Selection among competing alternatives is always interesting, but when organisms select between K+ and Na+ for transport across biological membranes, it is especially intriguing for a couple of reasons (Hille, 2001). First, the results are physiologically significant. Second, K+ and Na+ are nearly as similar as they could be while not being the same things. Perhaps the separation of isotopes of the same chemical species is more demanding. Despite decades of research, the question remains: how do K+-selective ion channels catalyze K+ movement but recognize the detailed molecular-scale differences of Na+ and discriminate against it? Suggestions based on channel size and coordination chemistry (Bezanilla and Armstrong, 1972; Eisenman and Horn, 1983) have been available for a long time, but the determination of a KcsA K+ channel crystal structure (Doyle et al., 1998) enabled molecularly specific modeling studies of this K+/Na+ selectivity. In the subsequent flood of computational studies, finding consistency in results and interpretations has proven challenging. Here, we describe our perspective on how molecular modeling has advanced our understanding of the specific chemical and structural design elements of biological molecules that enable selective ion transport.
Background
The simplest idea for K+/Na+ discrimination is just that these ions differ in size. Because permeation of the K+ ion is preferred over the slightly smaller Na+ ion, the possibility that K+/Na+ selectivity occurs because the channel is too narrow is quickly discarded. Instead, we must consider the energetic barriers that discourage Na+ ions from crossing the membrane (Bezanilla and Armstrong, 1972). Clearly, nature has capitalized on the size difference between the two ions to create energetic barriers for Na+, leaving K+ with a less resistive conduction pathway, but how?
Complications for explaining selectivity arise because the family of K+ channels exhibits a range of selectivities, from weakly selective HCN pacemaker K+ channels to strongly selective “maxi”-type K+ channels or their bacterial homologue, KcsA. Fig. 1 A illustrates the structure of a representative K+ channel (Zhou et al., 2001), the family of proteins primarily responsible for passive K+-selective transport. The narrowest region of the pore, referred to as the selectivity filter, is understood to impart K+ selectivity, although other portions of the channel also play a role. Diverse K+ channels typically share this filter architecture (Hille, 2001) according to sequence alignment (Shealy et al., 2003), x-ray structures of a variety of K+ channels, and a vast amount of physiological data. In light of this structural similarity, how does the variability in selectivity come about?
Figure 1.
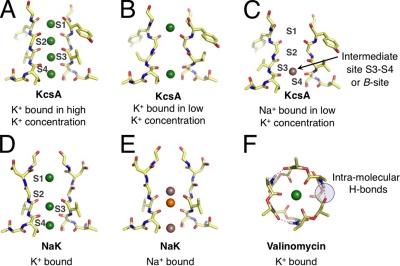
Representative binding modes of Na+ and K+ ions in structural motifs of K+-selective membrane transport molecules. (Note that only two units from the tetrameric selectivity filters are shown for clarity.) (A) The selectivity filter of KcsA (Zhou et al., 2001) adopts different configurations under conditions of high and (B) low K+ concentrations, presenting K+ with different sets of binding modes. (C) Under rare conditions when Na+ binds to the KcsA filter, Na+ prefers a binding site different from K+ (Nimigean and Miller, 2002; Shrivastava et al., 2002; Lockless et al., 2007; Thompson et al., 2009). (D) The bacterial NaK channel, which belongs to the family of CNG channels, has a selectivity filter architecture similar to KcsA, but is only weakly selective for K+. Initially, low temperature x-ray data suggested binding modes for K+ that are identical to Na+. (E) Newer higher resolution crystallographic studies show more variety in Na+ binding, attributing electron density at the S3 site to competitive binding of a contaminant (orange) with Na+, with other Na+-binding sites between planes of carbonyl or hydroxyl oxygens (Alam and Jiang, 2009). (F) In comparison to KcsA and the NaK channel, the K+-selective bacterial toxin molecule (Dobler, 1981), valinomycin, binds K+ differently, using six (or fewer) carbonyl oxygens, instead of eight (or fewer) as in KcsA and NaK.
The length of the selectivity filter with several evident ion-binding locations, and the known multiple-ion occupancy of the selectivity filter (Doyle et al., 1998; Åqvist and Luzhkov, 2000; Bernèche and Roux, 2001; Morais-Cabral et al., 2001; Zhou et al., 2001) provide yet further complications for explaining selectivity. Both kinetic modeling and free energy calculations show that multi-ion interactions within the filter affect the selective movement of K+ (Åqvist and Luzhkov, 2000; Morais-Cabral et al., 2001). In addition, K+ and Na+ have multiple binding modes within the dense dipole array of the carbonyl and hydroxyl-lined selectivity filter (Fig. 1, A–C). These underlie the complex behavior of K+ channels. For example, in the rare event that Na+ overcomes a barrier to entry into the filter, it obstructs K+ conductance (Nimigean and Miller, 2002; Thompson et al., 2009). This suggests that, in addition to barriers, the filter also presents Na+ with some binding modes that, in fact, compete with K+.
The goal of understanding ion selectivity extends beyond explaining the activities of highly selective K+ channels. CNG channels share the same basic architecture of the K+ channel selectivity filter (Fig. 1, D and E). Despite their similarity, the CNG channels exhibit no K+/Na+ selectivity (Alam and Jiang, 2009). How do the subtle differences between the CNG and K+ channels lead to the inability of CNG channels to select for K+? In contrast, the x-ray structure of a bacterial toxin (Dobler, 1981), valinomycin, bears little resemblance to the selectivity filters of K+ and CNG channels (Fig. 1 F). Nevertheless, valinomycin binds K+ ions selectively over Na+ ions and transports them, one by one, across the membrane. Furthermore, experiments show that the selectivity of valinomycin is sensitive to solvent properties. What explains the behavior of this ionophore?
Given the complexity in structure and the variability in selectivity of these biological molecules, it helps to focus on model systems where specific design principles may be isolated and their ramifications for selectivity directly tested. By determining ion-binding free energies from carefully specified structural and chemical elements, we can seek clear physical ideas to guide our understanding of design principles that give rise to macroscopic behaviors of ion transport molecules. The study of simplified binding site models, with implicit or explicit inclusion of environmental effects, provides a natural route for learning about the underpinnings of selective ion transport, even without proceeding to the selectivity of any specific transport case.
Whole molecule approaches such as electrophysiological or thermodynamic measurements, or computer simulations of entire channels, characterize in situ selectivity. These can also provide tests of mechanistic hypotheses, as illustrated with the experiments by Valiyaveetil et al. (2006) and molecular simulations by Fowler et al. (2008).
In light of these considerations, we present here progress that has resulted from studies that consider simplified model systems and how these studies, alongside traditional whole molecule observations, can advance our understanding of selective K+ movement across membranes. We begin our discussion by defining a thermodynamic measure of K+ selectivity over Na+.
Free energy of ion selectivity
The free energy difference, where Gx is the free energy for ion X, quantifies the thermodynamic preference for K+ versus Na+ in a medium, e.g., a solvent, a membrane protein, or an ionophore. Given additionally the free energy cost of replacing K+ with Na+ in bulk liquid water, the selectivity of the medium relative to liquid water for K+ over Na+ can be given in terms of the selectivity free energy,
| (1) |
This is simply a comparison of transfer free energies. If ΔΔG > 0, then the medium favors K+, if ΔΔG < 0, the medium favors Na+, and if ΔΔG = 0, then the medium exhibits no preference for either ion.
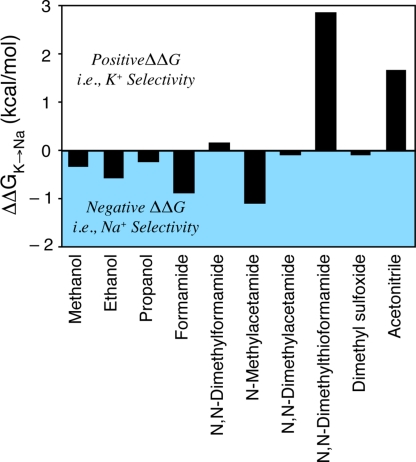
In a bulk liquid setting, the experimental free energy differences are unambiguous. For example, Fig. 2 displays the selectivity free energy for a variety of organic solvents relative to water as determined from experiments. As noted previously (Varma and Rempe, 2007; Bostick and Brooks, 2009; Asthagiri et al., 2010), some solvents display positive (K+) selectivity and some display negative (Na+) selectivity. The organic solvents composed of molecular analogues of the protein backbone, formamide and N-methylacetamide (NMA), are particularly interesting. These coordinate K+/Na+ with carbonyl oxygen atoms and provide Na+ selectivity. Also providing Na+ selectivity are the liquids that coordinate these ions with hydroxyl oxygen atoms, including methanol, ethanol, and propanol.
Figure 2.
Experimental estimates of selectivity free energy, for different organic solvents with respect to bulk liquid water (Cox and Parker, 1973; Marcus, 1983; Schmid et al., 2000; Yu et al., 2010a). For these estimates, is the partial molar Gibbs free energy for ion X in the specified medium relative to an ideal standard state, so that ΔG vanishes when ion–medium interactions vanish.
These simple observations alone tell us something important about K+ channels and valinomycin, which bind K+ selectively. Despite coordinating the cation (K+ or Na+) with carbonyl oxygen atoms (Fig. 1), the binding sites in these biological molecules, and the nonselective CNG channels, do not behave like liquid formamide or NMA. If they did, they would be Na+ selective. A model of K+/Na+ selectivity must, therefore, explain the design differences between the K+-selective sites of these molecules and the liquid states of their chemical analogues.
In contrast to liquid solutions, an ion-binding site in a channel is specified relative to the surrounding protein matrix and defined by the experimental crystal structure. Of course, the ion and the protein matrix, or ionophore matrix, jointly establish the binding configuration. Thermal motion, apparent in molecular dynamics simulation, blurs ideal geometrical consideration of a binding site. The inherent structure concept (Stillinger and Weber, 1982) eliminates that thermal blurring by cataloging basins of optimized configurations that are visited by the thermal motion. Inherent structure analysis has been widely implemented over recent decades (Rao and Karplus, 2010). Quasi-chemical theory, which is relevant to the ion selectivities discussed here (Asthagiri et al., 2010), follows directly from inherent structure concepts (Hummer et al., 1997). If the system presents more than one binding mode, perhaps including multi-ion binding, these additional complications can be treated by calculating a set of structurally constrained free energies (Rogers and Rempe, 2011).
Free energy differences, determined experimentally, are not as clear for binding sites as in liquids. For example, some experimental methods (Neyton and Miller, 1988; Lockless et al., 2007) probe the equilibrium thermodynamics of ion binding in K+ channels. Nonequilibrium electrophysiological techniques (Latorre and Miller, 1983; LeMasurier et al., 2001) might also be used to infer effective K+- and Na+-binding constants in K+ channel filters. These inferences rely on mechanistic assumptions about K+/Na+ binding in the filter—assumptions about the conformation of the filter, the number of binding sites and their characteristics, and about what other components such as water and additional ions might be doing. The determination of driving forces for selective ion binding, ΔΔG, is simplest in theoretical and computational settings where it is clear exactly which binding site, conformation, and ion/water occupancy is being considered. In these cases, simple theoretical models and hypotheses pertaining to the design of K+-selective complexes may be unambiguously evaluated in terms of ΔΔG.
When reconciling information from approaches based on simplified models or whole channels, it is important to keep in mind that the optimal binding sites of K+ and Na+ could be different. For example, in the K+ and NaK channels (Fig. 1), the K+ ion finds a position to organize eight ligands in the selectivity filters, whereas a Na+ ion optimally uses fewer ligands. Being distinct from each other, the optimal binding configuration for K+ can be a barrier configuration for Na+. The K+-binding regions—barriers for Na+—can be part of the larger explanation for the low permeability of Na+ (Bezanilla and Armstrong, 1972). Overlooking this important point can lead to discrepancies (Bucher and Rothlisberger, 2010) in correlating coordination structure and selectivity.
Here, we will analyze the design principles that have emerged to explain K+/Na+ selectivity. We start with a host–guest steric model (also known as “snug fit”), proposed decades before a K+ channel was crystallized, that has been used to explain the selectivity of both K+-selective ionophores and K+ channels.
A host–guest steric model
The first x-ray structure of a strongly selective K+ channel, KcsA (Doyle et al., 1998), produced a tangible picture of the molecular bases of K+ channel activity. Initial inspection of the structure supported an earlier hypothesis that the K+/Na+ selectivity in K+ channels might be attributable to a steric mechanism (Bezanilla and Armstrong, 1972). The specific positions of the K+ ions in the selectivity filter indicated that K+ ions preferred to occupy positions coordinated by eight carbonyl oxygen atoms (Fig. 1 A). Because of the relatively stronger van der Waals interactions of K+ compared with Na+ at this distance, a selective environment might result if the oxygen atoms of these sites maintained a cavity size that snugly fit the radius of K+ but did not collapse onto the smaller Na+ ion, as the central assumption to the steric model demands. Thus, a binding site designed to maintain a K+-specific cavity size would hypothetically provide the necessary energetic barrier to Na+ passage across the membrane.
Additional crystal diffraction studies produced KcsA structures that contained a K+ ion trapped in the water-filled central cavity of the structure, which also coordinated a cage of exactly eight water oxygen atoms. This finding reinforced the notion that the eightfold carbonyl-lined sites in the filter served as surrogate hydration shells for K+, mimicking the coordination geometry of K+ in the aqueous phase, and thus favoring its fast selective movement (Morais-Cabral et al., 2001; Zhou et al., 2001). This view of ion partitioning, however, eventually conflicted with new theoretical and experimental data (Varma and Rempe, 2006) showing that the most probable coordination number of both Na+ and K+ in liquid water is fewer than eight, and that coordination of Na+ or K+ with eight water molecules is unlikely in the aqueous phase (Bostick and Brooks, 2007; Varma and Rempe, 2007).
Molecular dynamics simulations of the whole channel embedded in a surrounding membrane (Guidoni et al., 1999; Shrivastava and Sansom, 2000; Bernèche and Roux, 2001) also demonstrated that fluctuations of the selectivity filter backbone atoms are larger than the size difference in Pauling radii between Na+ and K+ ions, which appeared to conflict with the host–guest steric mechanism of selectivity. Although this cavity-size argument does not satisfactorily explain K+/Na+ selectivity in the eightfold binding sites of K+ channels, quantum mechanical calculations subsequently demonstrated how this mechanism is operative in the bacterial ionophore molecule, valinomycin (Varma et al., 2008).
Valinomycin provides only six carbonyl ligands for ion coordination (Fig. 1 F). Experimental and simulation studies concur that the ionophore achieves K+/Na+ selectivity by physically resisting collapse onto the smaller Na+ ion. If the ligands are given complete freedom in simplified models, binding sites composed of six carbonyl ligands do not bind K+ selectively. Quantum mechanical studies of the whole molecule showed that geometric constraints on the carbonyl ligands in valinomycin are enforced through a combination of intramolecular hydrogen bonds and other structural features, including a specific ring size and the spacing between connected ligands. Competition with solvent can decrease the stability of valinomycin’s intramolecular hydrogen bonds, resulting in a selectivity that depends on solvent hydrogen-bonding strength. Although this solvent-dependent selectivity of valinomycin was observed experimentally decades ago (Eisenman et al., 1991), an explanation for the role of the surrounding environment arose only recently (Varma et al., 2008).
Other selectivity concepts matured alongside the steric mechanism for K+ channel selectivity (Bezanilla and Armstrong, 1972). We focus our discussion on the ideas developed by Eisenman and coworkers in the context of K+/Na+ selectivity.
The conventional field strength model
Studies by Eisenman on the effect of ligand chemistry, or field strength, on the ion selectivity of functionalized surfaces (Eisenman and Horn, 1983) led to two important conclusions: (1) “the primary physical variable controlling equilibrium cationic specificity is the field strength of the anion (i.e., the electronegative coordinating ligand),” and (2) “the primary factor controlling the magnitude of selectivity among ions at a given field strength is the amount of water admitted into the vicinity of the site.”
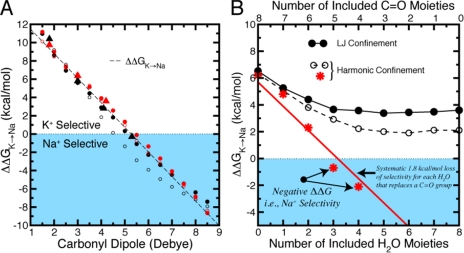
If this conventional ligand field strength model is applied to a K+/Na+-selective binding site, then decreasing the field strength of the ligand should increase selectivity for the larger K+ ion. Quantum mechanical calculations confirmed this trend for a hypothetical ion–ligand system in which substitution of ion-coordinating formamide by water (a carbonyl-to-water substitution) increases K+/Na+ selectivity (Varma and Rempe, 2007). The same behavior also occurred upon varying the ligand field strength in simulations (Fig. 3 A) of a simplified binding site of eight nonpolarizable linear ligands confined within a sphere of radius 3.5 Å around a central ion (Noskov et al., 2004; Thomas et al., 2007; Bostick et al., 2009).
Figure 3.
Dependence of K+/Na+ selectivity on ligand composition in eight-ligand binding site models where ligand distances are generically confined to be within a 3.5-Å radius of the central ion. (A) The conventional field strength trend observed by Eisenman is illustrated by the dependence of selectivity, ΔΔG, on the dipole moment of linear ligands. Results were taken from binding site models described previously (red triangles, Noskov et al., 2004; black triangles, Thomas et al., 2007; circles were calculated with standard [black] or modified [red] CHARMM parameters, Bostick et al., 2009). Filled circles show results from a half-harmonic boundary restraint, and open circles show the corresponding Lennard-Jones restraint. The conventional field strength trend (dashed line) is independent of these different restraints. (B) Dependence of K+/Na+ selectivity, ΔΔG, on incremental replacement of carbonyl-like dipolar groups with water molecules. In contrast to the trend in A, recent work predicted a systematic loss of selectivity for each water molecule that replaces a carbonyl group (∼1.8 kcal/mol per water using the CHARMM force field) (Noskov and Roux, 2006, 2007). The red line illustrates this trend toward Na+ selectivity, which supports the revised field strength model. Data from subsequent calculations (Bostick et al., 2009), using the same force field and either a Lennard-Jones (LJ, black solid lines/circles) or a half-harmonic (black dashed lines and open circles) confining potential, do not eliminate the K+ selectivity.
Although these results illustrate the promise of ligand field strength arguments in explaining K+ selectivity, it is important to remember that a successful model should explain the design differences between K+-selective binding sites of molecules and the liquid states of their chemical analogues (Fig. 2).
A revised field strength model
In computational investigations of KcsA, one study concluded that “the carbonyl groups coordinating the ion in the narrow pore are indeed very dynamic (‘liquid-like’) and that their intrinsic electrostatic properties control ion selectivity” (Noskov et al., 2004). By bringing ligand flexibility into consideration with a ligand field strength model, extended to include not only ion–ligand but also ligand–ligand interactions, this work demonstrated that a thermally fluctuating binding site could be selective. Although not a complete solution, the focus on simplified binding site models suggested a path for understanding the elusive selectivity problem.
Separating out electrostatic interactions in computations can be problematic (Onsager, 1939), however, because classical electrostatic interactions between charges are balanced non-uniquely by dispersion and core repulsion in force fields (MacKerell, 2005). Nevertheless, observing a correlation between strong ligand–ligand electrostatic interactions and high selectivity might suggest an enhancement of the K+ selectivity in binding sites composed of strongly dipolar carbonyls compared with the case of lower-dipole water ligands available in solution. Because this trend would contrast with the simple conventional ligand field strength model (see Figs. 2 and 3 A), we refer to this description as a revised field strength model.
A test of this revised field strength explanation of selectivity for weakly selective CNG (NaK) channels used free energy perturbation simulations (Noskov and Roux, 2006, 2007) in which eight linear carbonyl moieties around an ion were progressively replaced with molecules of a weaker ligand field strength, water. Those substitutions resulted in a loss and reversal of K+/Na+ selectivity (Fig. 3 B), which supported the view that increasing the ligand dipole moment or field strength (water to carbonyl) should enhance the free energy of K+ selectivity, ΔΔG.
Although attractive, this interpretation is difficult to reconcile with previously observed trends in simulations (Fig. 3 A) and experiments (Eisenman and Horn, 1983). Additionally, calculations (Fig. 3 B) using an identical methodology and force field to the initial works found that progressive substitution of linear carbonyl ligands with water molecules modifies the magnitude of K+ selectivity but does not abolish or reverse it. Reductions observed in the selectivity free energy were accounted for by differences in molecular geometry (e.g., size, shape, and charge distribution), which are controlled by the full balance of electrostatic, dispersion, and core repulsions in a force field model, rather than simply molecular dipole moment from electrostatic charge–charge interactions (Bostick et al., 2009). Although the disagreement in Fig. 3 B has not been resolved, more recent simulations showed that K+ selectivity persisted in water-based binding site models, even those composed purely of water (Bostick and Brooks, 2007, 2010; Varma and Rempe, 2007; Bostick et al., 2009).
Ligand composition clearly plays an important role in modulating selectivity (see Figs. 2 and 3), but a focus on chemical forces as a primary design principle does not account for differences in K+/Na+ selectivity between systems possessing comparable ligand composition and/or field strength. Although the conventional field strength model anticipates the effect of ligand type on ion binding, and the revised field strength model predicts K+ selectivity in a fluctuating binding site of strong dipolar ligands, several questions remain. Specifically, why do binding sites in K+ channels composed of carbonyl ligands select for K+, but liquids of carbonyl analogues, such as NMA and formamide, inherently select for Na+ (see Fig. 2 and Table I)? Furthermore, why do the chemically identical binding sites (i.e., sites S1, S2, and S3) of the K+ channel selectivity filter exhibit varying degrees of selectivity noted in whole channel simulations (Noskov et al., 2004), and why do NaK channels show reduced selectivity despite sharing two chemically identical binding sites with strongly selective K+ channels? The parsimonious conclusion is that the focus on ligand type is incomplete; the role of the surrounding matrix must be incorporated into a model that explains selectivity.
Table I.
K+/Na+ selectivity by experiment and calculation
| Method | Solvent | ΔΔG (kcal/mol) | Selective for |
| Formamidea | −0.9 | Na+ | |
| Experiment | Formamidebc | −0.7 | Na+ |
| NMAbd | −1.1 | Na+ | |
| Formamidec (CHARMM27) | 0.9 | K+ | |
| Formamidec (OPLS-AA) | 1.3 | K+ | |
| Pairwise Additive | NMAe (CHARMM22) | 1.6 | K+ |
| NMAcd (CHARMM27) | 2.8 | K+ | |
| NMAcd (CHARMM27/NBFIX) | 4.7 | K+ | |
| Formamidec (AMOEBA) | −1.3 | Na+ | |
| Polarizable | Formamidef (QM/MM) | −2.2 | Na+ |
| NMAd (Drude) | −0.3 | Na+ |
K+/Na+ selectivity of bulk liquid formamide and NMA as determined by experimental work, pairwise-additive (nonpolarizable) force field calculations, polarizable force field (AMOEBA and Drude) calculations, and QM/MM (hybrid quantum mechanical and molecular mechanical) calculations. Note that the pairwise-additive force field calculations display positive (K+) selectivity, whereas all other determinations display negative (Na+) selectivity.
A primary role for the surrounding matrix
Recent studies shed light on issues concerning variable selectivity in systems possessing comparable ligand composition or field strength. Using physiologically motivated ion-binding site models, several investigations suggested that external forces derived from the surrounding environment and exerted on the ion-ligated complex play a primary role in determining selectivity (Bostick and Brooks, 2007, 2009; Bostick et al., 2009; Thomas et al., 2007; Varma and Rempe, 2007, 2008; Varma et al., 2008). The most unambiguous demonstrations of these principles explicitly include electronic polarization (Guidoni and Carloni, 2002; Bostick and Brooks, 2010; Varma and Rempe, 2010) because the non-transferability of pairwise-additive force fields can lead to discrepancies (see Table I).
Returning to Fig. 2, we notice that ionic selectivities in bulk liquids, particularly for the carbonyl ligands, provide a control that illustrates K+/Na+ selectivity in the absence of macromolecular structure. Because selectivity differs between organic solvents and K+ channels, it must be concluded that the environment surrounding an ion-binding site is also necessary for explaining the selectivity of K+ channels. The challenge then centers on understanding physiological mechanisms that modulate binding site properties, and thus modulate selective ion binding.
Interactions between the ion–ligand complex and the remainder of the protein, membrane surfaces, other ions, or the solution can be modeled as external fields acting on the ion and its coordinating ligands. From the quasi-chemical development (Asthagiri et al., 2010) and other statistical mechanical theories (Bostick and Brooks, 2009), it is clear that these interactions need not be pair-decomposable. The effects of these interactions have been referred to in the literature as “topological” or “architectural” control (Bostick and Brooks, 2007, 2009, 2010; Thomas et al., 2007; Varma and Rempe, 2007; Varma et al., 2008; Bostick et al. 2009). Those interactions typically affect both the selectivity free energy and the structural attributes apparent in Fig. 1, such as the coordination number the ion–ligand complex adopts, the orientations of its ligands, or its deformability. In fact, recent crystallographic results confirm that the structures of channel-binding sites are not fixed; instead, the KcsA selectivity filter can take on radically different configurations depending on the surroundings (Cordero-Morales et al., 2006).
The constrained cavity size of valinomycin effected through intra-molecular hydrogen bonds, illustrates one particular form of architectural control (Varma et al., 2008). Constraints imposing over-coordination with respect to liquid solutions provide yet another example of architectural control (Bostick and Brooks, 2007; Varma and Rempe, 2007).
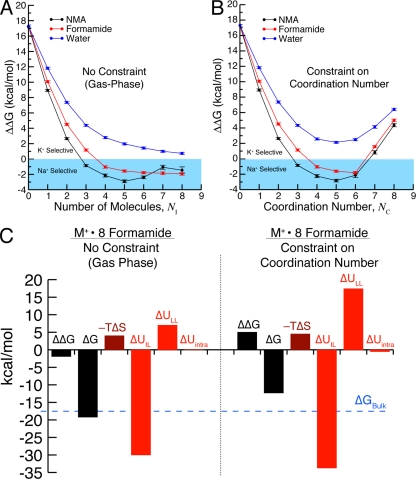
Simulations of binding site models illustrate the consequences of constraints on coordination (Fig. 4). Coordination by five or six carbonyl oxygen atoms typically provides the weak selectivity observed in bulk liquids (Fig. 2), whereas strong K+ selectivity is associated with seven and eight coordinate complexes that resemble K+ channel architectures (Figs. 1 A and 4 B). In the absence of external forces imposing over-coordination, carbonyl ligands naturally assume a lower coordination number around each ion and K+/Na+ selectivity is lost (Fig. 4 A). Quantum mechanical studies of a model binding site composed of diglycine molecules show the same trends with Na+ binding (Varma and Rempe, 2007). These results support the argument that an environment that achieves high coordination numbers results in K+/Na+ selectivity, whereas an environment that permits relaxed coordination numbers produces lower selectivity. Note that this does not suggest that selectivity, in general, is solely controlled by coordination number: other types of constraints on an ion-bound complex can also play a role in selectivity, and ligand chemistry remains an important modulatory factor.
Figure 4.
Results from binding site models demonstrating the effect of an external field, in the form of an ion coordination number constraint, on K+/Na+ selectivity. Calculations were performed using the AMOEBA polarizable force field (Bostick and Brooks, 2010). (A) ΔΔG versus the number of included molecules, NI, in gas-phase clusters around K+ and Na+ in the absence of a constraint on coordination. As NI increases, the observed selectivity approaches values expected for bulk liquids (for water, ΔΔG ≈ 0; for formamide/NMA, ΔΔG < 0). Note that the number of included molecules, NI, is not necessarily equal to the number of coordinating molecules, NC, because of the absence of a coordination constraint. (B) ΔΔG versus the number of molecules, NC, directly coordinating K+ and Na+. In agreement with quantum mechanical calculations (Varma and Rempe, 2007, 2008), ΔΔG is larger in the water-based models (blue) than in the carbonyl-based models (black and red). Because of the presence of an external field (half-harmonic confinement) that imposes a specific coordination number, K+ selectivity is observed for seven or more ligands. In the models that coordinate K+ or Na+ with carbonyl ligands, K+ selectivity is determined by the external field rather than the ligand identity (NMA, formamide, and water) because the binding site models are Na+ selective in the absence of the constraint on coordination number, NC. (C) Contributions from specified individual components of the selectivity free energy (Eq. 2) in models composed of eight formamide molecules: ligand–ligand interactions, ion–ligand interactions, intramolecular interactions, and entropy In the case considered here, the contribution from the external field, is negligible. In the absence of the field (left), the components yield net Na+ selectivity (ΔΔG < 0). When a field enforces eightfold coordination, the distribution of these individual components changes, producing net K+ selectivity (ΔΔG > 0). Thus, the redistribution of the individual components, and therefore the net K+ selectivity, is an effect of the applied external field.
To probe the energetic details giving rise to selectivity, the K+ selectivity free energy can be parsed in several ways (see, for example, Asthagiri et al., 2006; Bostick and Brooks, 2007; Varma and Rempe, 2007; Rogers and Rempe, 2011). One way is to decompose the selectivity free energy into contributions from different types of interactions in the force field. Using cases considered in Fig. 4 as an example, this decomposition can be written as (Bostick and Brooks, 2010),
| (2) |
Here, is the internal energy contribution from an imposed external field, and are contributions from ion–ligand interactions, ligand–ligand interactions, and intramolecular interactions, respectively, and is the entropic contribution. Decomposition of the selectivity free energy in this form shows several large contributions (Fig. 4 C), with the unfavorable ligand–ligand interactions () providing the largest contribution favoring K+ selectivity.
These results might be interpreted as suggesting that the ligand identity controls K+/Na+ selectivity through local interactions, as proposed by the revised field strength model. It is, however, important to note that each contribution in Eq. 2, including ligand–ligand interactions depends on the constraints imposed on the ion–ligated complex—in the case of Fig. 4, a constraint on coordination number (Bostick et al., 2009; Bostick and Brooks, 2010). In fact, without requiring the larger coordination number, these various contributions should combine to yield net Na+ selectivity, consistent with the experimental liquid solution results of Fig. 2. Fig. 4 C explicitly demonstrates the shift in contributions caused by imposing a constraint on coordination number for a test case of eight formamide molecules interacting with K+ and Na+.
Acknowledging the influence of the environment in ion binding, the question remains: how are those interactions involved in K+ channels? Protein composition and water occupancy help determine properties of the environment surrounding channel-binding sites. Significant environmental characteristics that could modulate these binding sites include, for example, steric and electrostatic interactions that define the flexibility of the local ion–ligand complex, and the distribution of functional groups that can hydrogen bond with the ion-coordinating ligands (Varma and Rempe, 2007, 2008).
During formation of an ionic binding site complex, ligands are extracted from the exterior environment and rearranged. Depending on characteristics of the environment, this ligand extraction step can be associated with high free energy penalties, even when the overall process of ion solvation is favorable. Computational studies (Varma and Rempe, 2007, 2008) demonstrate that alteration of these penalties affects the distribution of ligands in the ion complex. For example, reducing extraction penalties increases the stabilities of high coordination numbers, thus bringing down the energetic cost of transferring ions from water into over-coordinated binding sites. A significant reduction in this penalty for binding site formation can even drive up preferred coordination numbers. In contrast, an increased penalty can lower ion coordination preferences, as observed in highly concentrated salt solutions.
In biomolecules that bind ions in over-coordinated states, the free energy penalties for extracting ligands can be lowered by reducing direct favorable interactions of the ligands with atoms other than the ions themselves. In the case of highly selective K+ channels, a bioinformatics and molecular simulation analysis (Varma and Rempe, 2008) suggests that lowered ligand-extraction penalties that stabilize high coordinations are accomplished by a lowered density of free hydrogen bond donors near the carbonyl ligands of the binding sites, and by a constraint on the motions of selectivity filters and their neighboring environments.
Furthermore, a sequence alignment of K+ channels (Shealy et al., 2003) indicates specific chemical differences in hydrogen-bonding ability between the ion-binding neighborhoods of strongly and weakly selective channels. In weakly selective channels, the immediate neighborhoods of the ion-coordinating carbonyl ligands contain specific amino acids (hydrogen bond donors) that can form hydrogen bonds with those ligands. These donors are typically absent in strongly selective K+ channels. Hydrogen bond–donating groups in weakly selective K+ channels can reduce the availability of ligands to bind ions, resulting in reduced selectivities. In contrast, the presence of proximal hydrogen bond–accepting groups can enhance K+ selectivity by constraining optimal filter orientation, as predicted earlier (Varma and Rempe, 2007, 2008) and shown recently via electrophysiology and crystallography (Cheng et al., 2011). This provides one mechanism for the potassium channel family to exhibit a range of selectivities, despite sharing a common filter architecture (Varma and Rempe, 2007).
Alternatively, the observed diversity in selectivity exhibited by K+ channels could also arise from differences in the flexibilities of the ion–ligand complexes, which are influenced by the external environment as well (Bostick and Brooks, 2007; Varma and Rempe, 2007). Observations of large shifts in the crystal structures of KcsA selectivity filters with changes to the surrounding protein matrix lend support to this proposition (Cordero-Morales et al., 2006; Cheng et al., 2011).
Yet another environmental factor that can modulate a site’s selectivity is the availability of water molecules to a binding site. Because water molecules can serve as both hydrogen bond donors or acceptors, their presence provides external interactions that can reduce selectivity by either competing with ion-coordinating ligands for the ion, or by competing with the ion for ion-coordinating ligands (Bostick and Brooks, 2007; Varma and Rempe, 2007). This provides an explanation for the historical observations of the modulating effect of water exposure on ion selectivity (Eisenman and Horn, 1983).
In alignment with earlier predictions (Bostick and Brooks, 2007; Varma and Rempe, 2007), computer simulations of membrane-embedded whole channels (Fowler et al., 2008) indicated that the lack of K+ selectivity in CNG channels is accounted for by the combined effect of the enhanced exposure of binding sites to water and the enhanced flexibilities of the ion-binding sites in their selectivity filters, both of which foster lower coordination numbers for K+ and Na+.
Summary and conclusions
The advent of atomic resolution structures of ion-selective channels made possible the transition from black box models to molecular descriptions for considering the design principles underlying selectivity in a flexible selectivity filter. Simple theoretical models applied to interpret the complex behaviors observed in whole molecule experiments and molecular simulations led to suggestions that ligand type is of primary importance in determining selectivity. As discussed in this Perspective, this view is incomplete. Models that focus on chemical forces as a primary design principle do not explain why carbonyl ligands produce K+ selectivity in eightfold K+ channel–binding sites, but yield Na+ selectivity in liquid analogues (Fig. 2). Additionally, these models have not explained why chemically identical binding sites in the strongly selective KcsA channel show significantly different selectivities in simulations of whole channels. The same question applies to NaK channels, which lack selectivity despite sharing two chemically identical binding sites with strongly selective K+ channels. We are left with the conclusion that ion selectivity requires consideration of both ligand characteristics and the forces influencing binding site composition and structure.
Recent simulations support the view that interactions with the more distant environment of the membrane transport molecule can modify properties of the binding site and influence selective binding. In the case of K+ channels, restricted carbonyl motion or a decrease in their availability to the protein environment can drive up ion coordination numbers in the selectivity filter, leading to K+ selectivity. A snug fit as in valinomycin can be enforced by an environment that stabilizes intra-molecular hydrogen bonding, achieving selectivity without over-coordination through a constraint on cavity size. The importance of environmental controls on binding sites has since been recognized in several recent works (see, for example, Fowler et al., 2008; Miloshevsky and Jordan, 2008; Vora et al., 2008; Dixit et al., 2009; Dudev and Lim, 2009; Yu and Roux, 2009; Yu et al., 2009, 2010b; Roux, 2010; Rogers and Rempe, 2011). This Perspective provides a foundation necessary for understanding the more complex behavior of selective ion transport through membranes.
This Perspectives series includes articles by Andersen, Alam and Jiang, Nimigean and Allen, Roux et al., and Dixit and Asthagiri.
Acknowledgments
D.M. Rogers and S.B. Rempe acknowledge funding from the National Institutes of Health (NIH) through the NIH Road Map for Medical Research and the Sandia Laboratory-Directed Research and Development program. Sandia National Laboratories is a multi-program laboratory operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-AC04-94AL85000.
Footnotes
Abbreviations used in this paper:
- NMA
- N-methylacetamide
References
- Alam A., Jiang Y. 2009. Structural analysis of ion selectivity in the NaK channel. Nat. Struct. Mol. Biol. 16:35–41 10.1038/nsmb.1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åqvist J., Luzhkov V. 2000. Ion permeation mechanism of the potassium channel. Nature. 404:881–884 10.1038/35009114 [DOI] [PubMed] [Google Scholar]
- Asthagiri D., Pratt L.R., Paulaitis M.E. 2006. Role of fluctuations in a snug-fit mechanism of KcsA channel selectivity. J. Chem. Phys. 125:24701 10.1063/1.2205853 [DOI] [PubMed] [Google Scholar]
- Asthagiri D., Dixit P.D., Merchant S., Paulaitis M.E., Pratt L.R., Rempe S.B., Varma S. 2010. Ion selectivity from local configurations of ligands in solutions and ion channels. Chem. Phys. Lett. 485:1–7 10.1016/j.cplett.2009.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche S., Roux B. 2001. Energetics of ion conduction through the K+ channel. Nature. 414:73–77 10.1038/35102067 [DOI] [PubMed] [Google Scholar]
- Bezanilla F., Armstrong C.M. 1972. Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons. J. Gen. Physiol. 60:588–608 10.1085/jgp.60.5.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick D.L., Brooks C.L., III 2007. Selectivity in K+ channels is due to topological control of the permeant ion’s coordinated state. Proc. Natl. Acad. Sci. USA. 104:9260–9265 10.1073/pnas.0700554104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick D.L., Brooks C.L., III 2009. Statistical determinants of selective ionic complexation: ions in solvent, transport proteins, and other “hosts”. Biophys. J. 96:4470–4492 10.1016/j.bpj.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick D.L., Brooks C.L., III 2010. Selective complexation of K+ and Na+ in simple polarizable ion-ligating systems. J. Am. Chem. Soc. 132:13185–13187 10.1021/ja106197e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick D.L., Arora K., Brooks C.L., III 2009. K+/Na+ selectivity in toy cation binding site models is determined by the ‘host’. Biophys. J. 96:3887–3896 10.1016/j.bpj.2008.12.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D., Rothlisberger U. 2010. Molecular simulations of ion channels: a quantum chemist’s perspective. J. Gen. Physiol. 135:549–554 10.1085/jgp.201010404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W.W.L., McCoy J.G., Thompson A.N., Nichols C.G., Nimigean C.M. 2011. Mechanism for selectivity-inactivation coupling in KcsA potassium channels. Proc. Natl. Acad. Sci. USA. 108:5272–5277 10.1073/pnas.1014186108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Cuello L.G., Zhao Y., Jogini V., Cortes D.M., Roux B., Perozo E. 2006. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13:311–318 10.1038/nsmb1069 [DOI] [PubMed] [Google Scholar]
- Cox B.G., Parker A.J. 1973. Solvation of ions. XVII. Free energies, heats, and entropies of transfer of single ions from protic to dipolar aprotic solvents. J. Am. Chem. Soc. 95:402–407 10.1021/ja00783a015 [DOI] [Google Scholar]
- Dixit P.D., Merchant S., Asthagiri D. 2009. Ion selectivity in the KcsA potassium channel from the perspective of the ion binding site. Biophys. J. 96:2138–2145 10.1016/j.bpj.2008.12.3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobler M. 1981. Ionophores and Their Structures. John Wiley and Sons, Inc, New York: 379 pp [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Dudev T., Lim C. 2009. Determinants of K+ vs Na+ selectivity in potassium channels. J. Am. Chem. Soc. 131:8092–8101 10.1021/ja900168k [DOI] [PubMed] [Google Scholar]
- Eisenman G., Horn R. 1983. Ionic selectivity revisited: the role of kinetic and equilibrium processes in ion permeation through channels. J. Membr. Biol. 76:197–225 10.1007/BF01870364 [DOI] [PubMed] [Google Scholar]
- Eisenman G., Aqvist J., Alvarez O. 1991. Free energies underlying ion binding and transport in protein channels: free energy perturbation simulations of ion binding and selectivity for valinomycin. Faraday Transactions. 87:2099–2109 10.1039/ft9918702099 [DOI] [Google Scholar]
- Fowler P.W., Tai K., Sansom M.S.P. 2008. The selectivity of K+ ion channels: testing the hypotheses. Biophys. J. 95:5062–5072 10.1529/biophysj.108.132035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield A., Ren P., Ponder J.W. 2003. Ion solvation thermodynamics from simulation with a polarizable force field. J. Am. Chem. Soc. 125:15671–15682 10.1021/ja037005r [DOI] [PubMed] [Google Scholar]
- Guidoni L., Carloni P. 2002. Potassium permeation through the KcsA channel: a density functional study. Biochim. Biophys. Acta. 1563:1–6 10.1016/S0005-2736(02)00349-8 [DOI] [PubMed] [Google Scholar]
- Guidoni L., Torre V., Carloni P. 1999. Potassium and sodium binding to the outer mouth of the K+ channel. Biochemistry. 38:8599–8604 10.1021/bi990540c [DOI] [PubMed] [Google Scholar]
- Hille B. 2001. Ionic Channels of Excitable Membranes. Third Edition Sinauer Associates, Inc, Sunderland, MA, pp. 814 [Google Scholar]
- Hummer G., Pratt L.R., García A.E. 1997. Multistate Gaussian model for electrostatic solvation free energies. J. Am. Chem. Soc. 119:8523–8527 10.1021/ja971148u [DOI] [Google Scholar]
- Latorre R., Miller C. 1983. Conduction and selectivity in potassium channels. J. Membr. Biol. 71:11–30 10.1007/BF01870671 [DOI] [PubMed] [Google Scholar]
- LeMasurier M., Heginbotham L., Miller C. 2001. KcsA: it’s a potassium channel. J. Gen. Physiol. 118:303–314 10.1085/jgp.118.3.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless S.W., Zhou M., MacKinnon R. 2007. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 5:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Jr 2005. Empirical force fields for proteins: current status and future directions. Annu. Rep. Comput. Chem. 1:91–102 10.1016/S1574-1400(05)01007-8 [DOI] [Google Scholar]
- Marcus Y. 1983. Thermodynamic functions of transfer of single ions from water to nonaqueous and mixed solvents: part 1-Gibbs free energies of transfer to nonaqueous solvents. Pure Appl. Chem. 55:977–1021 10.1351/pac198355060977 [DOI] [Google Scholar]
- Miloshevsky G.V., Jordan P.C. 2008. Conformational changes in the selectivity filter of the open-state KcsA channel: an energy minimization study. Biophys. J. 95:3239–3251 10.1529/biophysj.108.136556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais-Cabral J.H., Zhou Y., MacKinnon R. 2001. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 414:37–42 10.1038/35102000 [DOI] [PubMed] [Google Scholar]
- Neyton J., Miller C. 1988. Potassium blocks barium permeation through a calcium-activated potassium channel. J. Gen. Physiol. 92:549–567 10.1085/jgp.92.5.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean C.M., Miller C. 2002. Na+ block and permeation in a K+ channel of known structure. J. Gen. Physiol. 120:323–335 10.1085/jgp.20028614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov S.Y., Roux B. 2006. Ion selectivity in potassium channels. Biophys. Chem. 124:279–291 10.1016/j.bpc.2006.05.033 [DOI] [PubMed] [Google Scholar]
- Noskov S.Y., Roux B. 2007. Importance of hydration and dynamics on the selectivity of the KcsA and NaK channels. J. Gen. Physiol. 129:135–143 10.1085/jgp.200609633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov S.Y., Bernèche S., Roux B. 2004. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 431:830–834 10.1038/nature02943 [DOI] [PubMed] [Google Scholar]
- Onsager L. 1939. Electrostatic interactions of molecules. J. Phys. Chem. 43:189–196 10.1021/j150389a001 [DOI] [Google Scholar]
- Rao F., Karplus M. 2010. Protein dynamics investigated by inherent structure analysis. Proc. Natl. Acad. Sci. USA. 107:9152–9157 10.1073/pnas.0915087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D.M., Rempe S.B. 2011. Probing the thermodynamics of competitive ion binding using minimum energy structures. J. Phys. Chem. B. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. 2010. Exploring the ion selectivity properties of a large number of simplified binding site models. Biophys. J. 98:2877–2885 10.1016/j.bpj.2010.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid R., Miah A.M., Sapunov V.N. 2000. A new table of the thermodynamic quantities of ionic hydration: values and some applications (enthalpy-entropy compensation and born radii). Phys. Chem. Chem. Phys. 2:97–102 10.1039/a907160a [DOI] [Google Scholar]
- Shealy R.T., Murphy A.D., Ramarathnam R., Jakobsson E., Subramaniam S. 2003. Sequence-function analysis of the K+-selective family of ion channels using a comprehensive alignment and the KcsA channel structure. Biophys. J. 84:2929–2942 10.1016/S0006-3495(03)70020-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava I.H., Sansom M.S.P. 2000. Simulations of ion permeation through a potassium channel: molecular dynamics of KcsA in a phospholipid bilayer. Biophys. J. 78:557–570 10.1016/S0006-3495(00)76616-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava I.H., Tieleman D.P., Biggin P.C., Sansom M.S.P. 2002. K+ versus Na+ ions in a K channel selectivity filter: a simulation study. Biophys. J. 83:633–645 10.1016/S0006-3495(02)75197-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillinger F.H., Weber T.A. 1982. Hidden structure in liquids. Phys. Rev. A. 25:978–989 10.1103/PhysRevA.25.978 [DOI] [Google Scholar]
- Thomas M., Jayatilaka D., Corry B. 2007. The predominant role of coordination number in potassium channel selectivity. Biophys. J. 93:2635–2643 10.1529/biophysj.107.108167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.N., Kim I., Panosian T.D., Iverson T.M., Allen T.W., Nimigean C.M. 2009. Mechanism of potassium-channel selectivity revealed by Na+ and Li+ binding sites within the KcsA pore. Nat. Struct. Mol. Biol. 16:1317–1324 10.1038/nsmb.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil F.I., Leonetti M., Muir T.W., Mackinnon R. 2006. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 314:1004–1007 10.1126/science.1133415 [DOI] [PubMed] [Google Scholar]
- Varma S., Rempe S.B. 2006. Coordination numbers of alkali metal ions in aqueous solutions. Biophys. Chem. 124:192–199 10.1016/j.bpc.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Varma S., Rempe S.B. 2007. Tuning ion coordination architectures to enable selective partitioning. Biophys. J. 93:1093–1099 10.1529/biophysj.107.107482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Rempe S.B. 2008. Structural transitions in ion coordination driven by changes in competition for ligand binding. J. Am. Chem. Soc. 130:15405–15419 10.1021/ja803575y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Rempe S.B. 2010. Multibody effects in ion binding and selectivity. Biophys. J. 99:3394–3401 10.1016/j.bpj.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Sabo D., Rempe S.B. 2008. K+/Na+ selectivity in K channels and valinomycin: over-coordination versus cavity-size constraints. J. Mol. Biol. 376:13–22 10.1016/j.jmb.2007.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora T., Bisset D., Chung S.H. 2008. Conduction of Na+ and K+ through the NaK channel: molecular and Brownian dynamics studies. Biophys. J. 95:1600–1611 10.1529/biophysj.107.126722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Roux B. 2009. On the utilization of energy minimization to the study of ion selectivity. Biophys. J. 97:L15–L17 10.1016/j.bpj.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Noskov S.Y., Roux B. 2009. Hydration number, topological control, and ion selectivity. J. Phys. Chem. B. 113:8725–8730 10.1021/jp901233v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Mazzanti C.L., Whitfield T.W., Koeppe R.E., II, Andersen O.S., Roux B. 2010a. A combined experimental and theoretical study of ion solvation in liquid N-methylacetamide. J. Am. Chem. Soc. 132:10847–10856 10.1021/ja103270w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Noskov S.Y., Roux B. 2010b. Two mechanisms of ion selectivity in protein binding sites. Proc. Natl. Acad. Sci. USA. 107:20329–20334 10.1073/pnas.1007150107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Morais-Cabral J.H., Kaufman A., MacKinnon R. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 414:43–48 10.1038/35102009 [DOI] [PubMed] [Google Scholar]