Abstract
The small mammalian mitochondrial DNA (mtDNA) is very gene dense and encodes factors critical for oxidative phosphorylation. Mutations of mtDNA cause a variety of human mitochondrial diseases and are also heavily implicated in age-associated disease and aging. There has been considerable progress in our understanding of the role for mtDNA mutations in human pathology during the last two decades, but important mechanisms in mitochondrial genetics remain to be explained at the molecular level. In addition, mounting evidence suggests that most mtDNA mutations may be generated by replication errors and not by accumulated damage.
The origin of mtDNA
The mitochondrial oxidative phosphorylation system is remarkable in its dependence on both nuclear- and mitochondrial DNA (mtDNA)-encoded subunits (Falkenberg et al., 2007). The involvement of two distinct genomes creates a demand for elaborate regulatory processes to coordinate gene expression in response to cellular demands for ATP synthesis (Falkenberg et al., 2007). Mitochondria are related to α-protobacteria, and the eukaryotic cell arose approximately two billion years ago by some type of fusion event between ancient cells related to α-protobacteria and archaebacteria (Yang et al., 1985; Lang et al., 1997; Andersson et al., 1998). Phylogenetic comparisons have shown that there is always cosegregation between the presence of mtDNA and a functional respiratory chain (Burger et al., 2003; Wallace, 2007). There is ongoing traffic of mtDNA fragments to the nucleus (Thorsness and Fox, 1990), and genes for many respiratory chain subunits have been transferred to the nucleus during evolution (Burger et al., 2003; Wallace, 2007). However, the genes for cytochrome b and cytochrome c oxidase subunit I are always maintained in mtDNA of the many organisms that have been studied to date (Wallace, 2007). The reason for the localization of these genes to mtDNA could be the hydrophobicity of the gene products, which may prevent mitochondrial import if the gene is relocated to the nucleus. Approximately 25% of the yeast mitochondrial proteome of ∼750–1,000 proteins is dedicated to maintenance and expression of mtDNA (Sickmann et al., 2003). This means ∼200–250 nucleus-encoded proteins are needed to express a handful of mtDNA-encoded proteins, and it is unclear why this costly arrangement has been maintained throughout evolution if the only reason is the hydrophobicity of certain gene products. An interesting alternate hypothesis proposes that mtDNA has been kept for a regulatory purpose and that the biogenesis of the oxidative phosphorylation system requires direct interactions between the respiratory chain subunits and mtDNA (Allen, 2003).
Transcription and replication of mammalian mtDNA
Mammalian mtDNA encodes 13 proteins that all are subunits of the oxidative phosphorylation system and 22 tRNAs and 2 ribosomal RNAs (rRNAs; Fig. 1). The transcription of mtDNA is polycistronic and is initiated at one main promoter on each strand, the light strand promoter (LSP) and heavy strand promoter (HSP). The existence of a second HSP dedicated to the transcription of the rRNA genes has been reported (Montoya et al., 1983; Martin et al., 2005); however, its existence has been questioned, as transcription from this putative promoter cannot be reconstituted in vitro with known components of the basal transcription machinery (Litonin et al., 2010). The steady-state levels of rRNAs are much higher than the levels of the downstream mRNAs, but this is, in principle, compatible with polycistronic transcription from a single HSP as the rRNAs are incorporated into ribosomes and therefore may be much more stable than the downstream mRNAs.
Figure 1.
Schematic representation of mammalian mtDNA. The double-stranded circular mammalian mtDNA molecule of ∼16.5 kb contains a single longer noncoding region, the displacement loop (D loop) region, harboring the promoters for transcription of both mtDNA strands (HSP and LSP) and the origin of leading strand replication (OH). The origin of lagging strand replication (OL) is embedded in a cluster of tRNA genes. The genes for the two rRNAs (12S and 16S rRNA), 13 mRNAs (ND1–6, ND4L, Cyt b, COI–III, ATP6, and ATP8), and 22 tRNAs (F, V, L1, I, M, W, D, K, G, R, H, S1, L2, T, P, E, S2, Y, C, N, A, and Q) are indicated by boxes. Illustration by Annika Röhl.
The basal machinery needed for transcription initiation of mtDNA has been fully reconstituted in vitro and consists of a set of three proteins: mitochondrial RNA polymerase (POLRMT), mitochondrial transcription factor B2 (TFB2M), and mitochondrial transcription factor A (TFAM; Falkenberg et al., 2002). Interestingly, POLRMT is most closely related to bacteriophage RNA polymerases (Shutt and Gray, 2006a) and in addition contains a large N-terminal extension that may have a role in coupling transcription to translation (Rodeheffer and Shadel, 2003). The paralogues TFB1M and TFB2M in mammalian cells are related to a highly conserved family of dimethyl adenosine methyltransferases (Falkenberg et al., 2002) that are present in bacteria, archaebacteria, and eukaryotes (mitochondria and the nucleus), where they dimethylate two highly conserved adenines at a stem-loop structure at the 3′ end of the small subunit rRNA (Shutt and Gray, 2006b). TFB1M was initially reported to have a role in mitochondrial transcription (Falkenberg et al., 2002; McCulloch and Shadel, 2003), but a conditional mouse knockout model has shown that it instead is an essential rRNA methyltransferase necessary for the integrity of the small subunit of the mitochondrial ribosome (Metodiev et al., 2009). TFB1M conditional knockout mice have severely impaired translation and are able to activate mitochondrial transcription, showing that there seems to be no functional redundancy between TFB1M and TFB2M (Metodiev et al., 2009). Biochemical studies further support the conclusion that TFB2M is essential for transcription, as it is a component of the catalytic site of POLRMT necessary for transcription initiation (Sologub et al., 2009; Litonin et al., 2010).
TFAM is not only a transcription factor but also functions as an mtDNA packaging protein (Larsson et al., 1998; Ekstrand et al., 2004). It belongs to the high mobility group domain proteins (Parisi and Clayton, 1991) and can wrap, bend, and compact DNA (Fisher et al., 1992; Kaufman et al., 2007). There is approximately one TFAM molecule/10–20 bp mtDNA in mouse tissues (Ekstrand et al., 2004; Pellegrini et al., 2009), and immunofluorescence experiments show a perfect colocalization between TFAM and mtDNA (Garrido et al., 2003), thus arguing that TFAM fully coats mtDNA in vivo. Genetic experiments in the mouse show that mtDNA levels in vivo are regulated according to levels of TFAM (Ekstrand et al., 2004). There are thousands of mtDNA copies in a mammalian somatic cell, and these genomes are packaged into mtDNA protein aggregates called nucleoids (Satoh and Kuroiwa, 1991; Kaufman et al., 2007; Bogenhagen et al., 2008). In budding yeast, a large number of proteins have been defined as components of the nucleoid based on a combination of biochemical experiments, showing that the studied protein can be cross-linked to and/or copurified with mtDNA, and genetic experiments, showing that the gene encoding the studied protein is essential for maintenance of mtDNA (Chen et al., 2005). This concept is not straightforward, as a protein may interact with mtDNA and be necessary for maintenance of mtDNA without having a role in packaging and organizing mtDNA. We propose that the definition of the nucleoid should focus on the role of the protein components in packaging and organizing mtDNA and that structural nucleoid proteins should comply with the following criteria: (a) the protein should interact with mtDNA (copurification or cross-linking); (b) the protein should have intrinsic biophysical properties consistent with packaging DNA; (c) the protein should be sufficiently abundant to coat mtDNA; (d) the protein should colocalize with mtDNA on high-resolution microscopy; and (e) the protein should be essential for maintenance of mtDNA in vivo. TFAM is currently the only mammalian protein that has been shown to fulfill all of these criteria. Definition of the structure of the nucleoid is a very fundamental question in mitochondrial genetics, and future studies are clearly warranted. Confocal microscopy experiments (Legros et al., 2004) have suggested that each nucleoid contains two to eight mtDNA molecules. This raises the interesting question of whether each nucleoid contains only one mtDNA genotype or whether there may be a mixture of mutant and wild-type mtDNA in a single nucleoid. Understanding this issue will provide novel insights into principles for mitotic segregation and maternal transmission of mtDNA, as discussed in the “Principles for maternal inheritance of mtDNA” section. The estimated size of the nucleoid in mammalian cells is ∼250 nm, which is at the resolution limit of confocal microscopy. It is important to apply novel high-resolution optic microscopy techniques to determine whether 250 nm is the true size of the nucleoids and how uniform the size is. It is currently unknown whether the nucleoids in mammalian cells are free in the mitochondrial matrix or whether they are attached to the inner mitochondrial membrane. Another fundamental question is whether all nucleoids have an equal protein composition and how a particular nucleoid is selected for transcription and/or replication of mtDNA. Future studies of the nucleoid are therefore likely to give very fundamental insights into mammalian mitochondrial biology.
Our understanding of the regulation of the basal transcription initiation machinery is rather limited despite the fact that there is a long list of nuclear transcription regulators that are proposed to have intramitochondrial isoforms, e.g., nuclear hormone receptors, orphan receptors, and other transcription factors (Shutt and Shadel, 2010). The mitochondrial transcription termination factors (MTERFs) are exclusively localized to mitochondria, and the first described member of this family is MTERF1, which has for >20 yr been implicated in transcription termination to enhance the production of rRNAs (Kruse et al., 1989; Yakubovskaya et al., 2010) and in transcription initiation (Martin et al., 2005). The paralogues MTERF2 and MTERF3 have roles in transcription initiation regulation (Park et al., 2007; Wenz et al., 2009a). Atomic structures of MTERF1 (Yakubovskaya et al., 2010) and MTERF3 (Spåhr et al., 2010) have unexpectedly shown that these proteins are very similar and contain repeated modules of a novel nucleic acid–binding fold, the MTERF motif.
Replication of mtDNA is dependent on POLRMT that forms the short RNA primers needed for initiation of mtDNA replication at the heavy and light strand replication origins (OH and OL; Clayton, 1991; Fusté et al., 2010). The minimal replisome needed for replication of mtDNA consists of the mtDNA polymerase (Pol-γ; Falkenberg and Larsson, 2009; Lee et al., 2009), the twinkle DNA helicase (Spelbrink et al., 2001), and the mitochondrial single-stranded DNA–binding protein (Falkenberg et al., 2007). The atomic structure for the Pol-γ was recently solved and showed that the complex is heterotrimeric and consists of one Pol-γ A subunit and two Pol-γ B subunits (Carrodeguas et al., 2001; Lee et al., 2009). The mode of mtDNA replication is controversial, as two competing models have been advocated: the strand-asymmetric (Clayton, 1982; Brown et al., 2005) and the strand-coupled replication modes (Holt et al., 2000; Yasukawa et al., 2006). According to the strand-asymmetric model, the leading strand replication is two thirds complete before lagging strand replication is initiated (Clayton, 1982; Brown et al., 2005). In contrast, the strand-coupled model argues that the replication of the leading and lagging strands is synchronous (Holt et al., 2000; Yasukawa et al., 2006). We have previously presented a more detailed discussion of these models (Falkenberg et al., 2007; Larsson, 2010), and only a few comments will be made here. There are many reported patients with high levels of mtDNA molecules with a single large deletion. The location of the deletion often varies between patients, but the deleted mtDNA molecule always retains OH, OL, and LSP, thus arguing that these sequences are essential for mtDNA replication (Moraes et al., 1991). Any valid model for mtDNA replication must therefore explain the role of these regulatory sequence elements in mtDNA replication. Recent data show that POLRMT specifically recognizes the stem-loop structure at OL and provides a primer for initiation of lagging strand mtDNA synthesis at this site (Fusté et al., 2010). A combination of biochemical and genetic approaches will be necessary to further clarify the mode of mtDNA replication in mammalian mitochondria.
Pathogenic mutations of mtDNA are often heteroplasmic
Pathogenic mtDNA mutations were first reported in human patients >20 yr ago (Holt et al., 1988; Wallace et al., 1988; Zeviani et al., 1988). These discoveries were important breakthroughs that led to a genetic basis for the classification of mitochondrial disease and revealed novel insights into mitochondrial genetics. A large number of pathogenic mtDNA mutations are known, and the reader is referred to specialized reviews on the associated clinical phenotypes (Larsson and Clayton, 1995; Munnich and Rustin, 2001; Zeviani and Di Donato, 2004).
Patients harboring pathogenic mtDNA mutations often have a mixture of wild-type and mutated molecules, a condition called heteroplasmy. The mutation load can vary widely between members of the same family and in different cells of particular tissues. This uneven distribution of mutated mtDNA leads to a mosaic respiratory chain deficiency in affected tissues. This mosaicism is likely an important determinant in disease pathophysiology, but few relevant animal models are available (Table I), and the mechanisms are therefore poorly understood. In an experimental study, mouse chimeras with mosaic respiratory chain deficiency in pyramidal neurons of the cerebral cortex were generated (Dufour et al., 2008). These mice developed clinical symptoms if they had a proportion of >20% respiratory chain–deficient neurons, and the defect caused mortality if there were >60–80% defective neurons (Dufour et al., 2008). Interestingly, a transneuronal degeneration phenomenon, whereby respiratory chain–deficient neurons induce death of neighboring neurons with normal respiratory chain function, was also observed (Dufour et al., 2008).
Table I.
Mouse models for mtDNA mutation
| Model | Experimental manipulation | Phenotype | References |
| mtDNA point mutation | |||
| Mutator mouse (POLGA D257A) | Knockin modification of DNA polymerase γ exonuclease domain | Impaired respiratory chain function, premature aging phenotypes | Trifunovic et al., 2004; Kujoth et al., 2005 |
| Trans-mitochondrial mice (CAPR) | Cytoplasmic transfer of chloramphenicol-resistant mtDNAs to ES cells | Growth retardation, myopathy, cardiomyopathy | Marchington et al., 1999; Sligh et al., 2000 |
| Trans-mitochondrial mouse (T6589C) | Cytoplasmic transfer of mtDNA with T6589C missense mutation to ES cells | Impaired respiratory chain function, growth retardation | Kasahara et al., 2006 |
| Trans-mitochondrial mouse (T6589C + 13885insCdelT) | Cytoplasmic transfer of mtDNA with T6589C, 13885insC, and 13885insCdelT mutation to ES cells | Decreased complex IV activity, myopathy, cardiomyophathy | Fan et al., 2008 |
| mtDNA deletion | |||
| Trans-mitochondrial mouse (ΔmtDNA4696) | Cytoplasmic transfer of deleted mtDNA (Δ7,759–12,454) to pronuclear stage embryo | Impaired respiratory chain function, renal failure, early mortality | Inoue et al., 2000 |
| Deletor mouse (Twinkle dup353–365) | Transgenic expression of mutant mtDNA helicase twinkle | Progressive decrease of respiratory chain function | Tyynismaa et al., 2005 |
| Mito-PstI mouse | Skeletal muscle–specific expression of mitochondrially targeted PstI restriction endonuclease | Impaired respiratory chain function in skeletal muscle, myopathy | Srivastava and Moraes, 2005 |
| mtDNA depletion | |||
| TFAM−/− | Knockout of TFAM in whole body | Respiratory chain deficiency, embryonic lethality | Larsson et al., 1998 |
| TFAM−/− heart | Knockout of TFAM in cardiomyocyte | Dilated cardiomyopathy | Wang et al., 1999 |
| TFAM−/− forebrain neuron | Knockout of TFAM in forebrain | Neurodegenerative symptoms | Sörensen et al., 2001 |
| TFAM−/− dopaminergic neuron | Knockout of TFAM in dopaminergic neuron | Parkinson’s disease symptoms | Ekstrand et al., 2007 |
| TFAM−/− pancreatic β cell | Knockout of TFAM in pancreatic β cell | Mitochondrial diabetes | Silva et al., 2000 |
| POLGA−/− | Knockout of POLGA | Respiratory chain deficiency, embryonic lethality | Hance et al., 2005 |
| MFN1−/− + MFN2−/− skeletal muscle | Skeletal muscle–specific knockout of MFN1 and MFN2, GTPases essential for mitochondrial fusion | Respiratory chain deficiency in skeletal muscle, muscle atrophy, early mortality | Chen and Chan, 2010 |
| TK2−/− | Knockout of thymidine kinase 2 (TK2) | Growth retardation, early mortality | Zhou et al., 2008 |
| TK2 H126N | Knockin modification of TK2 found in human mtDNA depletion syndrome patients (TK2 H126N) | Impaired respiratory chain function in brain, growth retardation, early mortality | Akman et al., 2008 |
| RRM2B−/− | Knockout of RRM2B gene, encoding p53-controlled ribonucleotide reductase (p53R2) | Growth retardation, renal failure, muscle atrophy, early mortality | Bourdon et al., 2007; Kimura et al., 2003 |
| RNASEH1−/− | Knockout of mitochondrial ribonuclease H1 (RNaseH1) | Embryonic lethality, respiratory chain deficiency | Cerritelli et al., 2003 |
| mtDNA increase | |||
| PAC-hTFAM | Transgenic expression of P1 artificial chromosome containing human TFAM | No apparent pathophysiology | Ekstrand et al., 2004 |
| TFAM-EGFP | Transgenic expression of TFAM-EGFP fusion protein | Improve mitochondrial disease phenotypes | Nishiyama et al., 2010 |
| CAG-hTFAM | Transgenic expression of mouse TFAM cDNA under CAG promoter | Improve cardiac failure after myocardial infarction, delay neuronal cell death | Ikeuchi et al., 2005; Hokari et al., 2010 |
| TWINKLE | Transgenic expression of mouse TWINKLE cDNA | No apparent pathophysiology | Tyynismaa et al., 2004 |
| Twinkle + hTFAM | Transgenic expression of mouse TWINKLE cDNA and human TFAM cDNA | Progressive respiratory chain dysfunction | Ylikallio et al., 2010 |
ES, embryonic stem.
Segregation of mutated mtDNA in somatic tissues
The mitotic segregation of heteroplasmic mtDNA mutations in somatic tissues and the principles for maternal transmission of mtDNA (the bottleneck phenomenon and the purifying selection) are cornerstones in mammalian mitochondrial genetics. Unfortunately, these phenomena are poorly understood at the cell biological level despite their fundamental importance. Increased molecular understanding of these processes would lead to new understanding of disease pathophysiology and would also improve genetic counseling in families with mitochondrial disease.
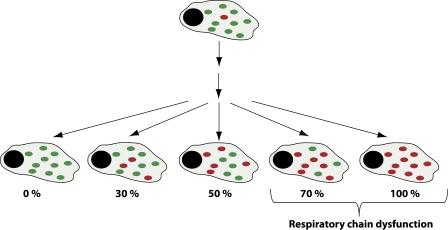
Replication of mtDNA is not linked to the cell cycle, and a particular mtDNA molecule may be replicated many times or not at all as a cell divides (Bogenhagen and Clayton, 1977). This mode of replication makes it possible for a single mutational event to expand clonally or be lost as the cell divides (Fig. 2). The turnover of mtDNA in differentiated tissues has not been extensively studied, and half-lives of ∼14 d have been reported in the rat brain and other tissues (Menzies and Gold, 1971). Thus, there is likely continuous replication of mtDNA in all tissues, which makes segregation of mtDNA mutations possible also in postmitotic cells. Heteroplasmic mtDNA mutations will only cause respiratory chain dysfunction if present above a certain threshold (Fig. 2), which varies depending on the type of mutation and the type of affected tissue. It is generally assumed that mitotic segregation of pathogenic mtDNA mutations is a largely random process and that high mutation levels may be selected against in rapidly dividing cells. However, there are also reports that seemingly neutral polymorphisms in mtDNA may undergo directional selection in certain tissues (Jenuth et al., 1997; Battersby and Shoubridge, 2001; Jokinen et al., 2010).
Figure 2.
Mitotic segregation of mtDNA. A single mutational event creates heteroplasmy in a cell, but the level of mutated mtDNA (red dots) is very low in comparison with normal mtDNA (green dots). There is no synchronization between cell division and mtDNA replication, and a particular mtDNA molecule may be replicated many times or not at all during a single cell cycle. Repeated cell division will lead to mitotic segregation of normal and mutated mtDNA, and accumulation of mutated mtDNA above a certain threshold level will lead to impaired respiratory chain function.
Mitochondria form a dynamic network undergoing continuous fusion and fission (Detmer and Chan, 2007; Hoppins et al., 2007). These processes ensure mixing of the gene product content of the mitochondrial network in a cell and are necessary for maintaining function, as exemplified by the finding that blocked fusion leads to loss of mtDNA (Chen and Chan, 2010). Respiratory chain deficiency leads to a fragmentation of the mitochondrial network in cell lines and mouse tissues (Duvezin-Caubet et al., 2006). Autophagocytosis has an important role in vivo to remove mitochondria during normal erythrocyte differentiation (Sandoval et al., 2008). Studies of mammalian cell lines have shown that Parkin, an E3-ubiquitin ligase mutated in juvenile-onset Parkinson’s disease, translocates to dysfunctional mitochondria to promote degradation by autophagocytosis (Narendra et al., 2008, 2009, 2010; Suen et al., 2010). The evidence for the existence of this mitochondrial quality control mechanism in mammals is at present based on forced expression of Parkin in transformed cell lines in combination with the use of rather drastic mitochondrial toxins (Narendra et al., 2008, 2009, 2010; Suen et al., 2010). Human patients with mitochondrial disease often have a deteriorating respiratory chain function and may accumulate mutated mtDNA with time (Larsson et al., 1990), thus questioning the efficiency of this proposed mitochondrial quality control mechanism. There is a clear need to verify that the mitochondrial quality control mechanism observed in cell lines is relevant for mammals in vivo.
Principles for maternal inheritance of mtDNA
Maternal transmission of mammalian mtDNA has been recognized for >35 yr (Hutchison et al., 1974; Giles et al., 1980). It came as a true surprise when it was observed that the mtDNA genotype could shift completely in a few generations in cows (Olivo et al., 1983). A similar rapid segregation was soon observed in many human pedigrees transmitting heteroplasmic pathogenic mtDNA mutations (Larsson et al., 1992). Heteroplasmic mice containing mtDNA from two different mouse strains, created by experimental manipulation of embryos, exhibited a similar rapid, apparently random, germline segregation of mtDNA (Jenuth et al., 1996). This segregation phenomenon is attributed to a bottleneck mechanism in the female germline, whereby only a small fraction of all mtDNA copies in the primordial germ cell is transmitted to the oocyte. Several studies have produced conflicting results proposing that the bottleneck for mtDNA transmission occurs at different stages in the maternal germline, and results are also conflicting about whether there is a concomitant mtDNA copy number reduction in primordial germ cells (Cao et al., 2007, 2009; Cree et al., 2008; Wai et al., 2008; Samuels et al., 2010).
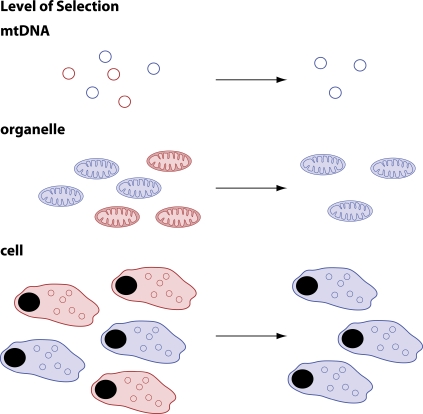
The transmission of mtDNA mutations through the mouse maternal germline is not neutral, but there is rather a strong purifying selection against deleterious mtDNA mutations (Fan et al., 2008; Stewart et al., 2008b). The mtDNA mutator mouse expresses a catalytic subunit of Pol-γ with deficient proofreading capacity (Table I), which leads to accumulation of high levels of acquired point mutations in mtDNA (Trifunovic et al., 2004). Maternal transmission of this apparently random set of point mutations generated in the mtDNA mutator mouse (Trifunovic et al., 2004) shows a strong selection against amino acid replacement mutations, whereas mutations affecting tRNA or rRNA coding genes are better tolerated (Stewart et al., 2008a,b). Surprisingly, the spectrum of mutations present in offspring to the mtDNA mutator mouse has clear similarities to the naturally occurring spectrum of mutations observed in human populations (Stewart et al., 2008a,b). These results suggest that the naturally occurring spectrum of mtDNA mutations in humans can be explained by mtDNA replication errors that have been subjected to purifying selection in the maternal germline. The molecular mechanism of purifying selection is not understood. Formally, such a selection could occur at different levels (Fig. 3). A mechanism for selection at the level of the mtDNA is not easy to envision, as it involves blocking replication or destroying mutant mtDNA genomes without the need for testing the function of the gene products encoded by the mutant genome. A more plausible mechanism is functional testing of mtDNA at the level of a single organelle (Shoubridge and Wai, 2008). Such a mechanism could involve fragmentation of the mitochondrial network during some stage of oocyte development to permit expression of individual mtDNA molecules and subsequent functional readout of respiratory chain function in single mitochondria (Stewart et al., 2008a). It is also possible that there is a competition between cells during germ cell development and that cells with high levels of mutated mtDNA will be disfavored or undergo apoptosis. Mutations of tRNA or rRNA genes are more likely than amino acid substitutions to escape the purifying selection in the mouse maternal germline, which is consistent with the observation that the majority of pathogenic mutations of human mtDNA affects tRNA genes, although they only occupy ∼9% of the genome (Stewart et al., 2008a). The bottleneck mechanism may be linked to the purifying selection, but it could also represent an independent protective mechanism. It is, of course, possible that a down-regulation of mtDNA copy number and fragmentation of the mitochondrial network may facilitate purifying selection in the maternal germline. However, the bottleneck mechanism could also prevent the spread of low levels of deleterious mtDNA mutations in maternal lineages (Stewart et al., 2008a). Available data on the nature of the bottleneck are mainly correlative, and it will be important to experimentally investigate possible mechanisms, e.g., by introducing pathogenic mtDNA mutations into mice concomitant with experimental manipulation of mtDNA copy number and mitochondrial dynamics in the female germline.
Figure 3.
Different levels at which purifying selection can occur in the maternal germline. (top) Genomes with mutations could be blocked from replication or selectively destroyed without the need for gene expression. (middle) A fragmented mitochondrial network would allow functional testing of individual mtDNA molecules. The presence of a mutated mtDNA molecule would result in a mitochondrion with deficient respiratory chain function, which, in turn, would lead to selection against and/or destruction of this mitochondrion. (bottom) Cells with high levels of mutated mtDNA may fail to compete with respiratory chain–competent cells and may be selected against or undergo apoptosis. The colors indicate mutant (red) and wild-type (blue) mtDNA (top); respiratory chain–deficient (red) and normal (blue) mitochondria (middle); and respiratory chain–deficient (red) and normal (blue) cells (bottom).
Aging and somatic mtDNA mutations
Patients with mitochondrial syndromes have a monoclonal expansion of mutated mtDNA leading to high levels of a single type of mutation, whereas aging is associated with polyclonal expansions leading to low levels of many different types of mtDNA mutations. It is important to emphasize that one should not expect fundamental differences in the pathophysiology of mtDNA mutations regardless of whether they occur in genetic syndromes or in aging. Important principles in mitochondrial genetics, e.g., mitotic segregation of heteroplasmic mtDNA mutations, are also applicable to aging. Furthermore, the consequence of an mtDNA mutation at the cellular level will not depend on whether the mtDNA mutation is spontaneous or acquired but will rather depend on the type of mtDNA mutation and how it impairs respiratory chain function and the consequences this impairment, in turn, has on specialized functions of the affected cell. The role of somatic mtDNA mutations in mammalian aging has recently been extensively reviewed (Krishnan et al., 2007; Larsson, 2010), and only some issues will be discussed here. It is well established that somatic mtDNA mutations in human aging undergo clonal expansion and cause a mosaic respiratory chain dysfunction in different tissues (Krishnan et al., 2007; Larsson, 2010). It has generally been assumed that age-associated somatic mtDNA mutations are caused by accumulated damage during the aging process (Wallace, 2001). However, an alternative hypothesis is that most of the mutations are created as replication errors during embryogenesis and then undergo clonal expansion in adult life (Larsson, 2010). Mosaic respiratory chain deficiency caused by clonal expansion of mtDNA mutations is ubiquitously observed in human aging, but the extent to which this aberration impairs organ function is unknown. The mtDNA mutator mice (Table I) develop a premature aging syndrome, which is caused by the accumulation of point mutations and the presence of linear deleted mtDNA molecules, which likely represent a continuously formed replication intermediate (Trifunovic et al., 2004; Bailey et al., 2009; Edgar et al., 2009). There has been one study arguing that a third type of mutation, circular mtDNA molecules with deletions, drives the phenotype of mtDNA mutator mice (Vermulst et al., 2008). However, this study has been refuted in several independent studies, in which sensitive PCR assays (Edgar et al., 2009; Kraytsberg et al., 2009) or deep sequencing (Williams et al., 2010; Ameur et al., 2011) could not detect any significant levels of circular mtDNA molecules with deletions in various tissues of mtDNA mutator mice. In addition, the biochemical phenotype in mtDNA mutator mice is fully consistent with the idea that point mutations of mtDNA create amino acid substitutions of respiratory chain subunits, which explains the observed decline in the stability of the respiratory chain complexes (Edgar et al., 2009).
Interestingly, the point mutation load is high already during embryogenesis in mtDNA mutator mice, but aging symptoms are not observed until early adult life, and there is no evidence for increased oxidative stress (Kujoth et al., 2005; Trifunovic et al., 2005). The results from the mtDNA mutator mouse thus lend some experimental support to the hypothesis that somatic mtDNA mutations created during embryogenesis may contribute to the creation of aging phenotypes in adult life.
Pathophysiological effects of mtDNA mutations
The studies of pathophysiological effects of altered mtDNA expression have to a large extent been dependent on the use of mouse models (Table I). It is important to emphasize that there is as yet no transfection system for mammalian mitochondria, and mouse models have therefore been developed by introducing naturally occurring mtDNA mutations into mouse embryos or by manipulating nuclear genes that control the maintenance and expression of mtDNA.
Expression of mtDNA is essential for oxidative phosphorylation (Larsson et al., 1998), and the most straightforward prediction is therefore that mtDNA mutations should have no other primary effects besides impairing cellular energy production. There are indeed a large number of different types of mtDNA mutations that have effects on respiratory chain function and ATP synthesis (Larsson and Clayton, 1995; Munnich and Rustin, 2001; Zeviani and Di Donato, 2004). Mutations of mtDNA have several downstream effects that are a consequence of deficient oxidative phosphorylation, and those will be further discussed in the next paragraphs.
It is important to remember that there are possibilities that mtDNA mutations could have effects unrelated to oxidative phosphorylation. Amino acid replacements of mtDNA-encoded proteins can create novel histocompatibility antigens in the mouse (Loveland et al., 1990; Morse et al., 1996). These antigens are derived by degradation of mtDNA-encoded respiratory chain subunits, and the corresponding peptides are released from mitochondria and reach the cell surface (Loveland et al., 1990; Morse et al., 1996). There is also evidence that mtDNA sequence variants may elicit an innate immune response against tumor cells (Ishikawa et al., 2010).
It was initially reported that mtDNA is not required for execution of apoptosis in cell lines (Jacobson et al., 1993). However, a subsequent study showed that depletion of mtDNA caused by knockout of TFAM in the mouse leads to increased in vivo apoptosis (Wang et al., 2001). Increased apoptosis and/or cell death has now been reported in a variety of mtDNA-depleted tissues in the mouse: the heart (Wang et al., 2001), pyramidal neurons (Sörensen et al., 2001), dopamine neurons (Ekstrand et al., 2007), and pancreatic β cells (Silva et al., 2000). In addition, there is extensive apoptosis in different tissues of the mtDNA mutator mouse (Kujoth et al., 2005; Niu et al., 2007). Human patients with genetic mitochondrial diseases often have extensive cell death in the nervous system (Alpers, 1931; Leigh, 1951; Oldfors et al., 1990). Cell loss is also a common problem in aging, and, thus, it is possible that the observed clonal expansion of somatic mtDNA mutations may contribute to cell loss in human aging (Krishnan et al., 2007; Larsson, 2010).
Patients with pathogenic mtDNA mutations often accumulate increased levels of mitochondria, e.g., manifested as ragged-red muscle fibers in skeletal muscle. Also, tissue-specific knockout mice with impaired mtDNA maintenance (Wang et al., 1999; Wredenberg et al., 2002), defective mitochondrial transcription (Park et al., 2007), or impaired mitochondrial translation (Metodiev et al., 2009) have increased mitochondrial mass in affected tissues. It is generally assumed that this accumulation of mitochondria is caused by activation of mitochondrial biogenesis in response to deficient energy production. Experiments in mice with mtDNA depletion in skeletal muscle have shown that activation of mitochondrial biogenesis at least partly can compensate for the deficient mitochondrial function and maintain close to normal overall ATP production in the affected tissue (Wredenberg et al., 2002). Similarly, forced expression of PGC1-α in mouse skeletal muscle induces mitochondrial biogenesis, which, in turn, protects against mitochondrial myopathy (Wenz et al., 2008) and age-related loss of muscle mass (Wenz et al., 2009b).
Increased production of reactive oxygen species (ROS) and oxidative damage are heavily implicated in human pathology and aging, but the role this type of damage plays is much debated (Bokov et al., 2004). This controversy is to some extent explained by the complex chemistry of ROS formation (Murphy, 2009) and uncertainties about the performance of methods used to measure ROS (Bokov et al., 2004; Murphy, 2009; Murphy et al., 2011). There are also some intriguing studies showing that a substantial increase of oxidative damage does not shorten lifespan in mice (Zhang et al., 2009). It is often argued that mitochondrial dysfunction increases ROS production (Wallace, 2005), but this concept has poor experimental support. On the contrary, it has been reported that ROS production and oxidative damage are not increased in different tissues of mice with depletion (Wang et al., 2001) or point mutations of mtDNA (Kujoth et al., 2005; Trifunovic et al., 2005). An impairment of respiratory chain function leads to a variety of metabolic consequences, e.g., deficient ATP production, increased NADH/NAD+ ratios, and alterations in Ca2+ homeostasis (Tavi et al., 2005; Aydin et al., 2009), which all may be more relevant for disease pathophysiology than ROS.
Future perspectives
During the last 25 yr, we have seen a remarkable increase in our understanding of mitochondrial genetics in mammals. We know today that maternal transmission of mtDNA is associated with a bottleneck phenomenon, which allows rapid shifts of genotypes between generations, and a purifying selection, which weeds out the most deleterious mutations. However, we still have no molecular understanding of these two processes despite their fundamental importance for mtDNA transmission. The different types of pathologies that are caused by mtDNA mutations are remarkable, and in many cases there is a reasonably good correlation between genotype and phenotype. However, the underlying pathophysiology is not understood in any depth. Important challenges for the future involve understanding the downstream effects of mitochondrial dysfunction on cell physiology in disease and aging.
Acknowledgments
This study was supported by Swedish Research Council grants and a European Research Council Advanced Investigator grant to N.G. Larsson. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea and funded by the Ministry of Education, Science, and Technology (grants 2010-0015769 and 2010-0001939).
Footnotes
Abbreviations used in this paper:
- ES
- embryonic stem
- HSP
- heavy strand promoter
- LSP
- light strand promoter
- mtDNA
- mitochondrial DNA
- MTERF
- mitochondrial transcription termination factor
- ROS
- reactive oxygen species
- rRNA
- ribosomal RNA
References
- Akman H.O., Dorado B., López L.C., García-Cazorla A., Vilà M.R., Tanabe L.M., Dauer W.T., Bonilla E., Tanji K., Hirano M. 2008. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum. Mol. Genet. 17:2433–2440 10.1093/hmg/ddn143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.F. 2003. Why chloroplasts and mitochondria contain genomes. Comp. Funct. Genomics. 4:31–36 10.1002/cfg.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpers B.J. 1931. Diffuse progressive degeneration of the gray matter of the cerebrum. Arch. Neurol. Psychiatry. 25:469–505 [Google Scholar]
- Ameur A., Stewart J.B., Freyer C., Hagström E., Ingman M., Larsson N.G., Gyllensten U. 2011. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 7:e1002028 10.1371/journal.pgen.1002028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S.G., Zomorodipour A., Andersson J.O., Sicheritz-Pontén T., Alsmark U.C., Podowski R.M., Näslund A.K., Eriksson A.S., Winkler H.H., Kurland C.G. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 396:133–140 10.1038/24094 [DOI] [PubMed] [Google Scholar]
- Aydin J., Andersson D.C., Hänninen S.L., Wredenberg A., Tavi P., Park C.B., Larsson N.G., Bruton J.D., Westerblad H. 2009. Increased mitochondrial Ca2+ and decreased sarcoplasmic reticulum Ca2+ in mitochondrial myopathy. Hum. Mol. Genet. 18:278–288 10.1093/hmg/ddn355 [DOI] [PubMed] [Google Scholar]
- Bailey L.J., Cluett T.J., Reyes A., Prolla T.A., Poulton J., Leeuwenburgh C., Holt I.J. 2009. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 37:2327–2335 10.1093/nar/gkp091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby B.J., Shoubridge E.A. 2001. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum. Mol. Genet. 10:2469–2479 10.1093/hmg/10.22.2469 [DOI] [PubMed] [Google Scholar]
- Bogenhagen D., Clayton D.A. 1977. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell. 11:719–727 10.1016/0092-8674(77)90286-0 [DOI] [PubMed] [Google Scholar]
- Bogenhagen D.F., Rousseau D., Burke S. 2008. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283:3665–3675 10.1074/jbc.M708444200 [DOI] [PubMed] [Google Scholar]
- Bokov A., Chaudhuri A., Richardson A. 2004. The role of oxidative damage and stress in aging. Mech. Ageing Dev. 125:811–826 10.1016/j.mad.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Bourdon A., Minai L., Serre V., Jais J.P., Sarzi E., Aubert S., Chrétien D., de Lonlay P., Paquis-Flucklinger V., Arakawa H., et al. 2007. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 39:776–780 10.1038/ng2040 [DOI] [PubMed] [Google Scholar]
- Brown T.A., Cecconi C., Tkachuk A.N., Bustamante C., Clayton D.A. 2005. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 19:2466–2476 10.1101/gad.1352105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Gray M.W., Lang B.F. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19:709–716 10.1016/j.tig.2003.10.012 [DOI] [PubMed] [Google Scholar]
- Cao L., Shitara H., Horii T., Nagao Y., Imai H., Abe K., Hara T., Hayashi J., Yonekawa H. 2007. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 39:386–390 10.1038/ng1970 [DOI] [PubMed] [Google Scholar]
- Cao L., Shitara H., Sugimoto M., Hayashi J., Abe K., Yonekawa H. 2009. New evidence confirms that the mitochondrial bottleneck is generated without reduction of mitochondrial DNA content in early primordial germ cells of mice. PLoS Genet. 5:e1000756 10.1371/journal.pgen.1000756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrodeguas J.A., Theis K., Bogenhagen D.F., Kisker C. 2001. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol. Cell. 7:43–54 10.1016/S1097-2765(01)00153-8 [DOI] [PubMed] [Google Scholar]
- Cerritelli S.M., Frolova E.G., Feng C., Grinberg A., Love P.E., Crouch R.J. 2003. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell. 11:807–815 10.1016/S1097-2765(03)00088-1 [DOI] [PubMed] [Google Scholar]
- Chen H., Chan D.C. 2010. Physiological functions of mitochondrial fusion. Ann. NY Acad. Sci. 1201:21–25 10.1111/j.1749-6632.2010.05615.x [DOI] [PubMed] [Google Scholar]
- Chen X.J., Wang X., Kaufman B.A., Butow R.A. 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 307:714–717 10.1126/science.1106391 [DOI] [PubMed] [Google Scholar]
- Clayton D.A. 1982. Replication of animal mitochondrial DNA. Cell. 28:693–705 10.1016/0092-8674(82)90049-6 [DOI] [PubMed] [Google Scholar]
- Clayton D.A. 1991. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 7:453–478 10.1146/annurev.cb.07.110191.002321 [DOI] [PubMed] [Google Scholar]
- Cree L.M., Samuels D.C., de Sousa Lopes S.C., Rajasimha H.K., Wonnapinij P., Mann J.R., Dahl H.H., Chinnery P.F. 2008. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 40:249–254 10.1038/ng.2007.63 [DOI] [PubMed] [Google Scholar]
- Detmer S.A., Chan D.C. 2007. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8:870–879 10.1038/nrm2275 [DOI] [PubMed] [Google Scholar]
- Dufour E., Terzioglu M., Sterky F.H., Sörensen L., Galter D., Olson L., Wilbertz J., Larsson N.G. 2008. Age-associated mosaic respiratory chain deficiency causes trans-neuronal degeneration. Hum. Mol. Genet. 17:1418–1426 10.1093/hmg/ddn030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M.F., Attardi G., Larsson N.G., et al. 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281:37972–37979 10.1074/jbc.M606059200 [DOI] [PubMed] [Google Scholar]
- Edgar D., Shabalina I., Camara Y., Wredenberg A., Calvaruso M.A., Nijtmans L., Nedergaard J., Cannon B., Larsson N.G., Trifunovic A. 2009. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 10:131–138 10.1016/j.cmet.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.G. 2004. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 13:935–944 10.1093/hmg/ddh109 [DOI] [PubMed] [Google Scholar]
- Ekstrand M.I., Terzioglu M., Galter D., Zhu S., Hofstetter C., Lindqvist E., Thams S., Bergstrand A., Hansson F.S., Trifunovic A., et al. 2007. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. USA. 104:1325–1330 10.1073/pnas.0605208103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg M., Larsson N.G. 2009. Structure casts light on mtDNA replication. Cell. 139:231–233 10.1016/j.cell.2009.09.030 [DOI] [PubMed] [Google Scholar]
- Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M. 2002. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31:289–294 10.1038/ng909 [DOI] [PubMed] [Google Scholar]
- Falkenberg M., Larsson N.G., Gustafsson C.M. 2007. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76:679–699 10.1146/annurev.biochem.76.060305.152028 [DOI] [PubMed] [Google Scholar]
- Fan W., Waymire K.G., Narula N., Li P., Rocher C., Coskun P.E., Vannan M.A., Narula J., Macgregor G.R., Wallace D.C. 2008. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 319:958–962 10.1126/science.1147786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.P., Lisowsky T., Parisi M.A., Clayton D.A. 1992. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 267:3358–3367 [PubMed] [Google Scholar]
- Fusté J.M., Wanrooij S., Jemt E., Granycome C.E., Cluett T.J., Shi Y., Atanassova N., Holt I.J., Gustafsson C.M., Falkenberg M. 2010. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 37:67–78 10.1016/j.molcel.2009.12.021 [DOI] [PubMed] [Google Scholar]
- Garrido N., Griparic L., Jokitalo E., Wartiovaara J., van der Bliek A.M., Spelbrink J.N. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 14:1583–1596 10.1091/mbc.E02-07-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles R.E., Blanc H., Cann H.M., Wallace D.C. 1980. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA. 77:6715–6719 10.1073/pnas.77.11.6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hance N., Ekstrand M.I., Trifunovic A. 2005. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 14:1775–1783 10.1093/hmg/ddi184 [DOI] [PubMed] [Google Scholar]
- Hokari M., Kuroda S., Kinugawa S., Ide T., Tsutsui H., Iwasaki Y. 2010. Overexpression of mitochondrial transcription factor A (TFAM) ameliorates delayed neuronal death due to transient forebrain ischemia in mice. Neuropathology. 30:401–407 10.1111/j.1440-1789.2009.01086.x [DOI] [PubMed] [Google Scholar]
- Holt I.J., Harding A.E., Morgan-Hughes J.A. 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 331:717–719 10.1038/331717a0 [DOI] [PubMed] [Google Scholar]
- Holt I.J., Lorimer H.E., Jacobs H.T. 2000. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 100:515–524 10.1016/S0092-8674(00)80688-1 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76:751–780 10.1146/annurev.biochem.76.071905.090048 [DOI] [PubMed] [Google Scholar]
- Hutchison C.A., III, Newbold J.E., Potter S.S., Edgell M.H. 1974. Maternal inheritance of mammalian mitochondrial DNA. Nature. 251:536–538 10.1038/251536a0 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Matsusaka H., Kang D., Matsushima S., Ide T., Kubota T., Fujiwara T., Hamasaki N., Takeshita A., Sunagawa K., Tsutsui H. 2005. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 112:683–690 10.1161/CIRCULATIONAHA.104.524835 [DOI] [PubMed] [Google Scholar]
- Inoue K., Nakada K., Ogura A., Isobe K., Goto Y., Nonaka I., Hayashi J.I. 2000. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat. Genet. 26:176–181 10.1038/82826 [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Toyama-Sorimachi N., Nakada K., Morimoto M., Imanishi H., Yoshizaki M., Sasawatari S., Niikura M., Takenaga K., Yonekawa H., Hayashi J. 2010. The innate immune system in host mice targets cells with allogenic mitochondrial DNA. J. Exp. Med. 207:2297–2305 10.1084/jem.20092296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M.D., Burne J.F., King M.P., Miyashita T., Reed J.C., Raff M.C. 1993. Bcl-2 blocks apoptosis in cells lacking mitochondrial DNA. Nature. 361:365–369 10.1038/361365a0 [DOI] [PubMed] [Google Scholar]
- Jenuth J.P., Peterson A.C., Fu K., Shoubridge E.A. 1996. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 14:146–151 10.1038/ng1096-146 [DOI] [PubMed] [Google Scholar]
- Jenuth J.P., Peterson A.C., Shoubridge E.A. 1997. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 16:93–95 10.1038/ng0597-93 [DOI] [PubMed] [Google Scholar]
- Jokinen R., Marttinen P., Sandell H.K., Manninen T., Teerenhovi H., Wai T., Teoli D., Loredo-Osti J.C., Shoubridge E.A., Battersby B.J. 2010. Gimap3 regulates tissue-specific mitochondrial DNA segregation. PLoS Genet. 6:e1001161 10.1371/journal.pgen.1001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara A., Ishikawa K., Yamaoka M., Ito M., Watanabe N., Akimoto M., Sato A., Nakada K., Endo H., Suda Y., et al. 2006. Generation of trans-mitochondrial mice carrying homoplasmic mtDNAs with a missense mutation in a structural gene using ES cells. Hum. Mol. Genet. 15:871–881 10.1093/hmg/ddl005 [DOI] [PubMed] [Google Scholar]
- Kaufman B.A., Durisic N., Mativetsky J.M., Costantino S., Hancock M.A., Grutter P., Shoubridge E.A. 2007. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 18:3225–3236 10.1091/mbc.E07-05-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Takeda S., Sagiya Y., Gotoh M., Nakamura Y., Arakawa H. 2003. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat. Genet. 34:440–445 10.1038/ng1212 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y., Simon D.K., Turnbull D.M., Khrapko K. 2009. Do mtDNA deletions drive premature aging in mtDNA mutator mice? Aging Cell. 8:502–506 10.1111/j.1474-9726.2009.00484.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan K.J., Greaves L.C., Reeve A.K., Turnbull D. 2007. The ageing mitochondrial genome. Nucleic Acids Res. 35:7399–7405 10.1093/nar/gkm635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse B., Narasimhan N., Attardi G. 1989. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 58:391–397 10.1016/0092-8674(89)90853-2 [DOI] [PubMed] [Google Scholar]
- Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., et al. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 309:481–484 10.1126/science.1112125 [DOI] [PubMed] [Google Scholar]
- Lang B.F., Burger G., O’Kelly C.J., Cedergren R., Golding G.B., Lemieux C., Sankoff D., Turmel M., Gray M.W. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 387:493–497 10.1038/387493a0 [DOI] [PubMed] [Google Scholar]
- Larsson N.G. 2010. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 79:683–706 10.1146/annurev-biochem-060408-093701 [DOI] [PubMed] [Google Scholar]
- Larsson N.G., Clayton D.A. 1995. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 29:151–178 10.1146/annurev.ge.29.120195.001055 [DOI] [PubMed] [Google Scholar]
- Larsson N.G., Holme E., Kristiansson B., Oldfors A., Tulinius M. 1990. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr. Res. 28:131–136 [DOI] [PubMed] [Google Scholar]
- Larsson N.G., Tulinius M.H., Holme E., Oldfors A., Andersen O., Wahlström J., Aasly J. 1992. Segregation and manifestations of the mtDNA tRNA(Lys) A—>G(8344) mutation of myoclonus epilepsy and ragged-red fibers (MERRF) syndrome. Am. J. Hum. Genet. 51:1201–1212 [PMC free article] [PubMed] [Google Scholar]
- Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18:231–236 10.1038/ng0398-231 [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Kennedy W.D., Yin Y.W. 2009. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 139:312–324 10.1016/j.cell.2009.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F., Malka F., Frachon P., Lombès A., Rojo M. 2004. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 117:2653–2662 10.1242/jcs.01134 [DOI] [PubMed] [Google Scholar]
- Leigh D. 1951. Subacute necrotizing encephalomyelopathy in an infant. J. Neurol. Neurosurg. Psychiatry. 14:216–221 10.1136/jnnp.14.3.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C.M., Temiakov D. 2010. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 285:18129–18133 10.1074/jbc.C110.128918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland B., Wang C.R., Yonekawa H., Hermel E., Lindahl K.F. 1990. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell. 60:971–980 10.1016/0092-8674(90)90345-F [DOI] [PubMed] [Google Scholar]
- Marchington D.R., Barlow D., Poulton J. 1999. Transmitochondrial mice carrying resistance to chloramphenicol on mitochondrial DNA: developing the first mouse model of mitochondrial DNA disease. Nat. Med. 5:957–960 10.1038/11403 [DOI] [PubMed] [Google Scholar]
- Martin M., Cho J., Cesare A.J., Griffith J.D., Attardi G. 2005. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 123:1227–1240 10.1016/j.cell.2005.09.040 [DOI] [PubMed] [Google Scholar]
- McCulloch V., Shadel G.S. 2003. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 23:5816–5824 10.1128/MCB.23.16.5816-5824.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies R.A., Gold P.H. 1971. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 246:2425–2429 [PubMed] [Google Scholar]
- Metodiev M.D., Lesko N., Park C.B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.G. 2009. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9:386–397 10.1016/j.cmet.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Montoya J., Gaines G.L., Attardi G. 1983. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 34:151–159 10.1016/0092-8674(83)90145-9 [DOI] [PubMed] [Google Scholar]
- Moraes C.T., Andreetta F., Bonilla E., Shanske S., DiMauro S., Schon E.A. 1991. Replication-competent human mitochondrial DNA lacking the heavy-strand promoter region. Mol. Cell. Biol. 11:1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse M.C., Bleau G., Dabhi V.M., Hétu F., Drobetsky E.A., Lindahl K.F., Perreault C. 1996. The COI mitochondrial gene encodes a minor histocompatibility antigen presented by H2-M3. J. Immunol. 156:3301–3307 [PubMed] [Google Scholar]
- Munnich A., Rustin P. 2001. Clinical spectrum and diagnosis of mitochondrial disorders. Am. J. Med. Genet. 106:4–17 10.1002/ajmg.1391 [DOI] [PubMed] [Google Scholar]
- Murphy M.P. 2009. How mitochondria produce reactive oxygen species. Biochem. J. 417:1–13 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.P., Holmgren A., Larsson N.G., Halliwell B., Chang C.J., Kalyanaraman B., Rhee S.G., Thornalley P.J., Partridge L., Gems D., et al. 2011. Unravelling the biological roles of reactive oxygen species. Cell Metab. 13:361–366 10.1016/j.cmet.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2009. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 5:706–708 10.4161/auto.5.5.8505 [DOI] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S., Shitara H., Nakada K., Ono T., Sato A., Suzuki H., Ogawa T., Masaki H., Hayashi J., Yonekawa H. 2010. Over-expression of Tfam improves the mitochondrial disease phenotypes in a mouse model system. Biochem. Biophys. Res. Commun. 401:26–31 10.1016/j.bbrc.2010.08.143 [DOI] [PubMed] [Google Scholar]
- Niu X., Trifunovic A., Larsson N.G., Canlon B. 2007. Somatic mtDNA mutations cause progressive hearing loss in the mouse. Exp. Cell Res. 313:3924–3934 10.1016/j.yexcr.2007.05.029 [DOI] [PubMed] [Google Scholar]
- Oldfors A., Fyhr I.-M., Holme E., Larsson N.-G., Tulinius M. 1990. Neuropathology in Kearns-Sayre syndrome. Acta Neuropathol. 80:541–546 10.1007/BF00294616 [DOI] [PubMed] [Google Scholar]
- Olivo P.D., Van de Walle M.J., Laipis P.J., Hauswirth W.W. 1983. Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature. 306:400–402 10.1038/306400a0 [DOI] [PubMed] [Google Scholar]
- Parisi M.A., Clayton D.A. 1991. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 252:965–969 10.1126/science.2035027 [DOI] [PubMed] [Google Scholar]
- Park C.B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., et al. 2007. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 130:273–285 10.1016/j.cell.2007.05.046 [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Asin-Cayuela J., Erdjument-Bromage H., Tempst P., Larsson N.G., Gustafsson C.M. 2009. MTERF2 is a nucleoid component in mammalian mitochondria. Biochim. Biophys. Acta. 1787:296–302 10.1016/j.bbabio.2009.01.018 [DOI] [PubMed] [Google Scholar]
- Rodeheffer M.S., Shadel G.S. 2003. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 278:18695–18701 10.1074/jbc.M301399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels D.C., Wonnapinij P., Cree L.M., Chinnery P.F. 2010. Reassessing evidence for a postnatal mitochondrial genetic bottleneck. Nat. Genet. 42:471–472 10.1038/ng0610-471 [DOI] [PubMed] [Google Scholar]
- Sandoval H., Thiagarajan P., Dasgupta S.K., Schumacher A., Prchal J.T., Chen M., Wang J. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 454:232–235 10.1038/nature07006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Kuroiwa T. 1991. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 196:137–140 10.1016/0014-4827(91)90467-9 [DOI] [PubMed] [Google Scholar]
- Shoubridge E.A., Wai T. 2008. Medicine. Sidestepping mutational meltdown. Science. 319:914–915 10.1126/science.1154515 [DOI] [PubMed] [Google Scholar]
- Shutt T.E., Gray M.W. 2006a. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 22:90–95 10.1016/j.tig.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Shutt T.E., Gray M.W. 2006b. Homologs of mitochondrial transcription factor B, sparsely distributed within the eukaryotic radiation, are likely derived from the dimethyladenosine methyltransferase of the mitochondrial endosymbiont. Mol. Biol. Evol. 23:1169–1179 10.1093/molbev/msk001 [DOI] [PubMed] [Google Scholar]
- Shutt T.E., Shadel G.S. 2010. A compendium of human mitochondrial gene expression machinery with links to disease. Environ. Mol. Mutagen. 51:360–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H.E., Schönfisch B., Perschil I., Chacinska A., Guiard B., et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 100:13207–13212 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.P., Köhler M., Graff C., Oldfors A., Magnuson M.A., Berggren P.O., Larsson N.G. 2000. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat. Genet. 26:336–340 10.1038/81649 [DOI] [PubMed] [Google Scholar]
- Sligh J.E., Levy S.E., Waymire K.G., Allard P., Dillehay D.L., Nusinowitz S., Heckenlively J.R., MacGregor G.R., Wallace D.C. 2000. Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc. Natl. Acad. Sci. USA. 97:14461–14466 10.1073/pnas.250491597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D. 2009. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 139:934–944 10.1016/j.cell.2009.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörensen L., Ekstrand M., Silva J.P., Lindqvist E., Xu B., Rustin P., Olson L., Larsson N.G. 2001. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J. Neurosci. 21:8082–8090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spåhr H., Samuelsson T., Hällberg B.M., Gustafsson C.M. 2010. Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain. Biochem. Biophys. Res. Commun. 397:386–390 10.1016/j.bbrc.2010.04.130 [DOI] [PubMed] [Google Scholar]
- Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. 2001. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 28:223–231 10.1038/90058 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Moraes C.T. 2005. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 14:893–902 10.1093/hmg/ddi082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.B., Freyer C., Elson J.L., Larsson N.G. 2008a. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Genet. 9:657–662 10.1038/nrg2396 [DOI] [PubMed] [Google Scholar]
- Stewart J.B., Freyer C., Elson J.L., Wredenberg A., Cansu Z., Trifunovic A., Larsson N.G. 2008b. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 6:e10 10.1371/journal.pbio.0060010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen D.F., Narendra D.P., Tanaka A., Manfredi G., Youle R.J. 2010. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. USA. 107:11835–11840 10.1073/pnas.0914569107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavi P., Hansson A., Zhang S.J., Larsson N.G., Westerblad H. 2005. Abnormal Ca(2+) release and catecholamine-induced arrhythmias in mitochondrial cardiomyopathy. Hum. Mol. Genet. 14:1069–1076 10.1093/hmg/ddi119 [DOI] [PubMed] [Google Scholar]
- Thorsness P.E., Fox T.D. 1990. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 346:376–379 10.1038/346376a0 [DOI] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., et al. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 429:417–423 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Trifunovic A., Hansson A., Wredenberg A., Rovio A.T., Dufour E., Khvorostov I., Spelbrink J.N., Wibom R., Jacobs H.T., Larsson N.G. 2005. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl. Acad. Sci. USA. 102:17993–17998 10.1073/pnas.0508886102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J.N., Holt I.J., Suomalainen A. 2004. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 13:3219–3227 10.1093/hmg/ddh342 [DOI] [PubMed] [Google Scholar]
- Tyynismaa H., Mjosund K.P., Wanrooij S., Lappalainen I., Ylikallio E., Jalanko A., Spelbrink J.N., Paetau A., Suomalainen A. 2005. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl. Acad. Sci. USA. 102:17687–17692 10.1073/pnas.0505551102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermulst M., Wanagat J., Kujoth G.C., Bielas J.H., Rabinovitch P.S., Prolla T.A., Loeb L.A. 2008. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 40:392–394 10.1038/ng.95 [DOI] [PubMed] [Google Scholar]
- Wai T., Teoli D., Shoubridge E.A. 2008. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 40:1484–1488 10.1038/ng.258 [DOI] [PubMed] [Google Scholar]
- Wallace D.C. 2001. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found. Symp. 235:247–263, discussion:263–266 10.1002/0470868694.ch20 [DOI] [PubMed] [Google Scholar]
- Wallace D.C. 2005. Mitochondria and cancer: Warburg addressed. Cold Spring Harb. Symp. Quant. Biol. 70:363–374 10.1101/sqb.2005.70.035 [DOI] [PubMed] [Google Scholar]
- Wallace D.C. 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76:781–821 10.1146/annurev.biochem.76.081205.150955 [DOI] [PubMed] [Google Scholar]
- Wallace D.C., Singh G., Lott M.T., Hodge J.A., Schurr T.G., Lezza A.M.S., Elsas L.J., II, Nikoskelainen E.K. 1988. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 242:1427–1430 10.1126/science.3201231 [DOI] [PubMed] [Google Scholar]
- Wang J., Wilhelmsson H., Graff C., Li H., Oldfors A., Rustin P., Brüning J.C., Kahn C.R., Clayton D.A., Barsh G.S., et al. 1999. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 21:133–137 10.1038/5089 [DOI] [PubMed] [Google Scholar]
- Wang J., Silva J.P., Gustafsson C.M., Rustin P., Larsson N.G. 2001. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc. Natl. Acad. Sci. USA. 98:4038–4043 10.1073/pnas.061038798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T., Diaz F., Spiegelman B.M., Moraes C.T. 2008. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 8:249–256 10.1016/j.cmet.2008.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wenz T., Luca C., Torraco A., Moraes C.T. 2009a. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 9:499–511 10.1016/j.cmet.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wenz T., Rossi S.G., Rotundo R.L., Spiegelman B.M., Moraes C.T. 2009b. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc. Natl. Acad. Sci. USA. 106:20405–20410 10.1073/pnas.0911570106 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Williams S.L., Huang J., Edwards Y.J., Ulloa R.H., Dillon L.M., Prolla T.A., Vance J.M., Moraes C.T., Züchner S. 2010. The mtDNA mutation spectrum of the progeroid Polg mutator mouse includes abundant control region multimers. Cell Metab. 12:675–682 10.1016/j.cmet.2010.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wredenberg A., Wibom R., Wilhelmsson H., Graff C., Wiener H.H., Burden S.J., Oldfors A., Westerblad H., Larsson N.G. 2002. Increased mitochondrial mass in mitochondrial myopathy mice. Proc. Natl. Acad. Sci. USA. 99:15066–15071 10.1073/pnas.232591499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskaya E., Mejia E., Byrnes J., Hambardjieva E., Garcia-Diaz M. 2010. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 141:982–993 10.1016/j.cell.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Oyaizu Y., Oyaizu H., Olsen G.J., Woese C.R. 1985. Mitochondrial origins. Proc. Natl. Acad. Sci. USA. 82:4443–4447 10.1073/pnas.82.13.4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T., Reyes A., Cluett T.J., Yang M.Y., Bowmaker M., Jacobs H.T., Holt I.J. 2006. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 25:5358–5371 10.1038/sj.emboj.7601392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikallio E., Tyynismaa H., Tsutsui H., Ide T., Suomalainen A. 2010. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 19:2695–2705 10.1093/hmg/ddq163 [DOI] [PubMed] [Google Scholar]
- Zeviani M., Di Donato S. 2004. Mitochondrial disorders. Brain. 127:2153–2172 10.1093/brain/awh259 [DOI] [PubMed] [Google Scholar]
- Zeviani M., Moraes C.T., DiMauro S., Nakase H., Bonilla E., Schon E.A., Rowland L.P. 1988. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology. 38:1339–1346 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ikeno Y., Qi W., Chaudhuri A., Li Y., Bokov A., Thorpe S.R., Baynes J.W., Epstein C., Richardson A., Van Remmen H. 2009. Mice deficient in both Mn superoxide dismutase and glutathione peroxidase-1 have increased oxidative damage and a greater incidence of pathology but no reduction in longevity. J. Gerontol. A Biol. Sci. Med. Sci. 64:1212–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Solaroli N., Bjerke M., Stewart J.B., Rozell B., Johansson M., Karlsson A. 2008. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum. Mol. Genet. 17:2329–2335 10.1093/hmg/ddn133 [DOI] [PubMed] [Google Scholar]