboro, NC) maintained in a 37°C warm room. Biofilm development on the glass coverslips was monitored at 4 and 28 h after the initial inoculation by CLSM analysis (Wen et al., 2006). Scans were taken in eight bits at a resolution of 512 × 512 pixels, and for each sample at least five regions were randomly selected for optical dissections. Post acquisition, image stacks were further analyzed by using MCID Elite 6.0 (Imaging Research, Inc., ON, Canada) to generate xyz, xz and yz reconstructions of biofilms. Quantitative analysis was carried out using image processing software COMSTAT (Heydorn et al., 2000; Klein et al., 2009). Biovolume and area occupied by bacteria in each layer were used to determine the differences between the wild-type and the mutant biofilms. The biovolume is defined as the volume (μm3) of the biomass per μm2 of substratum area.
RNA extraction
RNA extractions were carried out by using a hot phenol method, as previously described (Wen & Burne, 2004; Wen et al., 2006). For RNA of planktonic cells, both S. mutans wild-type UA159 and the LuxS-deficient mutant, TW26D, a LuxS-deficient mutant that has the whole coding sequence deleted and replaced by an erythromycin-resistance marker (Wen & Burne, 2002), were grown in 50 ml BHI and harvested at an optical density at 600 nm (OD600) ≅ 0.45 by centrifugation at 3800 g at 4°C for 5 min. The pellets were then quickly resuspended and treated with Qiagen RNAprotect (Qiagen, Inc., Valencia, CA) using the procedures recommended by the supplier. The treated cells were collected by centrifugation and stored at −80°C until RNA was extracted. For RNA from sessile populations, BMGS was used to grow bacterial cells on sterile glass slides that were deposited in 50-ml Falcon tubes. Adherent populations on the slides were transferred daily to fresh BMGS. After 3 days the biofilms were scratched off the slides with a sterile spatula and immediately treated with the RNAprotect reagent as described above. The treated bacterial cells were homogenized in the presence of glass beads using a BeadBeater (Biospect Products, Bartlesville, OK), and RNA was extracted with hot phenol (Wen & Burne, 2004; Wen et al., 2006), precipitated with isopropanol at −80°C overnight, and washed with 75% ethanol. To remove residual DNA, RNA samples were treated with RNase-free DNase I (Ambion, Inc., Austin, TX) and total RNA was retrieved with the Qiagen RNeasy purification kit. For reference RNA, 3 l of S. mutans UA159 were grown in BHI to OD600 ≅ 0.45, and total RNA was extracted using the same procedures described above. The purified RNA was then divided into aliquots and stored at −80°C until use.
Complementary DNA synthesis, labeling and hybridization
Microarray slides were obtained from the Pathogen Functional Genomics Resource Center (PFGRC) at the J. Craig Venter Institute (http://pfgrc.jcvi.org/). The slides have 70-mer oligonucleotides representing 1960 open reading frames of S. mutans UA159 printed on the surface of the microarray in four different grids. Array analysis was carried out using the procedures recommended by the PFGRC with minor modifications (Abranches et al., 2006; Wen et al., 2006). Briefly, for complementary DNA (cDNA) synthesis, RNA (10 μg) was denatured in the presence of 3 μg random hexamers at 70°C for 10 min. After briefly chilling on ice, 9 μl 5× first-strand buffer, 3 μl 0.1 M dithiothreitol, 2 μl 12.5 mM dNTP/aa-UTP with 1.5 : 1 ratio of aa-dUTP to dTTP, and 3 μl Superscript III reverse transcriptase were added. Reverse transcription reactions were carried out at 42°C overnight. Following completion of cDNA synthesis, 15 μl of 1 M NaOH was added to hydrolyze the RNA. The aminoallyl-labeled cDNA samples were then purified by using the QIAQuick PCR Purification kit (Qiagen, Inc.) and the samples were dried in a speed vacuum. The samples were then resuspended in 5 μl 0.1 M Na2CO3, pH 9.3, mixed with 5 μl of appropriately resuspended Cy dye (Amersham Biosciences, Piscataway, NJ), and Cy dye coupling was carried out in the dark for 2 h. For all conditions, the reference samples were coupled with Cy5 and both wild-type and the LuxS-deficient mutant samples were coupled with Cy3. After 2 h, the Cy dye-coupled samples were passed through Qiaquick PCR purification columns to remove uncoupled dyes and eluted in 100 μl elution buffer. The Cy3-labeled samples were mixed with a Cy5-labeled reference and the mixtures were dried in a speed vacuum. For hybridization, the Cy dye-coupled samples were resuspended in 55 μl hybridization buffer, and hybridization of microarray slides was carried out at 42°C overnight using a MAUI Hybridization System following the manufacturer’s instructions (BioMicro Systems, Salt Lake City, UT). Following the completion of hybridization, slides were washed and scanned at 530 nm and 650 nm by using an Axon GenePix 4000B scanner (Axon Instruments, Union City, CA). At least four separate array slides were performed on RNA isolated from four separate experiments.
Microarray data analysis
Array data were normalized with the TIGR Microarray Data Analysis System (http://www.tigr.org/software) and BRB Array Tools 3.01 (developed by Dr Richard Simon and Amy Peng Lam, National Cancer Institute, Bethesda, MD, http://linus.nci.nih.gov/BRB-ArrayTools.html) as described elsewhere (Abranches et al., 2006; Wen et al., 2006). Genes that were differentially expressed at statistical significance levels of P < 0.001 and P < 0.01 were then identified using the Group Comparison program, which was designed to identify the genes that are differentially expressed among groups of specimens collected from different types of tissues or under different experimental conditions, and cutoff was made: genes with a minimum ratio of 1.5 of the transcription level in the LuxS-deficient mutant relative to the wild-type strain were considered upregulated, and those with a ratio of 1.5 or above of the transcription level in the wild-type relative to the LuxS-deficient mutant were considered downregulated.
Real-time PCR
Gene-specific primers for real-time PCR were designed using Beacon Designer 3.0 (PREMIER Biosoft International, Palo Alto, CA) and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA) (Table 1). Real-time PCR were carried out with a Bio-Rad iCycler using procedures detailed elsewhere (Ahn et al., 2006; Wen et al., 2006), and the same RNA preparations that were isolated for DNA array analysis were used for these experiments.
Table 1.
Bacterial strains, plasmids and primers used in this study
| Strains/plasmid | Relevant characteristics | References |
|---|---|---|
| S. mutansUA159 | Wild-type | (Ajdic et al., 2002) |
| S. mutans TW26D | ΔluxS, Kanr | This study |
| S. mutans TW297.1 | Δsmu.44, Kanr | This study |
| S. mutans TW238 | Δsmu.423, Kanr | This study |
| S. mutans TW294.1 | Δsmu.46, Kanr | This study |
| S. mutans TW310 | Δsmu.299, Kanr | This study |
| pDL278 | Shuttle vector, Spr | (LeBanc & Lee, 1991) |
| pDL278:luxS | Shuttle carrying luxS, Spr | This study |
| S. mutans TW26DC | TW26D/pDL278:luxS, Ermr and Spr | This study |
| pAYBG754S | Shuttle vector with a gfp reporter, Kanr | (Yoshida & Kuramitsu, 2002) |
| Primers for mutagenesis | Sequence (5′–3′) | Sequence (5′–3′) | Flanking region amplified |
|---|---|---|---|
| luxS 5F | 55-aaatctgtcattgctgatggac | 53-atacccgagctcctactgagtagtc | 5′ of luxS for mutagenesis |
| luxS 3F | 35-agttcaaagctttcaacagtaacttc | 33-tgctagacgttgcatagcttgagc | 3′ of luxS for mutagenesis |
| smu.44 5F | 55-tgttggaaatccaccatatattacgtatc | 53-tgcacttctgaattctagtacattc | 5′ of smu.44 for mutagenesis |
| smu.44 3F | 35-ttcataacaggagaattcttagatac | 33-tccttcttcactaaagtgttgtac | 3′ of smu.44 for mutagenesis |
| smu.46 5F | 55-agatagtcttgttaacaaacatc | 53-agtttgccagaattcaatagcatc | 5′ of smu.46 for mutagenesis |
| smu.46 3F | 35-tagaacaccagaattctattaatcaac | 33-acgaccaccagaaccaacaac | 3′ of smu.46 for mutagenesis |
| smu.423 5F | 55-tcactattactgatgtccaaatg | 53-ttgttcaaatgcatgcgtattcat | 5′ of smu.423 for mutagenesis |
| smu.423 3F | 35-tgtaggtgcatgcactttttgttg | 33-ttcttcactcattttgatgtc | 3′ of smu.423 for mutagenesis |
| smu.299 5F | 55-atgattcatctcgatcaagcc | 53-tacatgtgaattcgtttcagcatc | 5′ of smu.299 for mutagenesis |
| smu.299 3F | 35-tagtaaatagctggaattcaggtgc | 33-tgaaaccagatcagctgttgac | 3′ of smu.299 for mutagenesis |
|
| |||
| Primers for qPCR | Forward (5′–3′) | Reverse (5′–3′) | Application |
|
| |||
| 16S-1 | rRNA cacaccgcccgtcacacc | cagccgcaccttccgatacg | 16S rRNA fragment, 160 bp |
| smu.1128 | aagggtgggttcggacttgg | gcaggcgagcttcaaacattac | ciaH fragment, 115 bp |
| smu.1528 | cggatgcgtgttgctcttactg | ggctgataaccaacggctgatg | atpD fragment, 164 bp |
| smu.1877 | ggtccttcacttggcata | ctgccattggtaagttcatccc | mannose-PST, EIIDMan, 79 bp |
| smu.1961 | ggaagccctttgacaacagc | gcattcatcaatctggttctat | levD fragment, 94 bp |
| smu.299 | acgatggagctaatggctatgc | agcgtaagcggcaaaacttg | smu.299 fragment, 140 bp |
| smu.423 | ggtggtggtatgattagatgtgc | ccagaccagcctcctaaagc | smu.423 fragment, 137 bp |
| smu.43 | agcaaccagttatcttagg | gtataatatagaatcccgaatagg | smu.43 fragment, 200 bp |
| smu.44 | tggcagactgggaaatataagc | ggcaaactcactcattgacaac | smu.44 fragment, 148 bp |
| smu.45 | gttacaggataccacggctgaag | accttgagttgccatagttcgtag | Smu.45 fragment, 96 bp |
| smu.46 | cacatagtgatgatgtccaaattg | gcatcagacttctttaaacttg | smu.46 fragment, 199 bp |
| smu.47 | ctgccacatagacgagaa | gtttacaattccacccacaa | smu.47 fragment, 124 bp |
| smu.610 | ttgccgatgaaacgaccactac | tcagcttccttactcgcactcc | spaP fragment, 115 bp |
| smu.78 | gggacttgggaagtacgagaag | aaacaagagctgctgcaccg | fruA fragment, 148 bp |
| smu.872 | gcggcttatgttacgggtacg | aaagcagtggtcgcaacaaaag | fruI fragment, 116 bp |
| smu.985 | cgccgtttatgtcaggaggtatc | ggcattggataagcaggcatagc | blgA fragment, 178 bp |
| smu.987 | acgactgcttctcaaacgaatg | ctgcttgttcacctgttgatgg | wapA fragment, 112 bp |
Kanr, Spr and Ermr, for kanamycin, spectinomycin and erythromycin resistance, respectively. Sequences underlined are restriction sites engineered for cloning.
Acid and hydrogen peroxide killing assays
The abilities of bacterial cells to withstand acid or hydrogen peroxide challenges were determined by using procedures described elsewhere (Wen & Burne, 2004; Wen et al., 2006). Both planktonic cultures and 3-day biofilms were used for these experiments. For planktonic cultures, bacterial cells were grown on BHI until mid-exponential phase (OD600 ≅ 0.45). Bacterial biofilms were grown on glass slides as described above, and brief sonication (30 s, twice at 10% energy level, with 2 min on ice between treatments) was used to de-chain and disperse biofilms using a Branson Digital Sonifier (model S-250D, Branson Ultrasonics Co, Danbury, CT). For acid killing, bacterial cells were washed once with 0.1 M glycine buffer, pH 7.0, and then incubated in 0.1 M glycine buffer, pH 2.8 for 45 min. For hydrogen peroxide challenge, bacterial cells were prepared similarly, and then incubated in 0.1 M glycine buffer, pH 7.0 containing 0.2% (weight/volume, or 58.8 mM, final concentration) hydrogen peroxide (Fisher Scientific, Pittsburgh, PA) for 90 min. Survival rate was determined by plating serial dilutions in triplicate.
RESULTS
LuxS-deficiency impairs biofilm development and alters biofilm architecture
Previously, we examined the impact of LuxS-deficiency on biofilm formation when S. mutans strains were grown under static conditions. The results showed that the LuxS-deficient mutants were less efficient at forming biofilms on either polystyrene or hydroxylapatite surfaces than the wild-type strain, and the defects appeared to be related to lack of biofilm development rather than to an impaired ability to adhere to the substratum (Wen & Burne, 2002, 2004). In this study, we used flowcells to support biofilm growth and analyzed the capacity of the LuxS-deficient strain to form biofilms on glass coverslips with constant medium flow as a shear force. Flowcells have proven to be an effective tool to study cell accumulation and biofilm structures under conditions that more closely mimic conditions on tooth surfaces, where saliva flow and mechanical shear force could have a significant impact on bacterial adherence, cell–cell interactions and biofilm maturation (Christensen et al., 1999; Egland et al., 2004; Rickard et al., 2006; Wen et al., 2006). Moreover, flow across biofilms should affect the accumulation of autoinducer signal. To allow visualization under CLSM, green fluorescence protein (GFP) was used to tag the S. mutans strains. Both strains UA159 and TW26D were transformed with plasmid pAYBG754S, which carries a promoterless gfp gene fused with the S. mutans glucosyltansferase B gene promoter (PgtfB) (Yoshida & Kuramitsu, 2002). The PgtfB::gfp system has been shown to be stable and to be useful for biofilm studies in S. mutans (Yoshida & Kuramitsu, 2002; Wen et al., 2006). Also, we have previously shown by reporter gene fusions that LuxS-deficiency does not have a significant impact on the expression of gtfB (Wen & Burne, 2004). No major differences were observed in the expression of GFP between the wild-type and the LuxS-deficient mutant TW26D (Z.T. Wen and R.A. Burne, unpublished data). Possession of GFP as a reporter has no major impact on bacterial growth, adherence or biofilm formation.
When grown in flowcells with only glucose as the supplemental carbohydrate source, S. mutans UA159 was able to bind to the coverslips in the flowcell chambers, but the number and size of microcolonies were limited and no significant biofilm accumulation was observed (data not shown). In contrast, adherent forms of S. mutans TW26D were not identified in flowcells when glucose was the only supplemental carbohydrate source (data not shown). To promote biofilm formation by S. mutans strains in the flowcell system, sucrose was included in the growth medium at a level of 2 mM (Tsumori & Kuramitsu, 1997; Hazlett et al., 1999; Ooshima et al., 2001). The presence of sucrose allows for the formation of extracellular glucans that serve as a scaffolding for adherence and biofilm formation. When sucrose was included in the growth medium (BMGS), both UA159 and TW26D were able to bind to the glass surface and form microcolonies within 4 h after inoculation. However, compared with the more evenly distributed microcolonies formed by the wild-type strain, the microcolonies of the LuxS-deficient mutant were sparse and more variable in size (Fig. 1). As the experiments went on, the wild-type strain continued to grow and accumulate, generating an average biomass of 1.86 (±0.39) μm3 per μm2 of substratum after 28 h. The LuxS-deficient mutant, however, had only limited accumulation of biofilms with an average biomass of 1.07 (±0.19) μm3 per μm2 (P = 0.00893) after 28 h. Similar results were also obtained with biofilms grown on glass slides (see Fig. S1). Evidently, growth under static conditions in an aerobic environment with 5% CO2 facilitated S. mutans biofilm formation dramatically, but the impact of LuxS-deficiency on the ability of S. mutans to form biofilms was also consistently demonstrated. The wild-type developed an average of 35.62 μm3 of biomass per μm2 of substratum surface after 24 h and 64.32 μm3 after 72 h. In contrast, the LuxS-deficient mutant, TW26D accumulated only 7.67 μm3 of biomass (P < 0.05) per μm2 of substratum after 24 h and 29.48 μm3 (P < 0.042) after 72 h. Consistent with our previous findings, the biofilms formed by the wild-type were tight, more evenly distributed, while biofilms of the LuxS-deficient mutant were loose, sporadic and had large gaps between cell clusters.
Figure 1.
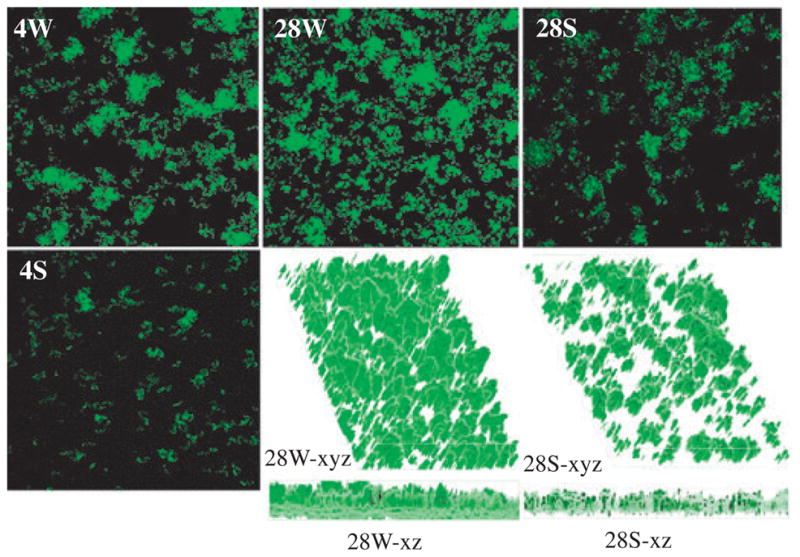
Confocal laser scanning microscopy analysis of Streptococcus mutans biofilms. The S. mutans UA159 and TW26D strains that carry a PgtfB::gfp reporter system were grown in flowcells in semi-defined biofilm medium with 18 mM glucose and 2 mM sucrose as the carbohydrate sources. Images 4S, 4W, 28S, 28W are representatives of compressed 4-h and 28-h biofilms of the LuxS-deficient mutant TW26D (S) and wild-type UA159 (W), respectively. The xyz and xz images are reconstructed three-dimensional structures. Dimensions of the regions displayed are 512 × 512 μm.
The LuxS-deficient mutant has an altered transcriptional profile
DNA microarrays were used to analyze the transcriptional profile of the LuxS-deficient mutants. When grown in BHI to mid-exponential phase, 11 genes were found to be altered by at least 1.5-fold (P < 0.001) in the LuxS-deficient mutant, with 10 downregulated and one upregulated (Table 2). At P < 0.01, 33 additional genes were identified as upregulated or downregulated as a result of LuxS-deficiency (see Table S1). Among the altered genes, several are known to be co-transcribed, including a mannose-specific PTS operon (smu.1877-79) (Abranches et al., 2003) and the inducible fructose-specific PTS operon (smu.870-72) (Wen et al., 2001). As expected, members of these operons were differentially expressed in the same manner in the LuxS-deficient mutant TW26D. To further validate the results of DNA array analysis, we also examined a subset of genes using real-time PCR with gene-specific primers (Table 1) and the same total RNA preparations used for DNA array analysis were used for these assays. As shown in Table 3, the results of real-time PCR were in strong agreement with those of DNA microarray analysis with respect to the trends of expression of the selected genes, with the exception of spaP and fruA. Contrary to the microarray data (see Table S1), but consistent with the real-time PCR results, fruA expression was previously found to be downregulated in response to LuxS-deficiency by more than 50% when analyzed by reporter gene fusions (Wen & Burne, 2004). Different from the microarray data, real-time PCR showed that the expression of spaP, which encodes the multifunctional adhesin of S. mutans, in TW26D was decreased by more than three-fold (Table 3). Downregulation of spaP in response to LuxS-deficiency was also recently reported by Merritt et al. (2003).
Table 2.
Upregulated and downregulated genes identified in planktonic cultures (P < 0.001)
| Locus | Description and putative function1 | Fold-difference2 | Parametric P value |
|---|---|---|---|
| SMU.1054 | Putative glutamine amidotransferase | 2.05 | 4.10E-04 |
| SMU.1347c | Conserved hypothetical protein, possible permease | −3.4 | 1.40E-05 |
| SMU.1363c | Putative transposase | −7.15 | 7.30E-05 |
| SMU.1735c | Acetyl-CoA carboxylase β subunit | −1.67 | 6.00E-04 |
| SMU.1739c | 3-oxoacyl-(acyl-carrier-protein) synthase | −2.03 | 7.90E-05 |
| SMU.1740c | 3-oxoacyl-acyl-carrier-protein reductase/3-ketoacyl-acyl carrier protein | −2 | 2.00E-04 |
| SMU.1741c | Malonyl-CoA (acyl-carrier-protein) transacylase | −1.94 | 6.60E-05 |
| SMU.1877 | PTS system, mannose-specific component IIAB | −3.63 | 2.00E-07 |
| SMU.1878 | PTS system, mannose-specific component IIC | −3.1 | 3.90E-05 |
| SMU.1879 | PTS system, mannose-specific component IID | −4.77 | 1.00E-07 |
| SMU.872 | Fructose-specific enzyme IIABC component | −7.67 | 2.70E-06 |
Description and putative function of the identified genes are based upon the published Streptococcus mutans database.
Defined as levels of expression in the LuxS-deficient mutant relative to those of the wild-type, with minus signs representing downregulation. Genes in italics represent those identified in both planktonic cultures and 3-day biofilm populations.
Table 3.
RealTime-PCR analysis of selected genes1
| Locus | Description and putative function | UA159 | TW26D | Fold-difference | Parametric P-value |
|---|---|---|---|---|---|
| Planktonic | |||||
| smu.1128 | Sensor/histidine kinase, CiaH | 8.17E5(1.13E4) | 2.62E5(1.93E4) | −3.12 | 0.01538 |
| smu.610 | Surface antigen SpaP | 1.39E6(1.06E5) | 3.98E5(2.89E4) | −3.49 | 0.00172 |
| smu.987 | Surface protein WapA | 4.52E6(1.54E6) | 5.91E6(2.1E6) | 1.32 | 0.02389 |
| smu.872 | Fructose-PTS, EllABCFru | 5.39E7(3.0E7) | 1.69E6(1.9E6) | −31.8 | 6.74E-07 |
| smu.78 | Exo-β-fructosidase, FruA | 4.63E5(1.79E4) | 1.98E5(7.66E3) | −2.34 | 6.45E-04 |
| smu.985 | β-glucosidase, BglA | 9.88E4(3.1E3) | 8.7E4(2.5E4) | −1.14 | 0.02897 |
| smu.364 | Putative glutamine synthetase | 3.59E7(2.03E6) | 2.53E7(3.5E6) | −1.43 | 0.04326 |
| smu.1877 | Mannose-PTS, EllDMan | 4.72E6(8.7E5) | 2.00E6(2.6E5) | −2.36 | 0.00869 |
| Biofilms | |||||
| smu.44 | DNA mismatch repair protein | 6.22E5(4.9E4) | 1.13E5(4.8E4) | −5.5 | 7.85E-04 |
| smu.45 | DNA mismatch repair protein | 2.44E5(1.3E4) | 1.32E4(1.9E3) | −18.48 | 5.60E-04 |
| smu.46 | LuxR-type regulator | 4.95E5(4.2E4) | 7.21E3(1.3E3) | −68.65 | 3.43E-04 |
| smu.47 | NHN-type endonuclease | 1.49E7(6.8E6) | 1.93E5(2.7E4) | −77.2 | 5.11E-04 |
| smu.299 | Bacteriocin precursor | 8.84E6(7.1E5) | 5.84E6(1.3E6) | −1.52 | 2.59E-04 |
| smu.423 | Mutacin-like protein | 1.60E5(1.1E5) | 8.77E5(1.1E5) | 5.48 | 2.38E-04 |
| smu.985 | β-glucosidase BglA | 2.27E5(9.2E4) | 4.2E5(4.4E4) | 1.85 | 4.51E-03 |
| smu.1528 | F-ATPase, β-subunit | 7.5E6(3.8E5) | 1.37E7(7.5E6) | 1.82 | 6.74E-03 |
| smu.1877 | Mannose-PTS, EllDMan | 3.55E6(3.38E5) | 1.02E6(4.0E5) | −3.48 | 2.67E-04 |
| smu.1961 | Fructose/mannose-PTS, LevD | 1.41E6(1.01E6) | 3.19E6(1.9E6) | 2.26 | 5.54E-03 |
| 16S-1 rRNA | 16S rRNA | 2.16E7(3.5E6) | 2.19E7(5.1E6) | 0.99 | 0.45 |
Results are expressed as copy numbers in 1 μg of total RNA. The data represent mean (± standard deviation) of no less than four independent experiments. See Table 2 for more information.
Recently, Sztajer et al. (2008) published a similar study on transcriptional profiling of the LuxS-deficient S. mutans when grown planktonically. In the study, Sztajer et al. used a slightly modified BM medium with 0.5% sucrose (BMS) as the supplemental carbohydrate sources to prepare the cultures for array analysis and the results revealed that the expression of 585 genes was affected in response to LuxS-deficiency. By cross-referencing the two transcriptional profiles, 16 of the 44 genes identified in the LuxS-deficient mutants in this study were also found to be differentially expressed in the Sztajer study (see Table S1). The level of LuxS expression in S. mutans has been shown to be growth-phase-dependent (Wen & Burne, 2004; Sztajer et al., 2008), indicative of an influence of environmental conditions on the production of LuxS. The fact that the cultures used for the microarray analyses were grown in two different media, BHI and BMS, could account for the lack of congruence of the two transcriptional profiles.
LuxS-deficiency affects a different group of genes in biofilms
To further characterize the impact of LuxS-deficiency on S. mutans biofilm formation, RNA was extracted from 3-day biofilms of S. mutans UA159 and TW26D grown on glass slides. DNA microarray analysis revealed that as compared with UA159, 27 genes were downregulated and 35 were upregulated in TW26D (P < 0.001, Tables 3–5). At a P < 0.01, 62 additional genes were found to be altered in TW26D, with 25 downregulated and 37 upregulated (see Table S2). Similar to planktonic cultures, a large number of affected genes were found to encode hypothetical proteins or conserved hypothetical proteins with no known functions. Multiple genes for sugar-specific enzyme II permeases of the PTS system and sugar metabolism were also differentially expressed in biofilm populations as a result of LuxS-deficiency, including a fructose/mannose-specific enzyme II (LevDEFGX) (Smu.1956/61c) (Zeng et al., 2006), a mannose-specific enzyme IID (Smu.1879) (Abranches et al., 2003) and members of the multi-sugar binding system (Smu.878/83) (Cote et al., 2000) (Tables 2–5). Different from planktonic cultures, however, the lev-DEFGX operon and the mannose-specific enzyme IID were shown to be upregulated and no major differences were found in expression of the inducible fructose-PTS operon (smu.870/2). Unlike in planktonic cultures, there were no significant changes in expression of either multifunctional adhesin SpaP or surface-associated protein WapA in biofilms when the wild-type and the LuxS-deficient mutant were compared.
Cross-referencing of the transcriptional profiles, sessile vs. planktonic (including data presented here and those recently published by the Sztajer group), revealed that 42 of the 124 genes identified in the LuxS-deficient mutant biofilms were also found to be altered in planktonic cultures of the deficient mutants (see Table S2) (Sztajer et al., 2008). Of the 42 consistently affected genes, four were shown to be reverted to the wild-type level of expression when synthetic AI-2 was included in the growth medium (see Table S2) (Sztajer et al., 2008). It is noted that in the recent study, Sztajer et al. carried out the transcriptional profiling using cultures grown on BMS, a condition similar to what we used [BM with glucose (18 mM) and sucrose (2 mM)] to grow biofilms. Therefore, it can be assumed that most, if not all, of these unmatched genes are likely biofilm-associated. A closer examination of these genes revealed that more than half of these unmatched genes encode hypothetical or conserved hypothetical proteins. The exact role of their products in S. mutans adherence and biofilm formation as well as their dependence on AI-2 signaling remains to be investigated. These results also suggest that LuxS-mediated signaling may affect a different set of genes in biofilms.
Mutation of genes encoding mutacin-like protein(s) had no impact on biofilm formation
Notably, several genes identified as differentially expressed in the LuxS-deficient mutant biofilms were found to encode a mutacin-like protein (smu.423), a bacteriocin precursor (smu.299) (Tables 3 and 4 and see Table S2) and a bacteriocin transport accessory protein (data not shown). Mutacins (bacteriocins produced by mutans streptococci) are known for their capabilities to kill or inhibit the growth of other closely related bacterial species in the oral flora (Chikindas et al., 1997), therefore are thought to play an important role in S. mutans establishment and persistence in the plaque and ultimately, the development of dental caries. To evaluate the role of these altered genes in S. mutans biofilm formation, isogenic mutants, TW310 and TW238 deficient of smu.299 and smu.423, respectively, were generated with a non-polar kanamycin-resistance element replacing the respective coding sequence. The deficiency of smu.299 and smu.423 was confirmed by DNA sequencing, and the non-polar effect of smu.299 and smu.423 mutation on the flanking genes was verified by real-time PCR using gene-specific primers (data not shown). The deficient mutants were grown on 96-well plates with either glucose or sucrose as the supplemental carbohydrate source. No major differences in the amount of biofilms were detected between the wild-type and the smu.299-deficient or smu.423-deficient mutants after 24 or 48 h of incubation under the conditions studied. Although the actual function of the selected genes in S. mutans biofilm formation remains unclear, especially when grown in a mixed-species consortium, alteration of expression of genes involving bacteriocin production and processing in response to LuxS-deficiency is consistent with the role of LuxS-mediated signaling in intra-and interspecies interactions.
Table 4.
Downregulated genes in 3-day biofilms (P < 0.001)1
| Locus | Description and putative function | Fold-difference | Parametric P value |
|---|---|---|---|
| SMU.09 | Conserved hypothetical protein | −2.67 | 1.6E-5 |
| SMU.12 | Conserved hypothetical protein | −2.80 | 3.9E-5 |
| SMU.528c | Conserved hypothetical protein | −1.52 | 3.1E-4 |
| SMU.730 | Conserved hypothetical protein | −1.71 | 8.2E-4 |
| SMU.1574c | Conserved hypothetical protein | −1.99 | 5.2E-5 |
| SMU.2129c | Conserved hypothetical protein | −1.89 | 2.7E-4 |
| SMU.735 | Hypothetical protein | −1.57 | 6.0E-4 |
| SMU.1895c | Hypothetical protein | −1.73 | 2.3E-4 |
| SMU.1896c | Hypothetical protein | −1.91 | 3.7E-5 |
| SMU.43 | Site-specific DNA-methyltransferase restriction-modification protein | −4.62 | 1.9E-5 |
| SMU.44 | Conserved hypothetical protein (possible DNA mismatch repair) | −6.67 | 1.9E-5 |
| SMU.46 | Hypothetic protein (putative response regulator) | −6.57 | 4.3E-4 |
| SMU.47 | Hypothetic protein (putative HNH-type endonuclease) | −4.41 | 1.0E-3 |
| SMU.127 | Putative acetoin dehydrogenase (TPP-dependent), E1 component α | −2.24 | 2.3E-6 |
| SMU.128 | Putative acetoin dehydrogenase (TPP-dependent), E1 component β | −2.79 | 2.9E-6 |
| SMU.129 | Putative dihydrolipoamide acetyltransferase | −2.99 | 3.8E-6 |
| SMU.130 | Putative dihydrolipoamide dehydrogenase | −2.79 | 4.6E-6 |
| SMU.131 | Putative lipoate-protein ligase | −3.33 | 3.0E-7 |
| SMU.132 | Putative hippurate amidohydrolase | −3.01 | 2.0E-7 |
| SMU.144c | Putative transcriptional regulator | −1.51 | 2.6E-4 |
| SMU.299c | Putative bacteriocin peptide precursor | −1.62 | 9.8E-4 |
| SMU.356 | Purine operon repressor | −2.18 | 6.4E-4 |
| SMU.732 | Possible inner membrane protein | −1.57 | 5.4E-4 |
| SMU.930c | Putative transcriptional regulator | −2.59 | 3.5E-4 |
| SMU.953c | Putative transcriptional regulator/aminotransferase | −1.55 | 2.7E-4 |
| SMU.1027 | Putative transcription regulator | −2.05 | 3.4E-4 |
| SMU.1452 | α-acetolactate synthase | −1.56 | 3.8E-4 |
See Table 2 for more information.
Smu.44 and Smu.46 are part of the LuxS-regulated stress response pathway
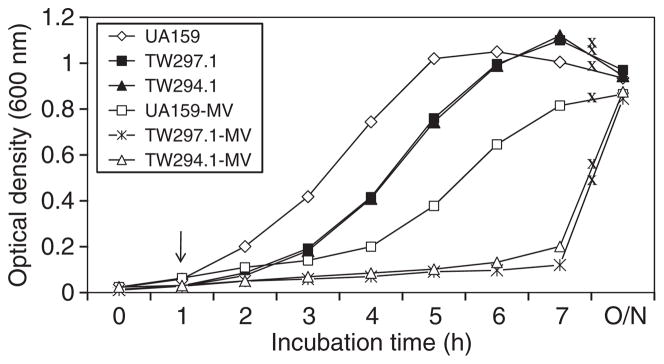
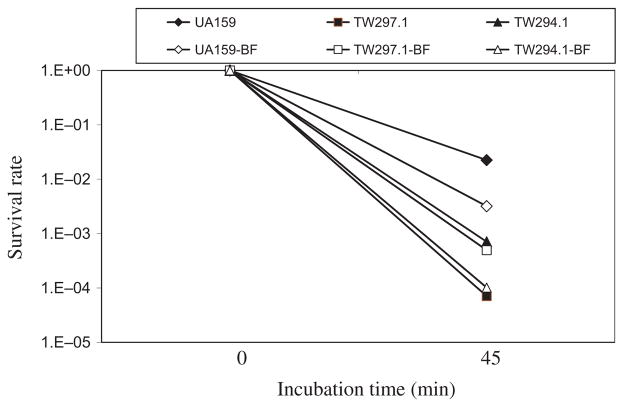
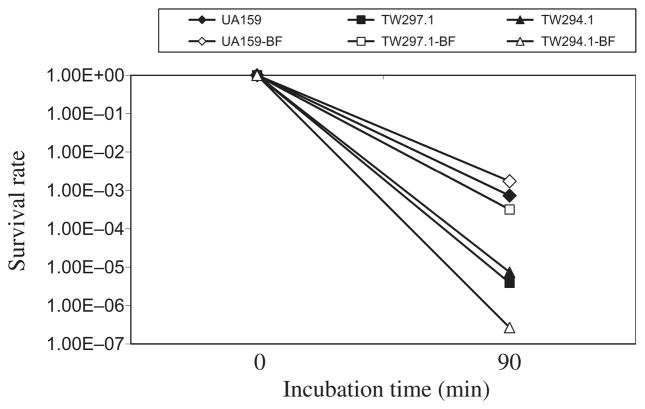
It was shown previously by Northern blot analysis that certain genes known to be involved in DNA repair were altered in response to LuxS-deficiency, including downregulation of recA, which could account perhaps in part for the weakened tolerance to acid killing (Wen & Burne, 2004). Of the genes identified as downregulated in the sessile populations, at least eight were also found to encode proteins with putative functions in nucleic acid metabolism and DNA repair, including a putative DNA mismatch repair protein (Smu.44) (Tables 3–5 and see Table S2). To evaluate the role of smu.44 in S. mutans biofilm formation, the coding sequence was deleted and replaced with a non-polar kanamycin-resistance element (Zeng et al., 2006). Deficiency of smu.44 did not show any major differences in growth rate when BHI or BMG was the growth medium (Fig. 2). When subjected to acid killing by incubating at pH 2.8 in 0.1 M glycine buffer for 45 min (Wen & Burne, 2004), the survival rate of the deficient mutant, TW297.1 was more than 2-logs lower than the wild-type UA159 (Fig. 3), suggesting that Smu.44 plays a role in tolerance of low pH. To determine whether sessile populations possessed similar features in acid tolerance, 3-day biofilms were grown on glass slides with glucose (18 mM) and sucrose (2 mM) as the carbohydrate sources and, following brief sonication to disperse the biofilms, these cells were washed and then subjected to acid killing assays as described above. Similar to the planktonic cultures, biofilms of the mutants deficient in smu.44 also showed a weakened tolerance to acid killing at pH 2.8 for 45 min (Fig. 3). To evaluate the ability of the deficient mutants to tolerate oxidative stress, methyl viologen (paraquat; Sigma, St Louis, MO) was included in the BHI broth (18 mM, final concentration) and its inclusion was found to dramatically inhibit the growth of strain TW297.1, as compared with the parental strain UA159 (Fig. 2). When challenged with hydrogen peroxide, a major endogenous source of oxidative stress in oral bacteria and a product often present in oral care products, the survival rate of the smu.44-deficient mutant was also about 2-log lower than strain UA159 (Fig. 4). Similar results were also obtained with 3-day biofilms (Fig. 4). When 96-well plates were used to grow biofilms with glucose as the supplemental carbohydrate source and measured using a spectrophotometer following crystal violet staining (Loo et al., 2000), the deficient mutant, TW297.1 was found to form as much as 62% less biofilm than the parent strain, UA159, although no significant difference was observed when sucrose was used as the carbohydrate source.
Table 5.
Upregulated genes in 3-day biofilms (P < 0.001)1
| Locus | Description and putative function | Fold-difference | Parametric P value |
|---|---|---|---|
| SMU.86 | Conserved hypothetical protein | 1.92 | 7.9E-4 |
| SMU.475 | Conserved hypothetical protein | 1.79 | 7.9E-5 |
| SMU.600c | Conserved hypothetical protein | 1.73 | 9.1E-5 |
| SMU.1404c | Conserved hypothetical protein | 1.77 | 9.3E-4 |
| SMU.68 | Hypothetical protein | 2.08 | 9.7E-4 |
| SMU.503c | Hypothetical protein | 1.67 | 3.8E-4 |
| SMU.618 | Hypothetical protein | 1.61 | 7.2E-4 |
| SMU.984 | Hypothetical protein | 1.68 | 4.7E-4 |
| SMU.1587c | Hypothetical protein | 1.96 | 1.9E-5 |
| SMU.67 | Putative acyltransferase | 1.78 | 2.6E-4 |
| SMU.84 | Putative tRNA pseudouridine synthase A | 1.68 | 1.7E-4 |
| SMU.101 | Putative sorbose PTS system, IIC component | 1.77 | 1.0E-4 |
| SMU.103 | Putative sorbose PTS system, IIA component | 1.86 | 7.4E-4 |
| SMU.148 | Putative alcohol-acetaldehyde dehydrogenase | 2.96 | 2.0E-7 |
| SMU.423 | Mutacin-like protein | 19.41 | 2.0E-3 |
| SMU.558 | Isoleucine-tRNA synthetase | 2.11 | 8.9E-5 |
| SMU.878 | Multiple sugar-binding ABC transporter, sugar-binding protein precursor | 1.94 | 3.3E-4 |
| SMU.881 | Sucrose phosphorylase, GtfA | 2.06 | 6.3E-4 |
| SMU.882 | Multiple sugar-binding ABC transporter, ATP-binding protein, MsmK | 2.04 | 5.8E-4 |
| SMU.883 | Dextran glucosidase DexB | 2.04 | 1.6E-4 |
| SMU.985 | β-glucosidase BglA | 1.73 | 6.1E-4 |
| SMU.1077 | Putative phosphoglucomutase | 1.59 | 1.2E-4 |
| SMU.1510 | Putative phenylalanyl-tRNA synthetase (β subunit) | 1.84 | 5.1E-4 |
| SMU.1511c | Putative acetyltransferase | 1.71 | 5.3E-4 |
| SMU.1531 | F1Fo membrane-bound proton-translocating ATPase, δ subunit | 1.74 | 2.8E-4 |
| SMU.1532 | F1Fo membrane-bound proton-translocating ATPase, b subunit | 1.67 | 5.2E-4 |
| SMU.1956c | PTS system, fructose/mannose-specific, LevX | 4.72 | 3.9E-4 |
| SMU.1957c | PTS system, fructose-specific IID component, LevG | 4.57 | 2.7E-4 |
| SMU.1958c | PTS system, fructose-specific IIC component, LevF | 4.15 | 2.9E-4 |
| SMU.1960c | PTS system, fructose-specific IIB component, LevE | 3.27 | 9.1E-4 |
| SMU.1879 | PTS system, mannose-specific component IID, | 2.02 | 1.9E-4 |
| SMU.1568 | Maltose/maltodextrin ABC transporter, sugar-binding protein MalX | 1.68 | 1.6E-5 |
| SMU.1569 | Maltose/maltodextrin ABC transporter, permease protein MalF | 1.62 | 7.8E-5 |
| SMU.1570 | Maltose/maltodextrin ABC transporter, MalG permease | 1.65 | 1.3E-5 |
| SMU.1586c | Putative threonyl-tRNA synthetase | 2.09 | 3.8E-4 |
See Table 2 for more information.
Figure 2.
Growth study. Streptococcus mutans strains were grown in brain–heart infusion broth with or without the addition of methyl viologen (MV, 18 mM, final concentration). Data presented here are representatives of more than four independent experiments. Arrow indicates the time when methyl viologen was added to the growth medium, and ‘x’ on growth curves illustrates disruption of measurement of the cultural optical density at 600 nm.
Figure 3.
Acid-tolerance assays. For acid tolerance, Streptococcus mutans strains were grown either in brain–heart infusion (BHI) broth until mid-exponential phase (OD600 ≅ 0.45) or in biofilm medium with glucose (18 mM) plus sucrose (2 mM) (BMGS) on glass slides for 3 days. Brief sonication was used to disperse biofilms. All cultures were washed once with 0.1 M glycine buffer, pH 7.0, and then subjected to acid killing at pH 2.8 for 45 min. The surviving cells were plated on BHI agar plates in triplicate, and expressed as survival rate over time. Data presented here are representatives of more than three independent experiments. Solid symbols represent planktonic cultures and open symbols represent biofilms (BF).
Figure 4.
Hydrogen peroxide killing assays. Streptococcus mutans cultures were prepared similarly as those for acid-tolerance assays. Following washes with 0.1 M glycine buffer, pH 7.0, the bacterial cells were incubated in glycine buffer containing 0.2% (volume/volume, 58 mM, final concentration) hydrogen peroxide for 90 min. The survival rates were determined by serial dilutions and plating on brain–heart infusion agar plates. Data presented here are representatives of more than three independent experiments. Solid symbols represent planktonic cultures and open symbols represent biofilms (BF).
The smu.46 gene is arranged in an apparent operon with smu.45 and smu.47 (see Fig. S2), which are predicted to code for another protein with some similarity to the DNA mismatch repair proteins and a putative NHN-type endonuclease (Hsia et al., 2004), respectively. Annotated as a hypothetical protein, the predicted product of the smu.46 gene shows features of the LuxR-family of regulatory proteins. To analyze the potential role of Smu.46 in S. mutans, a deficient mutant was generated using a strategy similar to the one described above. Similar to the smu.44-deficient mutants, loss of smu.46 did not cause any detectable differences in morphology and growth rate under the conditions studied. Addition of methyl viologen to the growth medium also displayed a significant inhibitory effect on growth by the deficient mutant, TW294.1, as compared with strain UA159 (Fig. 2). After incubation in buffer adjusted to pH 2.8 for 45 min or containing 0.2% hydrogen peroxide for 90 min, the survival rates were more than 1-log lower than the wild-type UA159 (Figs 3 and 4), suggesting that deficiency of smu.46 weakens the ability of the deficient mutants to tolerate acid- and oxidative stress. Similar results were also observed with sessile populations in acid and hydrogen peroxide tolerance (Figs 3 and 4) and in biofilm formation (data not shown). Apparently, the smu.46-deficient mutant possesses a phenotype that is similar to the strains lacking smu.44. One possible explanation is that smu.46-deficiency affects the expression of smu.44. To assess this possibility, real-time PCR was used to analyze the expression of smu.44 in TW294.1, but no major differences in messenger RNA in the wild-type and smu.46-deficient mutant were observed, nor was the expression of smu.45 and smu.47 altered (data not shown). These results suggest that Smu.46 likely exerts its regulatory function on other gene(s) involving acid and oxidative stress tolerance.
DISCUSSION
LuxS/AI-mediated signaling in several oral bacteria has been shown to play a critical role in biofilm formation (McNab et al., 2003; Merritt et al., 2003; Wen & Burne, 2004; Yoshida et al., 2005; Rickard et al., 2006). Consistent with our recent findings using scanning electron micrographs of biofilms formed by the LuxS-deficient mutant, TW26D on hydroxylapatite surfaces (Wen & Burne, 2004), the results on flowcells further support that LuxS-deficiency causes major defects in cell–cell interactions and biofilm maturation in S. mutans. In addition, the fact that the luxS mutant did not adhere well to the glass slides in the absence of sucrose, and formed fewer microcolonies than the wild-type, suggests that defects in initial adherence may be part of the reason the TW26D strain shows diminished biofilm forming capacity. In the absence of sucrose, SpaP (also called P1), a multi-functional adhesin that interacts with salivary agglutinin, is considered a major factor in S. mutans adherence and biofilm initiation on the tooth surface (Bowen et al., 1991; Crowley et al., 1999). Although no significant differences were identified in sessile populations between the wild-type and the LuxS-deficient mutant, real-time PCR analysis with total RNA extracted from planktonic cultures grown on BHI revealed that the expression of spaP was downregulated by more than threefold in response to LuxS-deficiency (Table 3), which is consistent with recent findings of Merritt et al. (2005). We have also used BIAcore assays to analyze the capacity of the LuxS-deficient mutant to bind to salivary agglutinin immobilized on a Pioneer F1 sensor chip (Oli et al., 2006). Preliminary data showed that, as compared with the wild-type strain, P1-mediated whole cell–ligand interaction of the LuxS-deficient mutant with salivary agglutinin was decreased by 17%, with the resonance signal for the wild-type at 938.95 ± 102.45 vs. 785.5 ± 84.5 for the LuxS-deficient mutant (P < 0.05). These results suggest that deficiency of LuxS and/or AI-2-mediated signaling affects the expression of the multifunctional adhesin, which consequently could modulate the ability of the deficient mutant to colonize the tooth surface in vivo. However, considering the fact that the flowcells were not coated with saliva, the impact of SpaP deficiency on bacterial adherence and biofilm initiation under the current conditions is probably limited.
It is evident that LuxS-deficiency in S. mutans causes substantial alterations in gene expression in both planktonic (Tables 2 and 3 and see Tables S1 and S2) (Sztajer et al., 2008) and sessile populations (Tables 3–5 and see Table S2). Based on the description and putative functions of the affected genes, LuxS-deficiency in S. mutans when grown in biofilms also affects nearly every aspect of cellular physiology and virulence, including carbohydrate and central metabolism (37), nucleic acid metabolism and DNA repair (8), amino acid and protein synthesis (6), stress responses (6), fatty acid synthesis (1), two component signal transduction (1), cell envelope (4), membrane transporters (5), transcription regulation (7), and bacteriocin and other cellular process (5). Of the genes affected by LuxS-deficiency in biofilms, at least 52 were found to code for proteins with a potential role in metabolic processes, which is consistent with findings in planktonic cultures of S. mutans (Sztajer et al., 2008) and some other bacteria (DeLisa et al., 2001; McNab et al., 2003; Winzer et al., 2003). Cross-referencing of the transcriptional profiles also provided evidence that of the genes consistently affected by LuxS-deficiency in both planktonic and sessile populations, only a few (smu.44, smu.103, smu.132 and smu.1511) were shown to respond to synthetic AI-2. However, it awaits further investigation how many of the remaining biofilm-associated genes are AI-2-dependent.
LuxS in S. mutans is shown to be differentially expressed at different growth phases (Wen & Burne, 2004; Sztajer et al., 2008), suggesting that changes in environmental conditions, such as pH, nutrient availability and cell density, and growth mode (planktonic vs. sessile) could have an impact on the transcription of LuxS, and consequently affect the AI-2 activity. Of note, the expression of dnaK and groEL was found in our earlier study to be upregulated in the LuxS-deficient mutant grown in BHI broth, especially in the late-exponential phase (OD600 ≅ 0.5) (Wen & Burne, 2004). The expression of recA was shown to be downregulated in early-exponential phase (OD600 ≅ 0.3), but was upregulated in late exponential phase. However, none of these genes were identified in our DNA microarray profiling. While efforts were made to use similar conditions to carry out the microarray analysis, differences may exist between the conditions when the cultures were harvested for total RNA extraction, contributing at least in part to the discrepancies observed. The differences in growth conditions and growth mode (planktonic vs. sessile) could also be attributed to some of the variations in the results presented here and those published by other groups (Merritt et al., 2005; Sztajer et al., 2008). In a recent study by Egland et al. (2004), it was found that expression of GFP in S. gordonii under the direction of amylase gene promoter was regulated in response to a signal produced by Veillonella atypica. However, only cells that were in close contact with V. atypica turned on the expression, suggesting that the signal concentration, as well as the proximity of bacterial species, are crucial for effective signal transduction in certain conditions. Although it remains unclear how the expression of LuxS is regulated when S. mutans is grown in the presence of a surface and in a high-density environment, the concentration and accessibility of AI-2 signal molecules in biofilms, a high cell-density community, may differ from planktonic cultures, attributing to the differences in gene transcription observed in the LuxS-deficient mutants when grown in planktonic vs. biofilm mode. However, the actual mechanism remains to be further investigated.
While the mechanisms are not yet fully understood, the intimate linkage between stress tolerance and biofilm formation is well documented (Li et al., 2002; Lemos et al., 2004, 2005; Wen & Burne, 2004; Abranches et al., 2006; Wen et al., 2006). Compared with the parental strain, one of the features of the LuxS-deficient mutants is the significantly weakened tolerance to low pH and acid killing (Wen & Burne, 2004). Of the genes affected by LuxS-deficiency, several have been shown to play a major role in acid tolerance, including downregulation of brpA and ciaH in planktonic cultures (Ahn et al., 2006; Wen et al., 2006), contributing at least in part to the increased susceptibility of the deficient mutants to acid stress. Proton extrusion by the membrane-associated F-ATPase is the primary mechanism employed by mutans streptococci to maintain intracellular pH homeostasis (Marquis, 1995). In contrast to planktonic cultures, the expression of the F-ATPase operon was found to be upregulated in TW26D when grown in biofilms (Tables 3 and 4). Interestingly, when analyzed for ATPase activity using an established method (Belli & Marquis, 1991), no significant differences were observed between the LuxS-deficient mutant biofilms and the wild-type biofilms (data not shown) (Wen & Burne, 2004). F-ATPase is a highly complex, membrane-associated protein, and as such, downregulation of genes that are known to be involved in fatty acid and phospholipids synthesis (smu.1734c/45c) (Table 2 and see Table S1) and protein secretion and membrane integrity (ffh) (Wen & Burne, 2004), which were identified in the planktonic populations but not in biofilms of the LuxS-deficient mutant, will likely have an impact on the conformation of and function in proton extrusion by the F-ATPase enzyme. Therefore, upregulation of F-ATPase expression may be a compensatory effort of the LuxS-deficient mutants to maintain pH homeostasis and combat defects in acid tolerance, especially when grown in biofilms, as a consequence of a change in either the activity or stability of the ATPase arising from alterations in the membrane or in cellular proteolysis.
Many stresses encountered by oral bacteria, such as acid and oxidative stress, can induce DNA damage (Lemos & Burne, 2008). Recent studies have shown that several enzymes of DNA repair systems, including recombination protein RecA, AP endonuclease Smx and UV repair excinuclease UvrA, are involved in the ability of S. mutans to survive challenges imposed by acid and low pH and hydrogen peroxide (Quivey et al., 1995; Hahn et al., 1999; Hanna et al., 2001; Faustoferri et al., 2005). In our previous study by Northern blotting analysis, the expression of recA, smx and nth was found to be altered as a result of LuxS-deficiency, and the most dramatic differences were observed in early-exponential phase (OD600 ≅ 0.3) of the LuxS-deficient mutants (Wen & Burne, 2004). In this study, DNA microarray analysis of the sessile populations revealed downregulation of several genes coding for proteins with putative function in nucleic acid metabolism and DNA repair, including smu.44 for a putative protein of DNA mismatch repair. Like Smx and UvrA (Hanna et al., 2001; Faustoferri et al., 2005), SMU.44-deficiency was found to affect the ability of the deficient mutants to endure acid and hydrogen peroxide killing. Although the underlying mechanism of SMU.46-mediated regulation awaits further investigation, strains deficient in SMU.46, a putative transcriptional regulator, also displayed a phenotype similar to the SMU.44-deficient mutant, TW297.1. Of note, downregulation of smu.44, smu.46 and recA in planktonic cultures were also recently reported by Sztajer et al. (2008) in transcriptional profiling of a LuxS-deficient S. mutans. In addition, inclusion of synthetic AI-2 to the growth medium was found to be able to restore the transcription of smu.44 to the level of the wild-type, suggesting its AI-2 dependence. As shown by real-time PCR, however, no major differences were identified in expression of either smu.44 or smu.46 between planktonic cultures and sessile populations of the wild-type UA159 (Z.T. Wen et al., data not shown). Neither one of these genes was identified in a recent DNA microarray analysis of genes associated with biofilm thickness by Shemesh et al. (2007). These results further suggest that DNA repair is part of the LuxS-regulated stress response circuit and that the weakened stress tolerance as a result of downregulation of enzymes like SMU.44 and SMU.46 is part of the underlying factors in biofilm formation by the LuxS-deficient mutants. Effort is currently directed to further characterization of the LuxS-regulon and their roles in biofilm formation by S. mutans.
Supplementary Material
Acknowledgments
We would like to thank NIDCR and JCVI for access to the S. mutans microarrays, Tim Vaught and the Optical Microscopy Facility at the University of Florida Brain Institute for their technical assistance with CLSM analysis, Cecilia Lopez at the University of Florida Department of Molecular Genetics and Microbiology for her expertise and support with DNA array analysis, R. Isoda for his technical assistance with the BIAcore assays. This work is supported by NIDCR grants DE13239 and DE12236 to R.A.B. and DE15501 to Z.T.W. and by the South Louisiana Institute of Infectious Disease Research.
References
- Abranches J, Chen YY, Burne RA. Characterization of Streptococcus mutans strains deficient in EIIABMan of the sugar phosphotransferase system. Appl Environ Microbiol. 2003;69:4760–4769. doi: 10.1128/AEM.69.8.4760-4769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Candella M, Wen TZ, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in the regulation of energy metabolism, biofilm development and competence in Streptococcus mutans. J Bacteriol. 2006;188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SJ, Wen ZT, Burne RA. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun. 2006;74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli WA, Marquis RE. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Schilling K, Giertsen E, et al. Role of a cell surface-associated protein in adherence and dental caries. Infect Immun. 1991;59:4604–4609. doi: 10.1128/iai.59.12.4606-4609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA. Oral streptococci…products of their environment. J Dent Res. 1998;77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Chikindas ML, Novak J, Caufield PW, Schilling K, Tagg JR. Microbially-produced peptides having potential application to the prevention of dental caries. Int J Antimicrob Agents. 1997;9:95–105. doi: 10.1016/s0924-8579(97)00040-x. [DOI] [PubMed] [Google Scholar]
- Christensen BB, Sternberg C, Andersen JB, et al. Molecular tools for study of biofilm physiology. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- Cote CK, Cvitkovitch D, Bleiweis AS, Honeyman AL. A novel β-glucoside-specific PTS locus from Streptococcus mutans that is not inhibited by glucose. Microbiology. 2000;146:1555–1563. doi: 10.1099/00221287-146-7-1555. [DOI] [PubMed] [Google Scholar]
- Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun. 1999;67:1201–1206. doi: 10.1128/iai.67.3.1201-1206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisa MP, Wu CF, Wang L, Valdes JJ, Bentley WE. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J Bacteriol. 2001;183:5239–5247. doi: 10.1128/JB.183.18.5239-5247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egland PG, Palmer RJ, Jr, Kolenbrander PE. Interspecies communication in Streptococcus gordonii–Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci USA. 2004;101:16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustoferri RC, Hahn K, Weiss K, Quivey RG., Jr Smx nuclease is the major, low-pH-inducible apurinic/apyrimidinic endonuclease in Streptococcus mutans. J Bacteriol. 2005;187:2705–2714. doi: 10.1128/JB.187.8.2705-2714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KP, Chung WO, Lamont RJ, Demuth DR. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect Immun. 2001;69:7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn K, Faustoferri RC, Quivey RG., Jr Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol Microbiol. 1999;31:1489–1498. doi: 10.1046/j.1365-2958.1999.01292.x. [DOI] [PubMed] [Google Scholar]
- Hanna MN, Ferguson RJ, Li YH, Cvitkovitch DG. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J Bacteriol. 2001;183:5964–5973. doi: 10.1128/JB.183.20.5964-5973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett KR, Mazurkiewicz JE, Banas JA. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect Immun. 1999;67:3909–3914. doi: 10.1128/iai.67.8.3909-3914.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol. 2004;186:3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Hsia KC, Chak KF, Liang PH, Cheng YS, Ku WY, Yuan HS. DNA binding and degradation by the HNH protein ColE7. Structure. 2004;12:205–214. doi: 10.1016/j.str.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Klein MI, Duarte S, Xiao J, Mitra S, Foster TH, Koo H. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Appl Environ Microbiol. 2009;75:837–841. doi: 10.1128/AEM.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–201. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- LeBanc D, Lee L. Replication function of pVA380-1. In: Dunny G, Cleary PP, McKay LL, editors. Genetics and Molecular Biology of Streptococci, Lactococci, and Enterococci. Washington, DC: American Society for Microbiology; 1991. pp. 235–239. [Google Scholar]
- Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology. 2008;154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Brown TA, Jr, Burne RA. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect Immun. 2004;72:1431–1440. doi: 10.1128/IAI.72.3.1431-1440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;7:95–107. [PubMed] [Google Scholar]
- Li YH, Lau PC, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol. 2002;184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo CY, Corliss DA, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis RE. Oxygen metabolism, oxidative stress and acid–base physiology of dental plaque biofilms. J Ind Microbiol. 1995;15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Qi F, Goodman SD, Anderson MH, Shi W. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect Immun. 2003;71:1972–1979. doi: 10.1128/IAI.71.4.1972-1979.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Kreth J, Shi W, Qi F. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol Microbiol. 2005;57:960–969. doi: 10.1111/j.1365-2958.2005.04733.x. [DOI] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Hayashi H, Shimizu T. The luxS gene is involved in cell–cell signaling for toxin production in Clostridium perfringens. Mol Microbiol. 2002;44:171–179. doi: 10.1046/j.1365-2958.2002.02863.x. [DOI] [PubMed] [Google Scholar]
- Oli MW, McArthur WP, Brady LJ. A whole cell BIAcore assay to evaluate P1-mediated adherence of Streptococcus mutans to human salivary agglutinin and inhibition by specific antibodies. J Microbiol Methods. 2006;65:503–511. doi: 10.1016/j.mimet.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Ooshima T, Matsumura M, Hoshino T, Kawabata S, Sobue S, Fujiwara T. Contributions of three glycosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J Dent Res. 2001;80:1672–1677. doi: 10.1177/00220345010800071401. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Faustoferri RC, Clancy KA, Marquis RE. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett. 1995;126:257–261. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Kuhner WL, Hahn K. Adaptation of oral streptococci to low pH. Adv Microb Physiol. 2000;42:239–274. doi: 10.1016/s0065-2911(00)42004-7. [DOI] [PubMed] [Google Scholar]
- Rickard AH, Palmer RJ, Jr, Blehert DS, et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- Shemesh M, Tam A, Steinberg D. Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology. 2007;153:1307–1317. doi: 10.1099/mic.0.2006/002030-0. [DOI] [PubMed] [Google Scholar]
- Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztajer H, Lemme A, Vilchez R, et al. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J Bacteriol. 2008;190:401–415. doi: 10.1128/JB.01086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumori H, Kuramitsu H. The role of the Streptococcus mutans glucosyltransferases in the sucrose-dependent attachment to smooth surfaces: essential role of the GtfC enzyme. Oral Microbiol Immunol. 1997;12:274–280. doi: 10.1111/j.1399-302x.1997.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Burne RA. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microbiol. 2002;68:1196–1203. doi: 10.1128/AEM.68.3.1196-1203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen ZT, Burne RA. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J Bacteriol. 2004;186:2682–2691. doi: 10.1128/JB.186.9.2682-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen ZT, Browngardt C, Burne RA. Characterization of two operons that encode components of fructose-specific enzyme II of the sugar:phosphotransferase system of Streptococcus mutans. FEMS Microbiol Lett. 2001;205:337–342. doi: 10.1111/j.1574-6968.2001.tb10969.x. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Baker HV, Burne RA. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J Bacteriol. 2006;188:2983–2992. doi: 10.1128/JB.188.8.2983-2992.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv Appl Microbiol. 2003;53:291–396. doi: 10.1016/s0065-2164(03)53009-x. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Kuramitsu HK. Streptococcus mutans biofilm formation: utilization of a gtfB promotergreen fluorescent protein (PgtfB::gfp) construct to monitor development. Microbiology. 2002;148:3385–3394. doi: 10.1099/00221287-148-11-3385. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Ansai T, Takehara T, Kuramitsu HK. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl Environ Microbiol. 2005;71:2372–2380. doi: 10.1128/AEM.71.5.2372-2380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Wen ZT, Burne RA. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol Microbiol. 2006;62:187–200. doi: 10.1111/j.1365-2958.2006.05359.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.