SUMMARY
Specific recognition of DNA by transcription factors is essential for precise gene regulation. In Wingless (Wg) signaling in Drosophila, target gene regulation is controlled by TCF, which binds to specific DNA sequences through a HMG domain [1]. However, there is considerable degeneracy in what constitutes a TCF binding site [2–5], raising the possibility that it is not sufficient for target location. Some isoforms of human TCF contain a domain termed the C-clamp that mediates binding to an extended sequence in vitro [6]. However, the significance of this extended sequence for the function of Wnt response elements (WREs) is unclear. In this report, we identified a new cis-regulatory element named the TCF Helper site (Helper site) that is essential for activation of several WREs. This motif greatly augments the ability of TCF binding sites to respond to Wg signaling. Drosophila TCF contains a C-clamp that enhances in vitro binding to TCF-Helper site pairs and is required for transcriptional activation of WREs containing Helper sites. A genome-wide search for clusters of TCF and Helper sites identified two new WREs. Our data suggest that DNA recognition by fly TCF occurs through a bipartite mechanism involving both the HMG domain and C-clamp, which enables TCF to locate and activate WREs in the nucleus.
Keywords: Wingless, TCF, C-clamp, Armadillo, Wnt
Results and Discussion
Helper sites are crucial for Wg signaling activation of WREs in the nkd locus
The Wg target gene naked cuticle (nkd) contains numerous clusters of potential TCF binding sites. However, chromatin immunoprecipitation studies revealed that TCF preferentially bind to two regions: in the first intron of nkd approximately 5 kb downstream of the transcription start site (TSS) (nkd-IntE) [7] and a region 10 kb upstream of the TSS (nkd-UpE) [5]. These regions contain functional WREs but several other TCF site clusters in the nkd locus not bound by TCF do not act as WREs [7] (data not shown). These data argue that TCF binding sites are not sufficient for TCF binding and WRE function.
To identify additional sequence information necessary for WRE function, systematic mutagenesis of the entire nkd-IntE with non-overlapping 10 bp substitutions was performed in Kc cells. Besides the three TCF sites that were already known [5], two other motifs adjacent to the TCF sites were identified that are required for full activation of the WRE (data not shown). Further mutagenesis revealed that both motifs (hereafter termed Helper sites) are 7 nucleotides (GCCGCCA). Simultaneous mutation of both motifs resulted in a 100-fold decrease in responsiveness of nkd-IntE when the Wg pathway was activated by expression of a stabilized form of Armadillo (Arm*; Figure S1A).
When the nkd locus was searched for additional clusters of TCF and Helper sites using Target Explorer (http://luna.bioc.columbia.edu/Target_Explorer/) [8], a second cluster was identified 10 kb upstream of the TSS in a WRE known as nkd-UpE2. As seen for nkd-IntE, mutation of these two Helper sites in nkd-UpE2 resulted in a drastic reduction in activation by Arm* in Kc cells (Figure S1A).
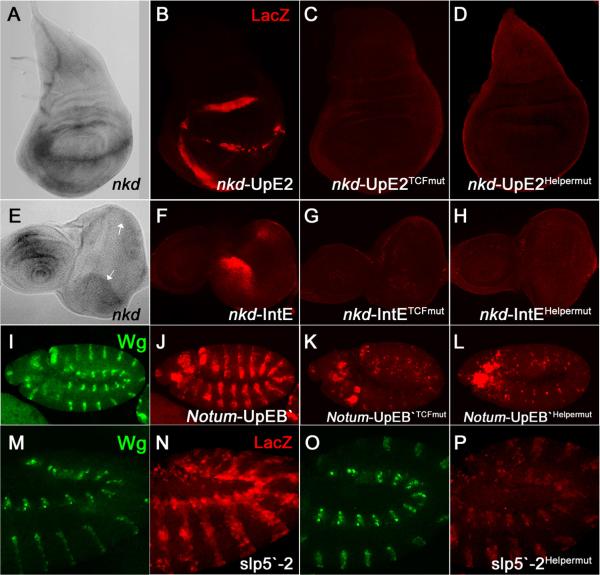
In transgenic fly reporter assays, both the nkd-IntE and nkd-UpE2 WREs are active in similar patterns as endogenous nkd [5] (Figures 1A, 1B, 1E, 1F). These reporters are activated by Wg signaling and mutation of their TCF sites abolishes activity [5] (Figures 1C and 1G). Strikingly, mutation of the Helper sites also completely abolished reporter gene expression (Figures 1D and 1H). Similar to the data shown for nkd-UpE2 in the wing imaginal disc (Figures 1A–D) and for nkd-IntE in the eye disc (Figures 1E–H), Helper sites were also required for the activity of these WREs in other imaginal discs as well (data not shown). These results demonstrate that Helper sites are indispensable for Wg responsiveness of nkd-WREs in a broad range of tissues.
Figure 1.
Helper sites are crucial for activation of several WREs.
(A, E) Endogenous nkd transcripts in the wing (A) and eye-antennal (E) imaginal discs of third instar larvae detected by in situ hybridization. The white arrows (E) indicate the dorsal and ventral regions of the presumptive eye where nkd is expressed. Confocal images of wing imaginal discs from P[nkd-UpE2-lacZ] flies (B–D), eye-antennal discs from P[nkd-IntE-lacZ] flies (F–H) and stage 11 embryos from P[Notum-UpEB`-lacZ] flies (I–L) or P[slp5`-2-lacZ] flies (M–P) immunostained for Wg and LacZ. The wild type reporters are active in patterns similar to the endogenous genes and are activated by Wg signaling (data not shown). Mutation of the TCF sites (C, G, K) or Helper sites (D, H, L, P) in these WREs significantly reduced the lacZ expression. Three to five independent transgenic lines for each construct were analyzed with similar results.
Functional Helper sites are also present in WREs from other Wg targets
A Target explorer search of the Wg target Notum (also called wingful, or wf) revealed TCF-Helper site clusters in a previously identified 2.2 kb WRE called wf-luc [9] upstream of the Notum TSS. Deletion analysis of wf-luc revealed two separable WREs (Figure S1B). Site-directed alterations of the predicted TCF or Helper sites in one of these WREs (Notum-UpEB`) greatly compromised Wg pathway responsiveness in Kc cells (Figure S1B).
In transgenic flies, the Notum-UpEB` WRE directed LacZ expression in a pattern consistent with activation by Wg signaling in a broad range of tissues (Figure I and J; data not shown). These expression patterns were largely TCF site- and Helper site-dependent (Figures 1K and 1L; data not shown).
Like nkd, Notum encodes a Wg feedback antagonist whose expression is activated by Wg signaling in many fly tissues throughout development [10–12]. However, most Wg target genes are regulated in a cell-specific manner at particular developmental stages [13, 14]. This raises the possibility that Helper sites are only found in broadly activated WREs. To address this issue, a WRE from the sloppy paired (slp) locus (slp5`-2) was examined. This WRE is directly activated by Wg signaling in the embryonic ectoderm and mesoderm (Figure 1N) [3]. slp5`-2 has four predicted Helper sites (Figure S1C). Mutation of the third Helper site in slp5`-2 caused a large reduction in reporter gene expression in Kc cells (Figure S1C) and in the epidermis and mesoderm of embryos (Figure 1P). These results extend the importance of Helper sites to tissue-specific targets of Wg signaling.
Helper sites may be a common strategy for WRE function in Drosophila. The consensus of the Helper site motif is GCCGCCR (Figure S1D). If one allows for one substitution from this consensus, then Helper sites are present near functional TCF sites in WREs from the Ultrabithorax (Ubx) and even-skipped (eve) loci [2, 4] as well as nkd-UpE1 [5]. Functional analysis of these putative Helper sites should help to refine the criteria of what constitutes a Helper site.
Helper sites augment TCF site-mediated transcriptional activation in response to Wg signaling
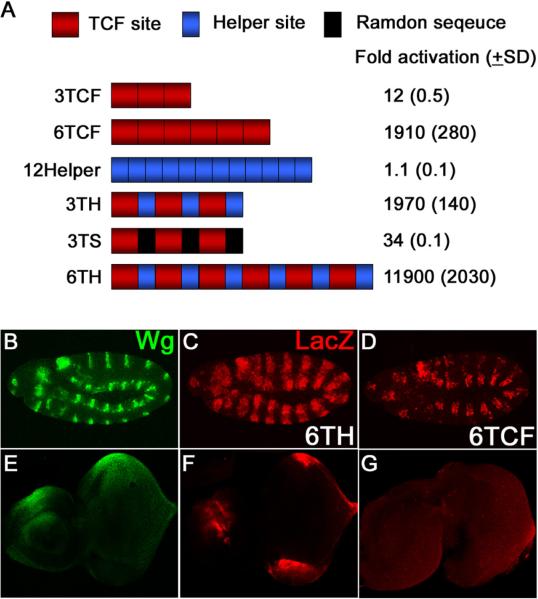
To learn more about the mechanism of how Helper sites function, a series of synthetic reporters were constructed (Figure 2A). Consistent with previous reports, multimerized TCF sites (3TCF or 6TCF) were substantially activated by Arm* (Figure 2A) [1, 15]. In contrast, Helper sites alone (three, six or twelve copies) had no response to Arm* (Figure 2A and data not shown). However, Helper sites potently augmented the ability of TCF sites to respond to Wg signaling. TCF/Helper site pairs in three (3TH) or six (6TH) copies showed a much greater activation by Arm* in Kc cells than constructs with the same number of TCF sites (Figure 2A). Replacing the Helper sites in 3TH with random sequence (3TS) reduced the fold activation 50-fold, arguing that the Helper site effect was not due to the spacing of the TCF sites.
Figure 2.
Helper sites have no activity by themselves but augment TCF site-mediated transcriptional activation by Wg signaling.
(A) TCF sites or Helper sites were multimerized and cloned upstream of a hsp70 core promoter/luciferase reporter and tested for activation by Arm*. TCF sites (3TCF and 6TCF) are responsive to Arm* but Helper sites alone (12Helper) are not. However, Helper sites adjacent to TCF sites greatly enhance activation by Arm* (3TCF vs. 3TH, 6TCF vs. 6TH). This effect is not observed when the Helper sites in 3TH are replaced by random sequence (3TS).
(B–G) Confocal images immunostained for Wg (green) and lacZ (red) from early stage 11 embryos (B–D) and late third instar eye-antennal imaginal discs (E–G). Embryos and discs containing a six TCF-Helper site tandem (6TH) cloned upstream of a hsp70 core promoter/lacZ reporter were double-stained for Wg and lacZ (B, C, E, F) while only the lacZ pattern is shown for the construct with six TCF sites (D, G). As with cultured cells, the presence of Helper sites greatly increased the ability of the reporters to be expressed in a pattern consistent with activation by Wg signaling.
In transgenic flies, a reporter construct containing six TCF sites (6TCF) had some expression in the embryonic epidermis (Figure 2D) and no detectable expression in eye-antennal imaginal discs (Figures 2G). A reporter with six copies of the Helper site did not exhibit significant expression in any tissue examined (data not shown). However, when Helper sites flank the TCF sites (6TH), a dramatic enhancement of reporter gene expression was observed in embryos and eye-antennal discs (Figures 2C and 2F). Similar results were observed in other imaginal discs (Figure S2). The results suggest that the presence of Helper sites markedly enhances the ability of TCF sites to respond to Wg signaling in many tissues.
Helper sites physically and functionally interact with the C-clamp domain of TCF
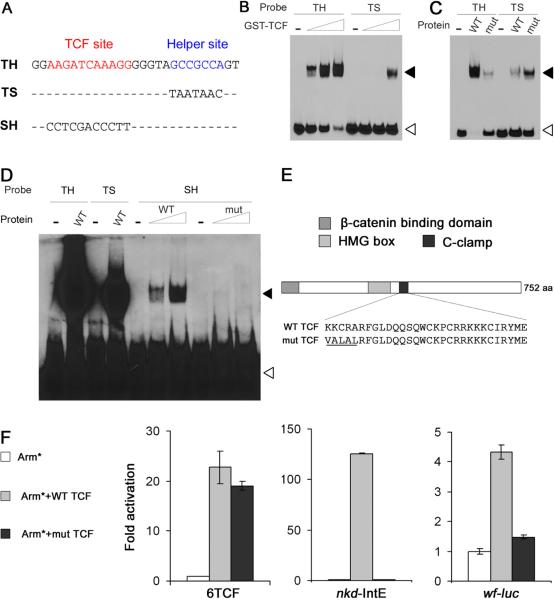
Drosophila TCF contains a Cysteine-rich domain called the C-clamp downstream of the HMG domain (Figure 3E). The presence of the C-clamp in human isoforms TCF-1E and TCF-4E enable them to bind in vitro to the classic TCF site plus an extended sequence (RCCG) [6]. This extended sequence is somewhat similar to the Helper site, raising the possibility that it interacts with the C-clamp. A fragment of TCF containing the HMG and C-clamp domains fused to glutathione-S-transferase (GST-TCF) was found to bind to an oligonucleotide containing a classic TCF site and a Helper site (TH in Figure 3A) in a electrophoretic mobility shift assay (EMSA; Figure 3B). An oligonucleotide containing only a TCF site was bound much less efficiently by GST-TCF (Figure 3B). No binding was observed with GST alone (data not shown). Competition assays showed that both the TCF site and Helper site are required for the specific binding of TCF to the TH probe (Figure S3A). A mutation in the C-clamp of GST-TCF (Figure 3E) weakened the affinity of the protein for the TH probe (Figure 3C), indicating that the enhanced binding of GST-TCF to the TH probe is C-clamp dependent.
Figure 3.
Helper sites physically and functionally interact with the C-clamp domain of TCF.
(A) The sequences of oligonucleotide probes used for the EMSA assays.
(B) Increasing concentrations of GST-TCF (0.1, 0.3 and 1 μM) were incubated with DNA probes (4nM). GST-TCF binds to the TH probe better than TS. White arrowheads indicate free probe and black arrowheads indicate the protein-DNA complexes.
(C) Binding of TH and TS oligonucleotides (4nM) to WT GST-TCF or C-clamp mutant proteins (1μM; mutated residues indicated in Figure 3E). The C-clamp mutant displays weaker affinity for the TH probe.
(D) TH and TS probes (20nM) were incubated with wild type GST-TCF (1μM). SH probes (20nM) were incubated with WT or C-clamp mutant GST-TCF (1 and 5μM). WT protein binds to SH with low affinity and this binding is C-clamp dependent.
(E) Cartoon of Drosophila TCF indicating the position of the HMG and C-clamp domains. The underlined amino acid sequence of the C-clamp indicates the five residues altered in the mutant.
(F) TCF rescue assays in Kc cells where endogenous TCF was depleted by treating dsRNA against the TCF 3`UTR. Each WRE reporter was co-transfected with Arm* and V5-tagged TCF expression plasmids. The activity of a 6TCF/luciferase reporter was efficiently rescued both by wild type and C-clamp mutant TCF. However, the C-clamp mutant didn't rescue the activity of the nkd-IntE/luciferase and wf-luc reporters. Luciferase activity in the presence of Arm* but without TCF expression was normalized to 1.0 for each reporter.
Can TCF bind to the Helper site independently of an adjacent TCF site? Under conditions containing high concentrations of both protein and probe, a weak interaction between GST-TCF and an Helper site only probe (SH) was observed (Figure 3D). Mutations in the C-clamp of GST-TCF abolished the interaction, implying that the C-clamp interacts directly with the Helper site. Consistent with this weak binding, multiple copies of the Helper site cannot mediate Wg activation of transcription (Figure 2A). These results support a model where high affinity DNA binding of TCF occurs through simultaneous HMG domain-TCF site and C-clamp-Helper site interactions.
To determine if the C-clamp is required for the activation of WREs containing functional Helper sites, Kc cells were depleted of endogenous TCF by RNAi and subsequently transfected with WRE reporter constructs and expression vectors for Arm* and either wild-type TCF or TCF containing the C-clamp mutation (the exogenous TCFs are not targeted by the TCF dsRNA used for RNAi). For all reporters examined, expression of wild-type TCF rescued the defect in Arm* responsiveness caused by TCF RNAi (Figure 3F). In stark contrast, the TCF C-clamp mutant rescued the activation of the 6TCF reporter (containing only TCF sites) but not that of the nkd-IntE and wf-luc reporters (Figure 3F). Western blots show that wild-type and C clamp mutant TCFs were expressed at similar levels (Figure S3B). These results demonstrate that the C-clamp of TCF is necessary for the activation of Helper site-dependent WREs by Wg signaling. These results are further supported by the previous finding that a missense mutation in the fifth position of the C-clamp (A374V; see Figure 3E) causes Wg signaling defects in fly embryos [1].
New WREs identified by genome-wide search for clusters containing both TCF sites and Helper sites
To identify new WREs through computational methods, Fly enhancer (http://genomeenhancer.org/fly) [16] was used to search the entire fly genome for clusters of TCF sites and Helper sites. To reduce the number of hits, stringent parameters were employed: the presence of two TCF sites (SSTTTGW) and two Helper sites (GCCGCC) within 100 bases. 97 clusters were identified. These positives were further prioritized by organization (alternating TCF and Helper sites), proximity of the TCF sites to Helper sites and phylogenetic conservation. After these secondary screens, seven clusters were selected for reporter assay analysis in Kc cells (Figure S4). Two positives out of seven clearly activated the expression of a reporter gene in response to Arm* (Figures S4B).
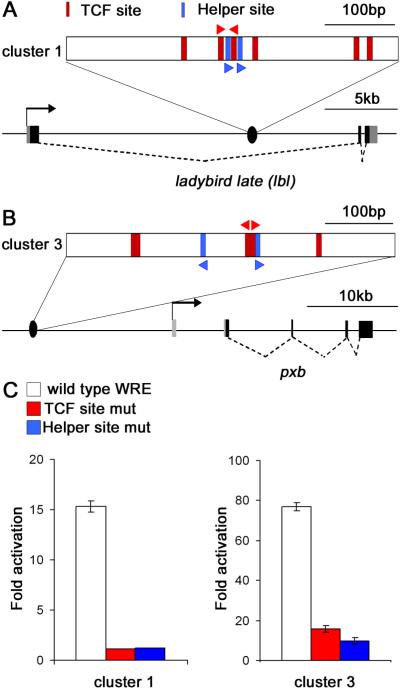
One positive (cluster 1) is located within the first intron of ladybird late (lbl) (Figure 4A), a gene known to be regulated by Wg signaling in muscle progenitors of Drosophila embryos [17]. Cluster 3 is found 15.2 kb upstream of pxb (Figure 4B), a gene that is expressed in an embryonic pattern consistent with activation by Wg signaling [18, 19]. Mutations in the TCF or Helper sites of both clusters resulted in a large reduction in Wg responsiveness (Figure 4C). These results further highlight the functional importance of Helper sties in WREs and illustrate how they can be used to facilitate identification of WREs in silico.
Figure 4.
A genome-wide search for TCF-Helper site clusters identified new WREs.
(A, B) Schematic diagram of the lbl (A) and pxb loci (B) showing the locations of the identified clusters. The gene structure for pxb was drawn based on the pxb-PB isoform. Red and blue triangles indicate the location and orientation of mutated TCF sites and the Helper sites, respectively (see Figure S4A).
(C) The fragments containing cluster 1 (478 bp) or cluster 3 (484 bp) were cloned upstream of a hsp70 core promoter/luciferase reporter. In Kc cells, both clusters activate luciferase expression when co-transfected with an Arm* expression plasmid. Mutation in the Helper sites or adjacent TCF sites significantly reduces the Arm*responsiveness of both reporters.
Bipartite recognition of DNA by TCF
Target location by transcription factors is critical for precise gene regulation. In order to find their appropriate targets among the vast excess of genomic chromatin, these proteins employ various strategies to enhance DNA recognition. For example, the p53 tumor suppressor binds DNA as a homotetramer, with each subunit contacting a quarter site. Thus a typical p53 binding site contains 20 bases of specific DNA information to mediate the recognition [20]. Many transcriptional factors enhance their specificity for DNA through cooperative binding with other transcription factors, e.g., HOX proteins and Extradenticle/Pbx [21, 22]. Drosophila TCF solves this problem by bipartite recognition of DNA. The binding of both the HMG domain and C-clamp to their respective DNA sites would effectively double the size of the TCF recognition site, enabling TCF to achieve high affinity and high specificity binding to WREs. In this way, fly TCF is similar to POU proteins, which contain two distinct DNA binding domains, a POU-specific domain and a homeodomain [23].
In all these above mentioned cases, a single DNA binding domain is not sufficient for recognition of transcriptional targets. This raises the question of how vertebrate TCFs solve the specificity problem since only a few TCF isoforms (TCF-1E and TCF-4E) contain a C-clamp [6]. For these isoforms, the presence of the C-clamp is necessary to activate an alternative promoter of the Lef-1 gene, and for dominant negative versions of TCF to suppress growth of a colorectal cancer cell line [6]. For all the TCF family members and isoforms that lack a C-clamp, we suggest the likelihood that additional mechanisms exist to enhance HMG domain binding to the classic TCF site to achieve high specificity DNA recognition.
Our knowledge of the Helper site consensus allowed us to locate WREs within target genes already known to be activated by Wg such as the nkd-UpE2 and Notum-UpEB` WREs (Figures 1 and S1). In addition, we were able to identify two new WREs in silico. However, several questions on the relationship of TCF sites and Helper sites remain unsolved. For example, the spacing and orientation of Helper sites in relation to nearby functional TCF sites vary significantly among the known TCF-Helper site pairs (Figure S4A). Further studies are needed to fully elucidate the parameters by which this motif enhances TCF binding and transcriptional activation in response to Wg signaling.
Supplementary Material
ACKNOWLEGMENTS
The authors are indebted to M. Waterman for sharing data concerning the C-clamp in vertebrate TCFs before publication. We also thank H. Li, Y. Ni, E. Anderson and C. Bhambhani for discussions and careful reading of the manuscript. This work was supported by an AHA predoctoral fellowship to M. V. C. and NIH grant GM08894 to K. M. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA Supplemental data include Experimental Procedures, four figures and one table, and can be found online at hhtp://www.current-biology.com/cgi/content/full/.
REFERENCES
- 1.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 2.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu SC, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 3.Lee HH, Frasch M. Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development. 2000;127:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- 4.Knirr S, Frasch M. Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol. 2001;238:13–26. doi: 10.1006/dbio.2001.0397. [DOI] [PubMed] [Google Scholar]
- 5.Chang JL, Chang MV, Barolo S, Cadigan KM. Regulation of the feedback antagonist naked cuticle by Wingless signaling. Dev Biol. 2008;321:446–454. doi: 10.1016/j.ydbio.2008.05.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker DS, Ni YY, Chang JL, Li J, Cadigan KM. Wingless signaling induces widespread chromatin remodeling of target loci. Mol Cell Biol. 2008;28:1815–1828. doi: 10.1128/MCB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sosinsky A, Bonin CP, Mann RS, Honig B. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 2003;31:3589–3592. doi: 10.1093/nar/gkg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmans R, Stadeli R, Basler K. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr Biol. 2005;15:1207–1211. doi: 10.1016/j.cub.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 10.Giraldez AJ, Copley RR, Cohen SM. HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev Cell. 2002;2:667–676. doi: 10.1016/s1534-5807(02)00180-6. [DOI] [PubMed] [Google Scholar]
- 11.Gerlitz O, Basler K. Wingful, an extracellular feedback inhibitor of Wingless. Genes Dev. 2002;16:1055–1059. doi: 10.1101/gad.991802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng W, Wharton KA, Jr., Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 13.Sanson B. Generating patterns from fields of cells. Examples from Drosophila segmentation. EMBO Rep. 2001;2:1083–1088. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadigan KM. Regulating morphogen gradients in the Drosophila wing. Semin Cell Dev Biol. 2002;13:83–90. doi: 10.1016/s1084-9521(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 15.Barolo S. Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene. 2006;25:7505–7511. doi: 10.1038/sj.onc.1210057. [DOI] [PubMed] [Google Scholar]
- 16.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagla T, Bellard F, Lutz Y, Dretzen G, Bellard M, Jagla K. ladybird determines cell fate decisions during diversification of Drosophila somatic muscles. Development. 1998;125:3699–3708. doi: 10.1242/dev.125.18.3699. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inaki M, Kojima T, Ueda R, Saigo K. Requirements of high levels of Hedgehog signaling activity for medial-region cell fate determination in Drosophila legs: identification of pxb, a putative Hedgehog signaling attenuator gene repressed along the anterior-posterior compartment boundary. Mech Dev. 2002;116:3–18. doi: 10.1016/s0925-4773(02)00119-3. [DOI] [PubMed] [Google Scholar]
- 20.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 21.Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 22.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Herr W, Cleary MA. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.