Abstract
Background
The UMLS Metathesaurus (UMLS-Meta) is currently the most comprehensive effort for integrating independently-developed medical thesauri and ontologies. UMLS-Meta is being used in many applications, including PubMed and ClinicalTrials.gov. The integration of new sources combines automatic techniques, expert assessment, and auditing protocols. The automatic techniques currently in use, however, are mostly based on lexical algorithms and often disregard the semantics of the sources being integrated.
Results
In this paper, we argue that UMLS-Meta’s current design and auditing methodologies could be significantly enhanced by taking into account the logic-based semantics of the ontology sources. We provide empirical evidence suggesting that UMLS-Meta in its 2009AA version contains a significant number of errors; these errors become immediately apparent if the rich semantics of the ontology sources is taken into account, manifesting themselves as unintended logical consequences that follow from the ontology sources together with the information in UMLS-Meta. We then propose general principles and specific logic-based techniques to effectively detect and repair such errors.
Conclusions
Our results suggest that the methodologies employed in the design of UMLS-Meta are not only very costly in terms of human effort, but also error-prone. The techniques presented here can be useful for both reducing human effort in the design and maintenance of UMLS-Meta and improving the quality of its contents.
Background
Ontologies — formal conceptualisations of a domain of interest in a machine-understandable format — are extensively used in bioinformatics. The most widely used ontology modelling language is the Web Ontology Language (OWL) [1] and its revision OWL 2 [2], which are World Wide Web Consortium (W3C) standards [3,4] The formal underpinning of OWL and OWL 2 is based on formal logic [5]. The key advantage of using logic over alternative representation mechanisms (e.g., semantic networks, frames, ER or UML diagrams) is that logic provides an unambiguous meaning to ontologies. As a result, ontologies can be used to process data (e.g., electronic patient records in the case of a medical application) in a more intelligent way. Prominent examples of biomedical OWL ontologies are the National Cancer Institute (NCI) Thesaurus [6,7], the Systematised Nomenclature of Medicine and Clinical Terms (SNOMED CT) [8], and the Foundational Model of Anatomy (FMA) [9]. These ontologies are gradually superseding the existing medical classifications and are becoming core platforms for accessing, gathering, and sharing biomedical knowledge and data.
SNOMED CT, NCI, and FMA describe partially overlapping domains. For example, both NCI and FMA describe the anatomy of the human heart; however, NCI describes the domain of human diseases, whereas FMA does not. Although the domains of interest of these ontologies may intuitively overlap, their vocabularies are most likely going to diverge. For example, NCI defines the entity “Myocardium” to describe the muscles that surround and power the human heart, whereas FMA uses the entity “Cardiac Muscle Tissue” to describe exactly the same set of muscles. This is because ontologies such as NCI and FMA have been independently developed and hence use different names and naming conventions for referring to their entities. Consequently, these ontologies, even if intuitively overlapping, are unrelated from a logical point of view. For example, if a data source described using FMA states that Mary Jones has suffered from an infarction affecting the “Cardiac Muscle Tissue”, and an NCI data source states that Paul Williams has suffered from an infarction affecting the “Myocardium” then a computer program would not be able to infer that both patients have suffered from the same condition.
To exchange or migrate data between ontology-based applications, it is crucial to establish correspondences (or mappings) between their ontologies. For example, a mapping between NCI and FMA should establish that the entities “Cardiac Muscle Tissue” and “Myocardium” are synonyms. Using this mapping, a computer program would then be able to migrate the data statement “Paul Williams has suffered from an infarction affecting the Myocardium” from an NCI-based application to an FMA-based application.
Ontology mappings are often conceptualised as tuples with the form 〈id, e1, e2,n, p〉 , where id is a unique identifier for the mapping, e1, e2 are entities in the vocabulary of the mapped ontologies, n is a numeric confidence measure between 0 and 1, and ϱ is a relation between e1 and e2,typically subsumption (i.e., e1 is more specific than e2), equivalence (i.e., e1 and e2 are synonyms), or disjointness (i.e., no individual can be an instance of both e1 and e2) [10]. Creating such mappings manually is often unfeasible due to the size and complexity of modern biomedical ontologies. For example, SNOMED CT in its version from January 2009 contains more than 300,000 entities, while NCI (version 08.05d) and FMA (version 2.0) contain around 79,000 and 67,000 entities, respectively. Since the number of potential mappings grows (at least) quadratically with the number of entities in the relevant ontologies, a tool would need to consider (at least) 10 billion candidate mappings between SNOMED CT and NCI.
Most existing automated mapping generation techniques are based on lexical algorithms (e.g., [11-17]). Some of these algorithms are rather sophisticated and may exploit the syntactic structure of the ontologies [12,13] , or access external knowledge sources (e.g., WordNet [18]) to look for synonyms of ontology entities and their lexical variations [14,15,17].
The growing number of available techniques has made the creation of mappings between real-world ontologies possible. The most comprehensive effort for integrating biomedical ontologies through mappings is the UMLS Metathesaurus (UMLS-Meta) [19], which is being used in many applications, including PubMed (a search engine for accessing citations of biomedical articles) and ClinicalTrials.gov (a registry of clinical trials conducted around the world).
Currently, the integration of new ontologies in UMLS-Meta combines lexical algorithms, expert assessment [14,15,20,21], and auditing protocols [22]. In its 2009AA version, UMLS-Meta integrates more than one hundred thesauri and ontologies, including SNOMED CT, FMA, and NCI, and contains more than 6 million entities. UMLS-Meta provides a list with more than two million unique identifiers (CUIs). Each CUI can be associated to entities belonging to different sources. Pairs of entities from different sources with the same CUI are synonyms and hence can be represented as an equivalence mapping. It has been noticed that UMLS-Meta, despite being carefully curated by domain experts, may contain errors [23-27]. Current auditing techniques aimed at detecting potential errors mostly rely on UMLS-Meta semantic network [28] — a “top level” semantic model grouping the entities from the UMLS-Meta sources into suitable semantic types. Semantic types are then organised into so-called semantic groups [29]. For example “Heart” is associated with the semantic type “Body Part, Organ, or Organ Component” and the semantic group “Anatomy”. Errors can then be detected by identifying incompatibilities in the assignment of such semantic types to entities in the sources. For example the UMLS-Meta “Globular Actin” has, among others, “Amino Acid, Peptide, or Protein” and “Cell Component” as semantic types, which belong to two different semantic groups “Chemicals & Drugs” and “Anatomy” respectively.
In this paper, we argue that UMLS-Meta’s current design and auditing methodologies could be significantly enhanced by taking into account the logic-based semantics of the ontology sources. We provide empirical evidence suggesting that UMLS-Meta in its 2009AA version contains a significant number of errors; these errors become immediately apparent if the rich semantics of the ontology sources is taken into account, manifesting themselves as unintended logical consequences that follow from the ontology sources together with the information in UMLS-Meta. We then propose general principles and specific logic-based techniques to effectively detect and repair such errors. We have evaluated our techniques using three widely-used and rich ontology sources, namely FMA, NCI and SNOMED CT, and obtained very encouraging empirical results; our techniques, however, are generic and therefore applicable to any other UMLS-Meta source that can be expressed in OWL. Furthermore, we believe that our novel techniques are complementary to current UMLS-Meta auditing methods [22-26] and studying how they can be effectively combined constitutes an interesting direction for future work.
Methods
The use of logic-based techniques requires the adoption of a formal representation of the mappings. This is crucial to reason unambiguously with the source ontologies and the corresponding mappings. Therefore, the first step in our research methodology has been to provide formal semantics to the mappings in UMLS-Meta. Once a coherent semantic framework has been identified, we can enable logical reasoning over the union of the source ontologies and their respective UMLS-Meta mappings and obtain logical consequences that were not derivable from any of them in isolation. Our main hypothesis is that such logical consequences can be used to identify errors in the mappings as well as to detect inherent incompatibilities between the source ontologies. To verify this hypothesis, we have identified three general principles, which describe precisely how these new logical consequences can be interpreted and exploited. Finally, we have designed a number of logic-based techniques that follow those general principles; these techniques are both semantically coherent and efficiently implementable in practice.
Logical representation of ontology mappings
A number of formal representations for ontology mappings have been proposed so far in the literature (e.g., [10,30,31]), and there is currently no consensus on which ones are more suitable for practical applications. In this research, we have adopted a pragmatic approach in which a set of mappings is given as an OWL 2 ontology. Mappings 〈 id, e1, e2, n, ϱ〉are directly represented as OWL 2 axioms of the form SubClassOf(e1 e2), EquivalentClasses(e1 e2), or DisjointClasses(e1 e2), for ϱ denoting subsumption, equivalence, and disjointness, respectively, and with id (the mapping id) and n (the confidence value) added as axiom annotations [2]. Such a representation seems semantically coherent, and allows us to reuse the extensive range of OWL-based algorithmic techniques and infrastructure that is currently available while preserving valuable information such as confidence values. In this setting, we can therefore restrict ourselves to consider the situation where OWL 2 ontologies O1 and O2 are integrated via a third OWL 2 ontology M, which contains the relevant mappings (e.g., O1 might be FMA, O2 NCI, and M UMLS-Meta). In the particular case of UMLS-Meta, we have processed the MRCONSO file from its distribution [32]. This file contains every entity in UMLS-Meta together with its concept unique identifier (CUI), its source vocabulary, its language, and other attributes not relevant for this work. Table 1 shows an excerpt from the rows in the MRSCONSO file associated to the CUI C0022417 (which represents the notion of “Joint”) with source vocabulary FMA, SNOMED CT or NCI.
Table 1.
An excerpt from the MRCONSO file for “Joint”
| CUI | Language | Source | Entity |
|---|---|---|---|
| FMA | Joint | ||
| Set_of_joints | |||
| C0022417 | ENG | SNOMED CT | Joint_structure |
| NCI | Joint | ||
| Articulation | |||
Table 1: An excerpt from the MRCONSO file for “Joint”
It follows from Table 1 that the notion of “Joint” is shared by FMA, SNOMED CT and NCI. In particular, FMA contains the entities Joint and Set_of_joints, NCI the entities Articulation and Joint, and SNOMED CT only the entity Joint_structure. All these entities have been annotated with the CUI C0022417 and therefore, according to UMLS-Meta’s intended meaning, they are synonyms. Then, for each pair of entities e and e′ from different sources and annotated with the same CUI, we have generated the OWL 2 mapping axiom EquivalentClasses(e e′). The axioms obtained for our example CUI are given in Table 2. We do not explicitly generate axioms involving entities from the same source because we are interpreting UMLS-Meta as a mapping theory, whose purpose is to integrate independently developed sources, rather than to model the domain (i.e., to add explicit content to each of the sources independently). Note, however, that the mappings from Table 2 do modify the contents of the ontology sources implicitly (for example, they imply that the entities Articulation and Joint from NCI are equivalent, even if they are not in the original source). In the following section, we argue that such implicit modifications of a source due only to the mappings are one of the main causes of logical errors.
Table 2.
Mappings between FMA, NCI and SNOMED CT
| Mapped Ontologies | Generated Mappings |
|---|---|
| EquivalentClasses(FMA:Joint NCI:Joint) | |
| FMA ∼ NCI | EquivalentClasses(FMA:Joint NCI:Articulation) |
| EquivalentClasses(FMA:Set_of_joints NCI:Joint) | |
| EquivalentClasses(FMA:Set_of_joints NCI:Articulation) | |
| FMA ∼ SNOMED CT | EquivalentClasses(FMA:Joint SNOMED:Joint_structure) |
| EquivalentClasses(FMA:Set_of_joints SNOMED:Joint_structure) | |
| SNOMED CT ∼ NCI | EquivalentClasses(SNOMED:Joint_ structure NCI:Joint) |
| EquivalentClasses( SNOMED:Joint_structure NCI:Articulation) | |
Table 2: Mappings between FMA, NCI and SNOMED CT. The prefixes “FMA:”, “NCI:” and “SNOMED:” are shown explicitly to emphasise that each ontology source uses a different namespace to refer to its entities. However, for simplicity, we will often obviate these prefixes in the text.
Proposed principles
We have identified three general principles, which describe how logic-based techniques can be applied to the integration of two ontology sources O1 and O2 using a third mapping ontology M.
1. The conservativity principle: Given an ontology source (say, O1) and the mappings M, the union O1∪M should not introduce new semantic relationships between entities from O1.
The conservativity principle is based on the purpose of M, which is to enable the interaction between O1 and O2, rather than to provide a new description of the domain. In the case of our previous example about “Joints”, UMLS-Meta contains two mappings establishing the equivalence between the entity Joint_structure from SNOMED CT and the FMA entities Joint and Set_of_joints respectively. As a consequence, UMLS-Meta implies that Joint is also equivalent to Set_of_joints. However, in FMA Joint neither subsumes, nor it is subsumed by Set_of_joints (see Figure 1). The conservativity principle suggests that the obtained mappings are in conflict and (at least) one of them is likely to be incorrect.
Figure 1.
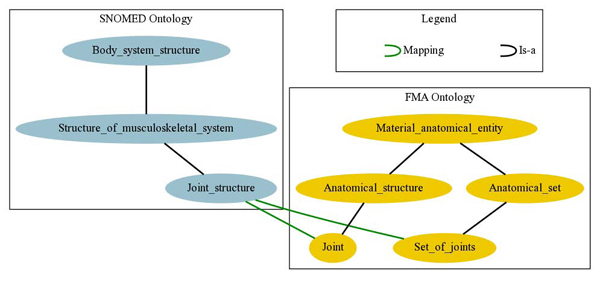
Conservativity principle violation between FMA and SNOMED CT mappings
2. The consistency principle: The ontology O1 ∪ O2 ∪M should be consistent and all the entities in its vocabulary should be satisfiable [5].
According to the consistency principle, the integration of well-established ontologies should not introduce logical inconsistencies, which are clear manifestations of a design error. These may be due to either erroneous mappings or to inherent incompatibilities between the source ontologies. In any case, in order for the integrated ontology to be successfully used in an application, these errors should be repaired by modifying either the source ontologies or the mappings.
For example, as shown in Figure 2, UMLS-Meta maps the FMA concept Protein to the NCI concept Protein, and the FMA concept Lymphokine to the NCI concept Therapeutic_Lymphokine. In FMA, Lymphokine is a Protein, whereas in NCI Therapeutίc_Lίmphokίne is a Pharmacologic_Substance. Furthermore, Pharmacologic_Substance and Protein are disjoint in NCI and hence the union of NCI, FMA and UMLS-Meta would imply that Lymphokine and Therapeutic_Limphokine are unsatisfiable (i.e.,there can be no instances of either entity).
Figure 2.
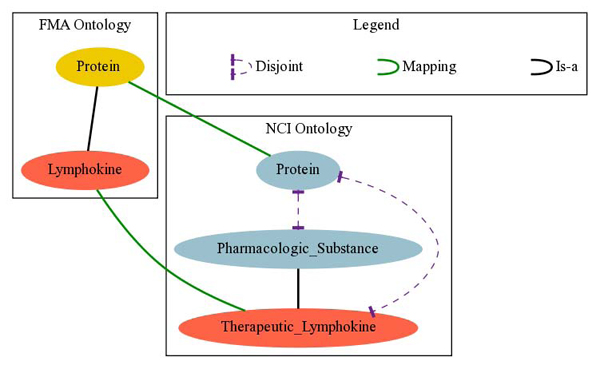
Consistency principle violation between FMA and NCI mappings
3. The locality principle: If two entities e1 and e2 from ontologies O1 and O2 are correctly mapped, then the entities semantically related to e1 in O1 are likely to be mapped to those semantically related to e2 in O2.
If the locality principle does not hold, then the following situations can be identified: (1) M may be incomplete and new mappings should be discovered, (2) the definitions of both concepts in their respective ontologies may be different or incompatible, or (3) the mapping between e1 and e2 may be erroneous.
Specific techniques
We next propose a collection of logic-based techniques based on each of these general principles. Our techniques exploit the following observations about UMLS-Meta ontology sources and mappings:
● Ob1: The OWL 2 ontology M that encodes the contents of UMLS-Meta only contains axioms of the form EquivalentClasses(e1 e2) where e1 is only mentioned in O1 and e2 is only mentioned in O2 (note that, as illustrated in Table 2, different ontology sources use different namespaces to refer to their entities). This observation is crucial to the design of techniques based on the conservativity principle.
● Ob2: UMLS-Meta ontology sources such as SNOMED CT, NCI and FMA contain both positive and negative information (e.g., if something is a “Protein”, then it is not a “Drug”). Logical inconsistencies can arise due to the simultaneous presence (either explicit or implicit) of two conflicting statements containing positive and negative information, e.g., the statement in FMA that lymphokine is a kind of protein and the (implicit) statement that lymphokine is not a kind of protein. This observation is important to design techniques based on the consistency principle.
● Ob3: The entities described in UMLS-Meta ontology sources such as SNOMED CT, NCI and FMA are “loosely interconnected”. Roughly speaking, this implies that the “meaning” of an entity in each of these ontologies only depends on a very small set of entities in the ontology that are “semantically related” to it. To formalise the notion of an entity being “semantically related” to another entity in an ontology, we use the logic-based ontology modularisation framework from [33]. This observation is important to design techniques based on either the consistency or the locality principles.
Techniques based on the conservativity principle
The conservativity principle can be directly expressed as the following reasoning problem. We say that an ontology source (say, O1) violates conservativity if there exists an OWL 2 axiom α such that O1 ∪M implies α, but O1 does not imply α. This problem is strongly related to the notion of conservative extension [34-36]. It is well-known that its computational complexity is very high even for lightweight ontology languages, and no practical algorithms currently exists.
As we describe next, however, in the case of UMLS-Meta we can exploit Observation Ob1 to significantly simplify the problem.
Let M contain only axioms of the form EquivalentClasses(e1 e2) where e1 is only mentioned in O1 and e2 is only mentioned in O2. Then, O1 violates conservativity if and only if there exist axioms EquivalentClasses(e1 e2) and EquivalentClasses(e′1 e2) in M, with e1 and e′1 different entities in O1, such that O1 alone does not imply the axiom EquivalentClasses(e1 e′1).
If this is the case, then the mappings EquivalentClasses(e1 e2) and EquivalentClasses(e′1 e2) from M are in conflict and one of them may be incorrect. In our previous example (recall Figure 1), the mappings EquivalentClasses( Joint_structure Joint) and EquivalentClasses( Joint_structure Set_of_joints) between SNOMED CT and FMA are likely to be in conflict.
In order to identify such conflicting mappings, it suffices to (syntactically) check in M whether two entities from one of the sources (e.g., Joint and Set_of_ joints from FMA) are mapped to the same entity in the other source (e.g., Joint_structure from SNOMED CT) and then check (semantically) whether these two entities were already equivalent with respect (only) to the former source. These checks can be performed efficiently in practice: the former is syntactic, and the latter involves a single semantic test using an ontology reasoner (e.g., Does FMA imply that Joint and Set_of_joints are equivalent?).
Techniques based on the consistency principle
Similarly to the conservativity principle, the consistency principle can also be easily formulated as a reasoning problem. Let us denote with sig(O) the vocabulary of an ontology O. We say that O1, O2 and M violate consistency if there is an entity e ∊ sig(O1) ∪ sig(O2) that is unsatisfiable with respect to O1 ∪O2 ∪ M (i.e., O1 ∪O2 ∪ M implies the axiom EquivalentClasses(e owl:Nothing)).
The obvious way to check consistency violation and identify sets of conflicting mappings is to use an ontology reasoner to check the satisfiability of each entity in the vocabulary of O1 ∪ O2 ∪ M and then apply state-of-the-art ontology debugging techniques to identify and disambiguate conflicts (e.g., see [30,37-41]). In our example from Figure 2, we could use a reasoner to identify that the concepts Lymphokine and Therapeutic_Limphokine are unsatisfiable with respect to the integration of FMA and NCI via UMLS-Meta. Then, debugging techniques could be used to identify the mappings and axioms from the source ontologies responsible for these errors.
This approach, however, can be computationally prohibitive; for example, the classification of the ontology obtained after integrating FMA and NCI using UMLS-Meta required almost 70 hours when using the reasoner HermiT [42] on a high performance server with 32GB of RAM; furthermore, the reasoner detected more than 15,000 unsatisfiable entities. Current debugging and repair techniques, which often rely on computing minimal sets of axioms responsible for each inconsistency, cannot be applied when the reasoning time is so prohibitive and the number of errors so high.
In our particular setting, unsatisfiable concepts can be caused by either erroneous mappings, or by inherent incompatibilities between the source ontologies. Our method relies on first identifying and disambiguating conflicting pairs of mappings (i.e., mappings that when occurring together make a concept unsatisfiable) and only then detecting and resolving incompatibilities between the sources.
Detecting conflicting mappings. We next exploit Observations Ob2 and Ob3 to define a simple heuristic technique, which is along the lines of those presented in [17,43,44].
The disjointness-based inconsistency heuristic is defined as follows. If e and e′ from O1 are mapped to f and f′ from O2 and O1 implies that e is subsumed by e′, but O2 implies that f and f′ are disjoint, then the consistency principle is violated (recall Figure 2). Note that the converse does not necessarily hold. This heuristic requires semantic tests to check whether e is subsumed by e′ in O1 and whether f and f′ are disjoint in O2. However, this heuristic requires only the classification of each of the source ontologies independently. (Note that many reasoners can produce also the implicit disjointness relationships between their entities as an additional output of classification with only a relatively short additional delay.)
Furthermore, by Observation Ob3 we can optimise even further and perform only the classification of the logic-based modules [33] for the mapped entities in each of the source ontologies. Consider, for example, the mappings between FMA and SNOMED CT. The logic-based module in FMA for the entities that are mapped to SNOMED CT contains only 10,204 entities (out of 67,000), and the corresponding module in SNOMED CT contains only 15,428 entities (out of 300,000). That is, extracting and classifying the modules instead of classifying the source ontologies as a whole results in a considerable simplification. Finally, the set of conflicting mappings is directly obtained and therefore there is no need to apply expensive debugging techniques to retrieve minimal sets of axioms responsible for the inconsistency.
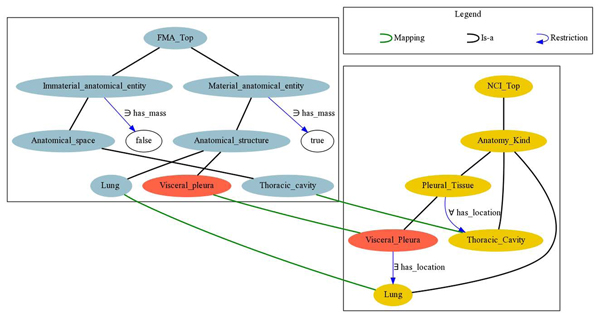
Detecting incompatibilities between the sources. Even if the mappings between two ontology sources are the intended ones, the sources may describe a particular aspect of the domain in incompatible ways. For example, consider Figure 3 describing the notion of “Visceral Pleura” in FMA and NCI. The three mappings between the entities “Visceral Pleura”, “Lung” and “Thoracic Cavity” in both ontologies are clearly the intended ones. However, their integration results in Visceral_Pleura becoming unsatisfiable. According to NCI, the visceral pleura is located in a lung; furthermore, it is a pleural tissue, which can only be located in the thoracic cavity. However, according to FMA the thoracic cavity is an immaterial anatomical entity, whereas the lung is a material anatomical entity. Finally, material and immaterial entities are disjoint, as implied by FMA. Therefore, the visceral pleura is located in some anatomical entity that is both material and immaterial, which leads to a contradiction.
Figure 3.
Unsatisfiability due to inherent incompatibilities between FMA and NCI
We believe that, in such cases, the ontology engineer must participate in the repair process. In order to detect these incompatibilities and help the user to repair them, we provide tool support in our prototype ContentMap [30], which we briefly describe later on.
Techniques based on the locality principle
The conservativity and consistency principles allow us to identify pairs of mappings in UMLS-Meta that are in mutual conflict. However, since UMLS-Meta does not assign a confidence value to each mapping, it is not clear how to disambiguate these conflicts (e.g., how to decide whether to map Joint_structure to Joint or to Set_of_joints). If there are too many conflicts, manual disambiguation becomes unfeasible. We propose to apply the locality principle in order to compute a confidence value for each conflicting mapping, which we can then exploit for (partially) automating the disambiguation process.
Computing confidence values. Assume that e from O1 is mapped via a mapping μ to f from O2. As already mentioned in Observation Ob3, the application of the locality principle relies on the well-known ontology modularisation framework from [33]. Therefore, if most of the entities in the module 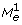 for e in O1 (i.e., those entities that are “semantically related” to e in O1) are also mapped to those in the module
for e in O1 (i.e., those entities that are “semantically related” to e in O1) are also mapped to those in the module 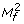 for f in O2, then we can assign a high confidence value to μ. Intuitively, such confidence values conf (μ) can be obtained by computing the ratio between the number of entities in the modules which are mapped via UMLS-Meta, and the total number of entities in the modules:
for f in O2, then we can assign a high confidence value to μ. Intuitively, such confidence values conf (μ) can be obtained by computing the ratio between the number of entities in the modules which are mapped via UMLS-Meta, and the total number of entities in the modules:
However, since the modules are of relatively small size, UMLS-Meta often does not contain enough mappings to obtain an accurate value. For example, UMLS-Meta maps Upper_Extremity from NCI and Arm from FMA, but none of the entities in the module for Upper_Extremity in NCI is mapped to an entity in the module for Arm in FMA. To address this issue, we use a lexical matching algorithm [11] to obtain additional lexical correspondences between entities in the modules and refine our confidence value.
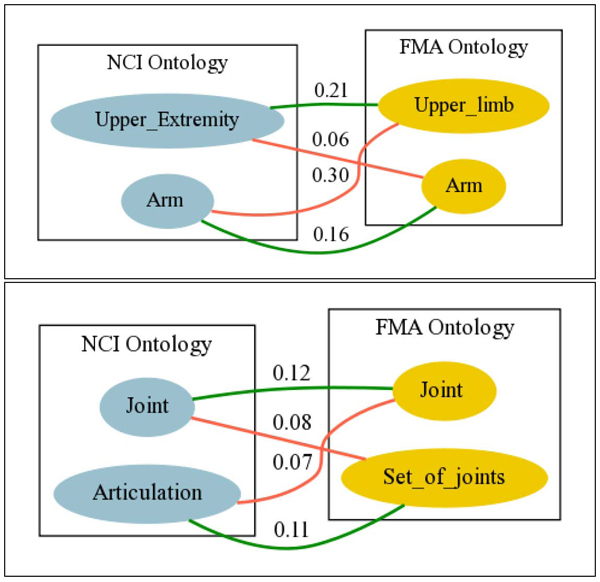
Disambiguating conflicts automatically. Figure 4 depicts sets of conflicting mappings obtained using the conservativity principle together with the confidence values we have obtained for each of them. Consider the mappings between NCI and FMA on the left-hand-side of the figure. Let μ1, μ2 represent the mappings respectively connecting Upper_Extremity in NCI to Upper_limb and Arm in FMA, and let μ3,μ4 represent those relating Arm in NCI to Upper_Limb and Arm from FMA, respectively. We can identify the following four conflicts:
Figure 4.
Combined cases of ambiguity
k1 = {μ1, μ2} k2 = {μ3, μ4} k3 = {μ1, μ3} k4 = {μ2, μ4}
In order to disambiguate all the conflicts between two source ontologies, we need to remove one mapping per conflict in such a way that the result of adding their confidence values is minimised. This is a standard diagnosis problem, for which practical algorithms are well-known [45]. In our example, the solution involves removing the mappings μ2 (with confidence 0.06) and μ3 (with confidence 0.30).
Resolving inherent incompatibilities between source ontologies
Finally, as already mentioned, conflicts due to inherent incompatibilities between source ontologies (and not to unintended mappings) may require the intervention of a domain expert. However, as seen in Figure 3, source incompatibilities are not always apparent and therefore domain experts need to be supported by suitable tools. To provide the required tool support, we have developed the ContentMap prototype [30]. ContentMap helps users to understand the semantic consequences of the integration by identifying and explaining the new entailments that hold in the merged ontology, but not in O1, O2 and M alone.
Furthermore, ContentMap proposes different minimal repair plans for those entailments that the user indicates are unintended and ranks them accoding to the impact of their application. These plans may involve both the deletion or modification of source ontology axioms, and thus expert intervention is required to select the most appropriate repair.
Implementation and empirical results
We have implemented our techniques and combined them in an integrated solution consisting of the following steps, which have been described in the Methods section:
1. Mapping extraction: given the ontology sources to be integrated, we extract the corresponding set of mappings expressed in OWL from UMLS-Meta’s MRCONSO file.
2. Conflict detection: given the OWL mappings previously extracted and the ontology sources in OWL format, we apply our techniques to compute sets of conflicting mappings.
3. Automatic conflict disambiguation: we apply our techniques based on the locality principle to compute a confidence value for each conflicting mapping; then, we apply repair methods to disambiguate each set of conflicting mappings.
4. Semi-automatic repair of source incompatibilities: we reason with the sources and the mappings resulting from automatic disambiguation, and then use ContentMap to detect and repair (with manual intervention) the remaining unintended entailments.
We have evaluated our techniques using UMLS-Meta version 2009AA and the corresponding versions of FMA, NCI and SNOMED CT, which contain 66,724, 78,989 and 304,802 entities, respectively. After translating the relevant parts of UMLS-Meta into OWL 2, we obtained 3,024 mapping axioms between FMA and NCI, 9,072 between FMA and SNOMED CT and 19,622 between SNOMED CT and NCI. The mapping extraction script from the MRCONSO file and the obtained OWL mappings are available online [46].
When reasoning over each of the source ontologies independently, all their entities were found satisfiable. However, after the respective integrations via UMLS-Meta mappings, we obtained a huge number of unsatisfiable entities, namely 5,015 when integrating FMA and NCI, 16,764 with FMA and SNOMED CT, and 76,025 with SNOMED CT and NCI. Thus, from a semantic point of view, the integration of these ontologies via UMLS-Meta is far from error-free.
In order to identify conflicts between the obtained UMLS-Meta mappings, we have then applied the conservativity and consistency principles. The conflict detection scripts, and the text files containing the obtained conflicts and the associated confidence values used for disambiguation can also be downloaded from [46].
● Using the principle of conservativity, we found 991 conflicting mapping between FMA and NCI, 2,426 between FMA and SNOMED CT and 9,080 between SNOMED CT and NCI.
● Using the disjointness-based inconsistency heuristic, we found 300 conflicting mapping pairs between FMA and NCI, 14,959 between FMA and SNOMED CT and 34,628 between SNOMED CT and NCI. Note that each of these conflicts will certainly lead to the unsatisfiability of an entity in the union of the respective source ontologies and UMLS-Meta mappings.
As discussed in the Methods section, the locality principle allows us to assign a confidence value to each conflicting mapping and then exploit this value to automatically disambiguate the identified conflicts. The automatic disambiguation process removed 570 (19%) of the mappings between FMA and NCI, 4,077 (45%) of those between FMA and SNOMED CT and 13,358 (63%) of those between SNOMED CT and NCI. The resulting disambiguated mappings are available online in OWL format (see Additional files 1, 2, 3) [46].
After automatic disambiguation, we found only 2 unsatisfiable entities when integrating FMA and NCI (recall example from Figure 3), 44 for FMA and SNOMED CT, and none for SNOMED CT and NCI. As already discussed, these errors are most likely due to inherent incompatibilities between the ontology sources; thus, expert assessment is required. Our tool ContentMap was then used to understand and repair these remaining inconsistencies. ContentMap proposed 19 repair plans for FMA and NCI and 372 FMA and SNOMED CT. We have inspected the top-ranked repair plans and found them intuitive and reasonable from a modelling perspective. For example, the repair plans for FMA and NCI suggested the deletion/modification of the NCI axiom
SubClassOf(Pleural_Tissue allValuesFrom (has_location Thoracic_Cavity)) among others (recall example from Figure 3). A link to ContentMap’s binaries is provided in [46].
Despite the large number of deprecated mappings during automatic disambiguation, the integration of our source ontologies via UMLS-Meta still results in a considerable number of (possibly intended) new logical consequences. For example, after repairing all inconsistencies using ContentMap, the integration between FMA and SNOMED CT still results in 973 new subsumption relationships between FMA entities and 586 new subsumption relationships between SNOMED CT entities. Further manual revision (e.g., using again our tool ContentMap) would be necessary to determine which ones among these new consequences are deemed unintended.
Discussion and conclusion
When integrating ontology sources via UMLS-Meta, the contents of the ontology sources should be taken into account. Several authors have proposed different kinds of structural analysis of SNOMED CT, FMA and NCI to evaluate their compatibility [47,48]. In this paper, we have argued that the rich logic-based semantics of these ontology sources should also be considered. Our specific contributions can be summarised as follows:
1. We have provided empirical evidence suggesting that, by taking into account the semantics of the sources, one can detect a significant number of additional errors in UMLS-Meta as well as many inherent incompatibilities between the sources’ description of particular domains.
2. We have proposed three general principles which describe how logic-based techniques can be applied to audit the integration of ontology sources. These principles are generic and could be applied to ontologies in any domain. The consistency principle has been used in a similar formulation in related literature and the principle of locality is (at least implicitly) behind the design of many structural heuristics for ontology matching. To the best of our knowledge, however, the principle of conservativity is novel and has not been used in the context of ontology mapping.
3. We have proposed and implemented logic-based techniques based on these principles. Those based on the conservativity and locality principles are novel with the latter exploiting state of the art ontology modularisation techniques.
4. We have adapted state of the art repair techniques to our setting by taking into account the confidence values obtained using the locality principle.
5. We have implemented an integrated solution that combines all our techniques in a systematic way, and evaluated it over UMLS-Meta with encouraging empirical results. Our techniques, however, could be used in contexts other than UMLS-Meta.
We consider that our results naturally complement those in [47,48], as well as current auditing methodologies in UMLS-Meta [22-26].
For future work, we plan to improve our techniques in several ways. First, our automatic disambiguation is rather aggressive, in the sense that a significant number of UMLS-Meta mappings are discarded in order to prevent logical errors. We plan to explore, on the one hand, how the disambiguation process can be relaxed to include as many of the original mappings in UMLS-Meta as possible and, on the other hand, how to improve the accuracy of the disambiguation confidence metrics based on the locality principle.
Second, our techniques rely on the information contained in UMLS-Meta’s MRCONSO file. UMLS-Meta, however, provides other sources of information. The semantic types and groups of UMLS-Meta could be given a logic-based interpretation (at least partially) and then used to enhance our heuristics.
Furthermore, the file AMBIGSUI.RRF, which contains a list of ambiguous and redundant terms, could be used for filtering out obvious logical errors.
Third, different ontology sources may describe the same concept at different levels of granularity, which may lead to incompatibilities when mapping those sources; this is a well-known (and open) issue in ontology matching. Although our logic-based heuristics can help identifying errors caused by such incompatibilities, they do not tackle this issue directly, which we leave for future research.
Finally, we aim at seeking feedback from domain experts concerning both the automatic and the tool-assisted disambiguation processes; this feedback could provide us with precision and recall values for our techniques.
List of abbreviations
FMA: Foundational Model of Anatomy; NCI: National Cancer Institute; SNOMED CT: Systematised Nomenclature of Medicine and Clinical Terms; CUI: Concept Unique Identifier; OWL: Ontology Web Language; and UMLS: Unified Medical Language System
Competing interests
The authors declare that they have no competing interests.
Authors contributions
EJ and BCG conceived the project and led the development of the research. EJ also implemented the proposed techniques and performed their evaluation. IH and RB contributed to the discussions, supervised the progress of the project, and participated in the writing of the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Description of data: Disambiguated UMLS2009AA equivalence mappings (2454 from 3024) between FMA and NCI ontologies. File Format: OWL (Ontology Web Language)
Description of data: Disambiguated UMLS2009AA equivalence mappings (4995 from 9072) between FMA and SNOMED CT ontologies. File Format: OWL (Ontology Web Language)
Description of data: Disambiguated UMLS2009AA equivalence mappings (6264 from 19622) between NCI and SNOMED CT ontologies. File Format: OWL (Ontology Web Language)
Contributor Information
Ernesto Jiménez-Ruiz, Email: ernesto.jimenez.ruiz@gmail.com.
Bernardo Cuenca Grau, Email: berg@comlab.ox.ac.uk.
Ian Horrocks, Email: ian.horrocks@comlab.ox.ac.uk.
Rafael Berlanga, Email: berlanga@uji.es.
Acknowledgements
We would like to thank the the anonymous referees of the SWAT4LS workshop and special issue for their useful suggestions. This work has been partially funded by the Spanish National Research Program (TIN2008-01825/TIN). Ernesto Jimenez-Ruiz was supported by the PhD Fellowship Program of the Generalitat Valenciana (BFPI06/372). Bernardo Cuenca Grau is supported by a Royal Society University Research Fellowship.
This article has been published as part of Journal of Biomedical Semantics Volume 2 Supplement 1, 2011: Semantic Web Applications and Tools for Life Sciences (SWAT4LS), 2009. The full contents of the supplement are available online at http://www.jbiomedsem.com/supplements/2/S1
References
- Horrocks I, Patel-Schneider PF, van Harmelen F. From SHIQ and RDF to OWL: the making of a Web Ontology Language. Journal Web Semantics. 2003;1:7–26. [Google Scholar]
- Cuenca Grau B, Horrocks I, Motik B, Parsia B, Patel-Schneider P, Sattler U. OWL 2: The Next Step for OWL. J. Web Semantics. 2008;6(4):309–322. [Google Scholar]
- Patel-Schneider P, Hayes P, Horrocks I. Web Ontology Language OWL Abstract Syntax and Semantics. W3C Recommendation. 2004. http://www.w3.org/TR/owl-semantics/
- Motik B, Patel-Schneider P, Cuenca-Grau B. OWL 2 Web Ontology Language Direct Semantics. W3C Recommendation. 2009. http://www.w3.org/TR/owl2-semantics/
- Baader F, Calvanese D, McGuinness DL, Nardi D, Patel-Schneider PF , editor. The Description Logic Handbook: Theory, Implementation, and Applications. Cambridge University Press; 2003. [Google Scholar]
- Hartel FW, de Coronado S, Dionne R, Fragoso G, Golbeck J. Modeling a description logic vocabulary for cancer research. Journal of Biomedical Informatics. 2005;38(2):114–129. doi: 10.1016/j.jbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Golbeck J, Fragoso G, Hartel FW, Hendler JA, Oberthaler J, Parsia B. The National Cancer Institute’s Thésaurus and Ontology. J. Web Sem. 2003;1:75–80. [Google Scholar]
- Spackman K. SNOMED RT and SNOMED CT. Promise of an international clinical ontology. M.D. Computing. 2000;17 [PubMed] [Google Scholar]
- Mejino JLV, Jr, Rosse C. Symbolic modeling of structural relationships in the Foundational Model of Anatomy. In Proc. of First International Workshop on Formal Biomedical Knowledge Representation (KR-MED 2004) 2004. pp. 48–62.
- Euzenat J. Semantic Precision and Recall for Ontology Alignment Evaluation. In Proceedings of the 20th International Joint Conference on Artificial Intelligence, IJCAI. 2007. pp. 348–353.
- Stoilos G, Stamou GB, Kollias SD. A String Metric for Ontology Alignment. In Proc. of the International Semantic Web Conference (ISWC) 2005. pp. 624–637.
- Euzenat J, Valtchev P. Similarity-Based Ontology Alignment in OWL-Lite. In Proceedings of the 16th Eureopean Conference on Artificial Intelligence, ECAI. 2004. pp. 333–337.
- David J. AROMA results for OAEI 2008. In Proceedings of the 3rd International Workshop on Ontology Matching (OM), Volume 431, CEUR Workshop Proceedings. 2008.
- Huang KC, Geller J, Halper M, Cimino JJ. Piecewise Synonyms for Enhanced UMLS Source Terminology Integration. In Proceedings of the 2007 American Medical Informatics Association (AMIA) Annual Symposium. 2007. pp. 339–343. [PMC free article] [PubMed]
- Huang KC, Geller J, Halper M, Perl Y, Xu J. Using WordNet synonym substitution to enhance UMLS source integration. Artif. 2009;46(2) doi: 10.1016/j.artmed.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tang J, Li Y, Luo Q. RiMOM: A Dynamic Multistrategy Ontology Alignment Framework. IEEE Transactions on Data and Knowledge Engineering. 2009;21(8):1218–1232. doi: 10.1109/TKDE.2008.202. [DOI] [Google Scholar]
- Jean-Mary YR, Shironoshita EP, Kabuka MR. Ontology matching with semantic verification. J. Web Semantics. 2009;7(3):235–251. doi: 10.1016/j.websem.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GA. WordNet: a lexical database for English. Commun. ACM. 1995;38(11):39–41. doi: 10.1145/219717.219748. [DOI] [Google Scholar]
- Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(Database issue) doi: 10.1093/nar/gkh061. http://dx.doi.org/10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001. pp. 17–21.http://view.ncbi.nlm.nih.gov/pubmed/11825149 [PMC free article] [PubMed]
- Mougin F, Burgun A, Bodenreider O. Using WordNet to Improve the Mapping of Data Elements to UMLS for Data Sources Integration. AMIA Symp. 2006. pp. 574–578. [PMC free article] [PubMed]
- Geller J, Perl Y, Halper M, Cornet R. Special Issue on Auditing of Terminologies. Journal of Biomedical Informatics. 2009;42(3):407–411. doi: 10.1016/j.jbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Cimino JJ. Auditing the Unified Medical Language System with Semantic Methods. Journal of the American Medical Informatics Association (JAMIA) 1998;5:41–51. doi: 10.1136/jamia.1998.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino JJ, Min H, Perl Y. Consistency across the hierarchies of the UMLS Semantic Network and Metathesaurus. Journal of Biomedical Informatics. 2003;36(6):450–461. doi: 10.1016/j.jbi.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Mougin F, Bodenreider O, Burgun A. Analyzing polysemous concepts from a clinical perspective: Application to auditing concept categorization in the UMLS. Journal of Biomedical Informatics. 2009;42(3):440–451. doi: 10.1016/j.jbi.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrey CP, Geller J, Halper M, Perl Y. The Neighborhood Auditing Tool: A hybrid interface for auditing the UMLS. Journal of Biomedical Informatics. 2009;42(3):468–489. doi: 10.1016/j.jbi.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-Yepes A, Jimenez-Ruiz E, Berlanga R, Rebholz-Schuhmann D. Reuse of terminological resources for efficient ontological engineering in Life Sciences. BMC Bioinformatics. 2009;10(Suppl 10):S4. doi: 10.1186/1471-2105-10-S10-S4. http://www.biomedcentral.com/1471-2105/10/S10/S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray A. In: Proc 13th Annu Symp Comput App Med Care. LC K, editor. IEEE Computer Society Press; 1989. The UMLS Semantic Network. pp. 503–507. [Google Scholar]
- Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. Journal of Biomedical Informatics. 2003;36(6):414–432. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Ruiz E, Cuenca Grau B, Horrocks I, Berlanga R. Proc. of European Semantic Web Conference (ESWC), Volume 5554 of LNCS. Springer-Verlag; 2009. Ontology Integration Using Mappings: Towards Getting the Right Logical Consequences. pp. 173–187. [Google Scholar]
- Borgida A, Serafini L. Distributed Description Logics: Assimilating Information from Peer Sources. J. Data Semantics. 2003;1:153–184. full_text. [Google Scholar]
- UMLS Reference Manual. http://www.ncbi.nlm.nih.gov/books/NBK9676/
- Cuenca Grau B, Horrocks I, Kazakov Y, Sattler U. Just the Right Amount: Extracting Modules from Ontologies. In Proc. of WWW 2007. 2007. pp. 717–727. full_text.
- Lutz C, Wolter F. In: Proceedings of the 21th Conference on Automated Deduction (CADE-21), Volume 4603 of Lecture Notes in Artificial Intelligence. Pfenning F, editor. Springer-Verlag; 2007. Conservative Extensions in the Lightweight Description Logic EL. pp. 84–99. [Google Scholar]
- Lutz C, Walther D, Wolter F. Conservative Extensions in Expressive Description Logics. In Proc. of the 20th International Joint Conference on Artificial Intelligence (IJCAI 2007) 2007.
- Cuenca Grau B, Horrocks I, Kazakov Y, Sattler U. Modular Reuse of Ontologies: Theory and Practice. JAIR. 2008;31:273–318. [Google Scholar]
- Kalyanpur A, Parsia B, Sirin E, Hendler JA. Debugging unsatisfiable classes in OWL ontologies. J. 2005;3(4):268–293. [Google Scholar]
- Kalyanpur A, Parsia B, Sirin E, Cuenca Grau B. proc. of the 2nd European Semantic Web Conference (ESWC 2006), Volume 4011 of LNCS. Springer; 2006. Repairing Unsatisfiable Concepts in OWL Ontologies. pp. 170–184. [Google Scholar]
- Schlobach S, Huang Z, Cornet R, van Harmelen F. Debugging Incoherent Terminologies. J. Autom. Reasoning. 2007;39(3):317–349. doi: 10.1007/s10817-007-9076-z. [DOI] [Google Scholar]
- Kalyanpur A, Parsia B, Horridge M, Sirin E. 6th International Semantic Web Conference, Volume 4825 of Lecture Notes in Computer Science. Springer; 2007. Finding All Justifications of OWL DL Entailments. pp. 267–280. [Google Scholar]
- Horridge M, Parsia B, Sattler U. International Semantic Web Conference, Volume 5318 of Lecture Notes in Computer Science. Springer; 2008. Laconic and Precise Justifications in OWL. pp. 323–338. [Google Scholar]
- Motik B, Shearer R, Horrocks I. Hypertableau Reasoning for Description Logics. Journal of Artificial Intelligence Research. 2009;36:165–228. [Google Scholar]
- Meilicke C, Stuckenschmidt H, Tamilin A. Supporting Manual Mapping Revision using Logical Reasoning. In Proceedings of the Twenty-Third AAAI Conference on Artificial Intelligence, AAAI. 2008. pp. 1213–1218.
- Meilicke C, Stuckenschmidt H. Third International Conference on Web Reasoning and Rule Systems, RR, Volume 5837 of Lecture Notes in Computer Science. Springer; 2009. An Efficient Method for Computing Alignment Diagnoses. pp. 182–196. [Google Scholar]
- Reiter R. A Theory of Diagnosis from First Principles. Artif. Intell. 1987;32:57–95. doi: 10.1016/0004-3702(87)90062-2. [DOI] [Google Scholar]
- Online empirical results and implementation sources. http://krono.act.uji.es/people/Ernesto/umlsassessment
- Bodenreider O, Zhang S. Comparing the Representation of Anatomy in the FMA and SNOMED CT. In AMIA Annual Symposium. 2006. pp. 46–50. [PMC free article] [PubMed]
- Bodenreider O. Proceedings of the Third International Conference on Knowledge Representation in Medicine, KR-MED, Volume 410 of CEUR Workshop Proceedings. CEUR-WS.org; 2008. Comparing SNOMED CT and the NCI Thesaurus through Semantic Web Technologies. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of data: Disambiguated UMLS2009AA equivalence mappings (2454 from 3024) between FMA and NCI ontologies. File Format: OWL (Ontology Web Language)
Description of data: Disambiguated UMLS2009AA equivalence mappings (4995 from 9072) between FMA and SNOMED CT ontologies. File Format: OWL (Ontology Web Language)
Description of data: Disambiguated UMLS2009AA equivalence mappings (6264 from 19622) between NCI and SNOMED CT ontologies. File Format: OWL (Ontology Web Language)