Abstract
Summary:CompleteMOTIFs (cMOTIFs) is an integrated web tool developed to facilitate systematic discovery of overrepresented transcription factor binding motifs from high-throughput chromatin immunoprecipitation experiments. Comprehensive annotations and Boolean logic operations on multiple peak locations enable users to focus on genomic regions of interest for de novo motif discovery using tools such as MEME, Weeder and ChIPMunk. The pipeline incorporates a scanning tool for known motifs from TRANSFAC and JASPAR databases, and performs an enrichment test using local or precalculated background models that significantly improve the motif scanning result. Furthermore, using the cMOTIFs pipeline, we demonstrated that multiple transcription factors could cooperatively bind to the upstream of important stem cell differentiation regulators.
Availability: http://cmotifs.tchlab.org
Contact: sekwon.kong@childrens.harvard.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Gene expression is a complex process that is coordinately regulated by interactions of multiple transcription factors (TFs) and other proteins that form promoter and enhancer complexes. The recent development of next-generation sequencing technologies has made it possible to determine in vivo and in vitro bindings of diverse TFs on a genomic scale (i.e. ChIP-Seq). One immediate question is whether the peak regions are functionally associated with the transcriptional regulation of target genes. An indication of such functional associations is the presence of loci occupied by multiple TFs or highly conserved sequences across species (Wasserman and Sandelin, 2004). Hence, an important step towards understanding functional binding events is to study the presence of DNA sequence motifs in the context of nearby TFs, genomic features and epigenetic modifications.
There are several challenges in finding motifs in peak regions identified from TF binding experiments. In the case where only a subset of ChIP-Seq peaks has common motifs, de novo discovery tools might not be optimal. Scanning for known motifs is faster and capable of detecting motifs that appear less frequently; however, the relatively short (4–25 bp) nature of protein-binding motifs can produce non-significant results. Many tools such as GimmeMOTIFs (van Heeringen and Veenstra, 2010), Tmod (Sun et al., 2010) and MoAn (Valen et al., 2009) have been deployed to identify sequence motifs, while RSAT (Thomas-Chollier et al., 2008) and Galaxy (Goecks et al., 2010) provide more comprehensive means to analyse regulatory sequences in general. Each method has its own strength in identifying potential transcription factor binding site (TFBS); however, it is crucial to combine results from complementary tools (Tompa et al., 2005) and provide an intuitive interface that streamlines peak annotation, filtering, motif discovery and summary.
CompleteMOTIFs (cMOTIFs) was developed to provide biologists an easy to use yet comprehensive web tool for ChIP-seq data analysis. First, peak regions are annotated with comprehensive genomic information such as known genes, conservation scores and repeating sequence elements. Second, Boolean logic operations such as intersection and union can combine multiple datasets from different TF bindings or histone modification states. Using the annotation and Boolean operations, users can filter and combine peak lists from one or multiple experiments, facilitating the study of cooperative TF binding events (Farnham, 2009). Third, combining the top 10 ranked motifs from three de novo methods and known motif scanning using background models helps to identify novel or known TFBS of interest as well as possible co-factor binding sites.
2 IMPLEMENTATION
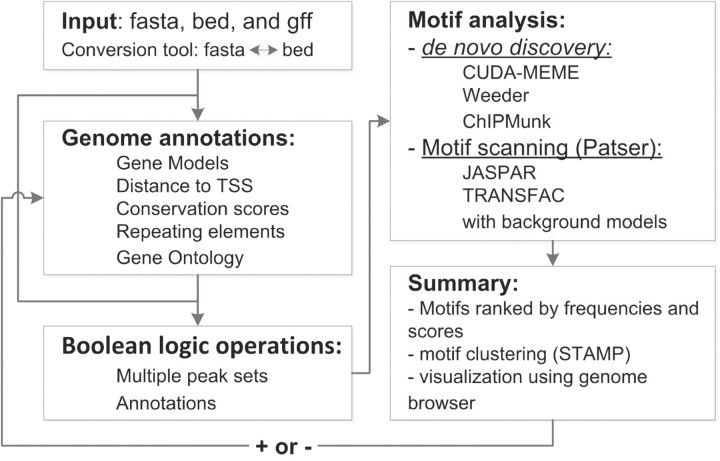
The cMOTIFs pipeline was built using MySQL, Perl, CGI and R statistical language, and the overview of workflow is illustrated in Figure 1.
Fig. 1.
Workflow of analysis pipeline. A subset of peak sequences based on the presence (+) or absence (−) of specific motifs can be easily defined for in-depth iterative analysis.
Input format: the pipeline accepts DNA sequences in FASTA format or genomic coordinates in BED or GFF formats. Format conversion tools are provided for human (hg18, hg19), mouse (mm9) and rat (rn4) genomes.
Genomic annotation: cMOTIFs provides annotations from UCSC Genome Bioinformatics including gene annotations (RefSeq and Ensembl), multispecies conservation scores (PhastCons) and repeating sequence elements (RepeatMasker), which enable users to select regions based on any combination of annotation criteria. The GOstats R library (Falcon and Gentleman, 2007) was integrated to the pipeline to summarize enriched Gene Ontology categories for the peak-associated genes. For instance, users can select peaks, which are highly conserved, contain no repeating sequence elements, and are located within 10 kb of transcription start sites of known genes, for subsequent motif discovery processes.
Boolean logic operations with multiple datasets: eight operations are provided to perform intersection, union, subtraction, common merge, common region, merge by distance, unique and extension on multiple genomic-region files (more details on the web site). For instance, to identify possible multiple transcription factor binding loci (MTL), users can merge neighbouring motif-enriched regions from different TF experiments.
Motif discovery: we incorporated three existing de novo discovery algorithms [MEME (Bailey and Elkan, 1994), Weeder (Pavesi et al., 2004) and ChIPMunk (Kulakovskiy et al., 2010)] and a motif scanning method [Patser (Hertz and Stormo, 1999)] with position-specific scoring matrices (PSSM) from databases including TRANSFAC (public version 7.0) (Matys et al., 2003) and JASPAR (version October 12, 2009) (Bryne et al., 2008) to prioritize overrepresented motifs. The pipeline also accepts user-defined PSSM for motif scanning. The Computer Unified Device Architecture (CUDA) library enhanced MEME performance (Liu et al., 2010). A suffix-tree-based exhaustive enumeration algorithm, Weeder (v1.4.2), and an iterative algorithm that combines greedy optimization with bootstrapping, ChIPMunk, have been included. The Patser [version 3b, (Hertz and Stormo, 1999)] is used to scan motifs from JASPAR and TRANSFAC databases. After the original Patser scanning, each target sequence is shuffled N times (default N = 1000) while maintaining (mono, di or tri) nucleotides frequency to create a random background model based on user input sequences or pre-compiled upstream sequences for each species. A permutation P-value is calculated from the frequency of random motif occurrences with the false discovery rate calculation (Storey and Tibshirani, 2003). This procedure allows estimating the significance of a motif occurrence in a target sequence compared with a set of randomly shuffled sequences. The current version of the pipeline allows up to a total of 500 000 bases for motif discovery, and this can be increased to 5 million bases if users choose to run ChIPMunk alone.
Motif summary: the top 10 motifs from the above four methods are ranked with corresponding scores and frequencies, and summarized with the STAMP tool by clustering motifs based on similarities (Mahony and Benos, 2007), and loci can be saved as BED format for further visual inspection and iterative analysis. Results are confidentially stored in users' personal accounts.
3 RESULTS AND DISCUSSION
We demonstrated our pipeline with a previously published ChIP-Seq dataset, which mapped binding locations of 13 TFs involved in embryonic stem cell differentiation (Chen et al., 2008). For 12 of the 13 TFs (except for E2f1), the pipeline successfully identified the reported binding motifs as the top-ranked motifs. Notably, the dinucleotide shuffling improved motif rankings from Patser (see Supplementary Material). The pipeline facilitates identification of multiple TF-binding loci. For instance, after processing the top 500 peaks from Nanog, Oct4 (encoded by Pou5f1 gene) and Sox2 TF profiles, we found a subset of peaks enriched with known binding motifs in JASPAR. Using Boolean logic, we merged neighbouring motif ‘+’ regions within 500 bp and identified 35 MTL, each of which was occupied by Nanog, Oct4 and Sox2, suggesting collaborative binding from all three. These illustrate some examples of complex gene expression regulations that can be analysed efficiently with the pipeline (see Supplementary Material for the detail).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Kulakovskiy for sharing ChIPMunk source codes for our pipeline.
Funding: Charles H. Hood Foundation and NHLBI (HL098166) to S.W.K.
Conflict of Interest: none declared.
REFERENCES
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Kulakovskiy IV, et al. Deep and wide digging for binding motifs in ChIP-Seq data. Bioinformatics. 2010;26:2622–2623. doi: 10.1093/bioinformatics/btq488. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. CUDA-MEME: Accelerating motif discovery in biological sequences using CUDA-enabled graphics processing units. Pattern Recognit. Lett. 2010;31:2170–2177. [Google Scholar]
- Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, et al. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, et al. Tmod: toolbox of motif discovery. Bioinformatics. 2010;26:405–407. doi: 10.1093/bioinformatics/btp681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Valen E, et al. Discovery of regulatory elements is improved by a discriminatory approach. PLoS. Comput. Biol. 2009;5:e1000562. doi: 10.1371/journal.pcbi.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeringen SJ, Veenstra GJ. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq636. [Epub ahead of print, November 15, 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.