Abstract
We investigated whether genetic polymorphisms in the promoter region of the pro-apoptotic beta-2 adrenergic receptor gene (ADRB2) influence treatment-induced changes in ADRB2 expression in leukemia cells and response to chemotherapy. The ADRB2 promoter region was genotyped in germline DNA from 369 children with acute lymphoblastic leukemia (ALL). For 95 patients, sufficient RNA was available before and after in vivo treatment to assess treatment-induced gene expression changes in ALL cells. After treatment, the median ADRB2 mRNA expression was 9-fold lower in leukemia cells of patients who ultimately relapsed compared to patients who remained in continuous complete remission. Polymorphisms in the ADRB2 promoter were significantly linked to methotrexate induced up-regulation in ADRB2 gene expression in ALL cells. Moreover, the ADRB2 promoter haplotype was significantly related to early treatment response in 245 uniformly treated children with ALL. We conclude that germline polymorphisms in ADRB2 are linked to the antileukemic effects of ALL chemotherapy.
Keywords: pharmacogenetics, promoter gene polymorphism, treatment-induced gene expression changes
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease caused by genetic abnormalities, including non-random chromosomal translocations that produce aberrant gene fusions or inappropriate expression of oncogenes resulting in different genetic subtypes with differing prognoses. This disease was the first disseminated cancer for which chemotherapy achieved cures in the majority of childhood cases (1–3). Using largely empirical strategies to improve the efficacy of combination chemotherapy over the past twenty years, long term disease-free survival now exceeds 80% for childhood ALL (4). Unfortunately, little is known about why treatment fails in up to 20% of patients; thus, new insights into the biology and treatment response of this disease are needed if cure rates are to be further improved. Pharmacogenomic studies have revealed the genetic basis for inherited differences in many drug effects (5;6). We have recently shown that ALL cells of different genetic or lineage subtypes share common pathways of genomic response to the same treatment (7), and that the nature of changes in gene expression after treatment has the potential to identify patients whose ALL cells respond aberrantly to drug treatment.
In the current study, we found changes in gene expression in ALL cells following in vivo treatment with methotrexate and mercaptopurine that were related to treatment outcome (disease relapse vs. continuous complete remission) using a cohort of uniformly treated patients. The beta-2 adrenergic receptor gene (ADRB2) was one of the significant genes with the highest discriminating weight to distinguish patients who relapsed from those who remained in continuous complete remission. This is consistent with previous findings showing the beta-adrenergic receptor as a regulator of apoptosis (8;9). Furthermore, we identified genetic polymorphisms of the ADRB2 promoter region that regulate treatment-induced changes in ADRB2 expression, are associated with treatment-induced changes in ADRB2 expression in ALL cells, and are significantly linked to early treatment failure. Thus, using treatment-induced changes in gene expression in ALL cells (in vivo), we identified polymorphisms in a gene not previously linked to treatment outcome in human cancer.
Results
Treatment-specific changes in gene expression differed in patients who relapsed
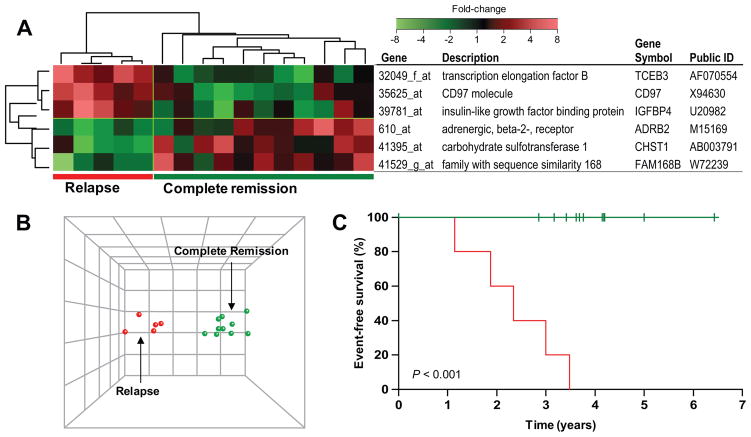
The relation between changes in gene expression after in vivo treatment and clinical outcome was initially assessed in patients treated with low-dose methotrexate plus mercaptopurine (LDMTX plus MP, n = 16), because this was the largest group investigated using the same gene expression array with long clinical follow-up (median: 11.9 years, range: 2.4 – 12.2). Demographic and biological characteristics of the five patients who relapsed (two hematological, two CNS and one ocular relapse) and the 11 patients who remained in continuous complete remission (CCR) are summarized in Table 1. ALL lineage was the only variable significantly associated with relapse in this exploratory group of patients, which was therefore included as a covariate in the outcome analysis. Using a Cox proportional hazard regression model, with lineage as a covariate, we identified 145 gene probe sets that were related to relapse (P < 0.05), based on changes in gene expression after treatment. Permutation analysis indicated that the smallest P-value was achieved with 87 probe sets (P = 0.028). Linear discriminant analysis was performed on these 87 probe sets, to rank them by their ability to discriminate the five patients who relapsed from the 11 patients who remain in complete remission. Hierarchical clustering using the six genes with the highest discriminating power, clearly separated patients according to outcome, as depicted in Figure 1A. The principal component analysis (PCA) illustrates the degree of discrimination of treatment outcome (Figure 1B), and the Kaplan-Meier estimates of event-free survival depict the clinical outcome for these two groups (Figure 1C; P < 0.001). The other up-front treatment groups of this first analysis done in patients treated on St. Jude Total 13B protocol did not include any early relapse events (before two years after diagnosis) and were therefore not analyzed.
Table 1.
Characteristics of patients in the initial treatment outcome analysis.
| n | Age at diagnosis2 | Sex | Race | Lineage | DNA ploidy | Molecular translocations | |
|---|---|---|---|---|---|---|---|
| Relapse | 5 | 6.8 | Male: 4 Female: 1 |
White: 4 Black: 0 Other: 1 |
B: 2 T: 3 |
Hyperdiploid: 1 Other: 4 |
TEL-AML: 0 ND2: 5 |
| Complete remission | 11 | 5.3 | Male: 9 Female: 2 |
White: 8 Black: 1 Other: 2 |
B: 11 T: 0 |
Hyperdiploid: 1 Other: 10 |
TEL-AML: 4 ND2: 7 |
| P-value1 | P = 0.74 | P = 1.0 | P = 1.0 | P = 0.018 | P = 0.52 | P = 0.26 |
P-value by Fisher’s exact test except Age by Wilcoxon’s rank sum test,
Median in years, 2ND =none detected
Figure 1. Gene expression changes that discriminate treatment outcome.
(A) Expression changes of the six most discriminating genes by linear discriminant analysis (LDA) in patients who remained in complete remission (n =11) and in patients who relapsed (n = 5). (B) Principal component analysis (PCA) plot of 16 patients who received LDMTX/MP, using the six most significant genes predictive of relapse (P = 0.028). (C) Event-free survival curves estimated by Kaplan-Meier analysis.
The six most discriminating genes include a transcription factor (TCEB3), an insulin like growth factor protein (IGFBP4) a gene involved in keratan sulfate biosynthesis (CHST1), a hypothetical protein (FAM168B), and two G protein-coupled receptors (CD97 and ADRB2) (P < 0.009, Table 2). Because ADRB2 is annotated as a “positive regulator of apoptosis” (8;9), we focused further studies on this gene.
Table 2.
Genes identified by Cox proportional hazard regression analysis.
| Probe set ID | Identifier | Gene name | Gene symbol | Median FC CCR | Median FC Relapse B-lineage | Median FC Relapse T-lineage | P-value1 | Weight by LDA |
|---|---|---|---|---|---|---|---|---|
| 32049_f_at | AF070554 | transcription elongation factor B (SIII) | TCEB3 | −2.3 | +2.9 | +1.7 | 0.0005 | 0.295 |
| 35625_at | X94630 | CD97 molecule | CD97 | −1.1 | +1.6 | +1.5 | 0.0009 | 0.207 |
| 41395_at | AB003791 | carbohydrate sulfotransferase 1 | CHST1 | −1.2 | −2.9 | −4.3 | 0.0045 | 0.193 |
| 41529_g_at | W72239 | family with sequence similarity 168 | FAM168B | +1.3 | −1.2 | +1 | 0.0074 | 0.186 |
| 39781_at | U20982 | insulin-like growth factor binding protein 4 | IGFBP4 | −1.1 | +1.6 | +2.8 | 0.0085 | 0.175 |
| 610_at | M15169 | adrenergic, beta-2-, receptor, surface | ADRB2 | +1.1 | −3.1 | −4.3 | 0.0005 | 0.167 |
P-value determined by t-test.
FC: Fold-change; (−) indicating a decrease in expression, (+) indicating an increase in expression after treatment with LDMTX/MP; CCR: continuous complete remission; LDA: linear discriminant analysis
Patients who ultimately relapsed down-regulated expression of the ADRB2 gene in their leukemia cells after antimetabolite treatment (five of five patients, median change: 3.5-fold down-regulation), in contrast to patients who remained in complete remission, in whom ADRB2 expression increased in the majority of patients (8 of 11 patients, median: 2.3-fold up-regulation, P = 0.005,). Moreover, the level of ADRB2 expression in post-treatment samples, determined by log2 signal intensity on the expression arrays, was approximately 9-fold higher in ALL cells from patients who remained in continuous remission compared to patients who relapsed (median of 10.2 vs. 7.1; P = 0.003). The ADRB2 mRNA expression level in the diagnostic samples was the same in patients who relapsed and those who remained in remission (median of 9.37 vs. 8.01; P = 0.27, Wilcoxon rank sum, Supplemental Figure 1).
ADRB2 promoter haplotype and treatment-induced changes in expression
The ADRB2 promoter activity is reported to be located primarily within ~550 bp upstream of the translational start site (10) and our PCR fragment covers this region and about 142 bp downstream of the start codon. Sequencing analysis revealed six variable sites in this span of 694 bp in ADRB2 gene. All of the six SNPs (single nucleotide polymorphism) have been previously identified (nucleotide -468C/G, -367T/C, -47T/C, -20T/C, 46G/A [codon 16], 79C/G [codon 27] relative to the first nucleotide of the start codon). Because several promoter SNPs and coding region SNPs in ADRB2 have been associated with altered expression (using luciferase reporter activity) or function (receptor density) of the beta-2 adrenergic receptor (10–12), we elected to sequence this region of genomic DNA from 369 ALL patients including the 95 patients in whom we had determined the magnitude of change in ADRB2 expression in leukemia cells after in vivo treatment with mercaptopurine and/or methotrexate, having observed that the expression of this gene increased in about 80% of patients and decreased or was unchanged in about 20% (Supplementsl Figure 2).
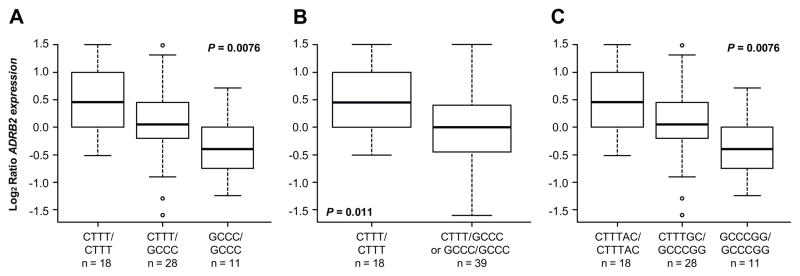
During the period of sequencing a newer updated version of the Affymetrix array became available. Most of the genes interrogated on these two versions of the Affymetrix array are the same, but the exact oligonucleotide probes are not identical. We determined that the sensitivity is about 7-fold higher based on the Affymetrix’ present call ratio on the new U133A compared to the old U95A array and this underestimation is even more amplified in the logratio data. Of note, ADRB2 expression signals from diagnosis and post-treatment cells are correlated, the ratio is not identical. Unfortunately, the relation of the U133 and U95 data is not linear especially in the low signal range therefore logratios were computed from the same array version for the pre-treatment and post-treatment analysis of any given patient. As shown in Figure 2, our analysis of the 57 patients treated uniformly with upfront HDMTX revealed a significant association between the treatment-induced changes in ADRB2 expression (obtained from the updated HG-U133A array) and the ADRB2 haplotype for these four promoter SNPs. Patients who have at least one ADRB2 allele with the CTTT haplotype at these four promoter SNP loci (in high LD) have significantly greater up-regulation of ADRB2 expression after HDMTX treatment than those who have only the GCCC haplotype. The difference in ADRB2 expression was expressed as fold-change of post treatment signal intensity relative to the pre-treatment signal intensity. The first panel (Figure 2A) shows three promoter groups (CTTT homozygotes, CTTT/GCCC heterozygotes, and GCCC homozygotes; P = 0.013, Kruskal-Wallis test), whereas the middle panel (Figure 2B) shows patients with one or two CTTT alleles vs. those without a CTTT allele (i.e., GCCC/GCCC; P = 0.004). It is known that the codon 27 SNP (nt79) is in strong LD with the four promoter SNPs (as also observed in our patients, see Figure 2 above). As expected, results were the same when we included the codon 27 SNP in this analysis, and compared patients who have either the five SNP GCCCG allele as homozygous for this halpotype the CTTTC/GCCCG heterozygous allele or the CTTTC allele. The codon 16 SNP (nt46) was in weaker LD with the other four promoter SNPs and the codon 27 SNP and was not significantly associated with diagnostic or post-treatment expression level or with gene expression change after treatment. Similarly, when we included both coding SNPs in this analysis, and compared patients who have the six SNP GCCCGC allele homozygous for this haplotype vs. those without this allele, the GCCCGC homozygous allele had the least up-regulation, with the heterozygous trending to be intermediate and those without the GCCCGC allele having the greatest up-regulation (Figure 2C; P = 0.0076, Kruskal-Wallis test).
Figure 2. Treatment induced changes in ADRB2 gene expression, by ADRB2 promoter haplotypes/diplotypes.
Treatment induced changes in ADRB2 gene expression in 57 patients with ALL, by ADRB2 promoter haplotypes/diplotypes. Box and whisker plots show the medians (horizontal line), interquartile ranges (box), and ranges (whiskers) of changes in ADRB2 gene expression, excluding outliers which are shown as circles. (A) The haplotypes shown are, CTTT/CTTT or CTTT homozygotes (n = 18), CTTT/GCCC or CTTT/GCCC heterozygotes (n = 28), GCCC/GCCC or GCCC homozygotes (n = 11); P = 0.0076, Kruskal-Wallis test). (B) The diplotypes shown are, CTTT/CTTT or CTTT/GCCC (n = 46), GCCC/GCCC or GCCC homozygotes (n = 11); P = 0.0104, t-test). (C) The diplotypes shown are, CTTT/CTTT or CTTT/GCCC (n = 46), GCCC/GCCC or GCCC homozygotes (n = 11); P = 0.010, t-test).
We also analyzed ADRB2 expression levels before HDMTX treatment and whether this differed by ADRB2 genotype or haplotype, to determine whether basal promoter activity is influenced by individual SNPs or combination of SNPs: the initial mRNA expression in ALL cells did not differ in the different genotype or haplotype groups (P = 0.20), consistent with prior transfection studies in other tissues (15).
ADRB2 promoter haplotype is associated with initial treatment response
Given the significant association between ADRB2 haplotype and the change in ADRB2 expression post-treatment (Figure 2), and the association between changes in ADRB2 expression and treatment outcome (Figure 1), we genotyped a large number of patients with ALL treated on a single protocol, to determine whether the GCCC ADRB2 haplotype was associated with a poor treatment response.
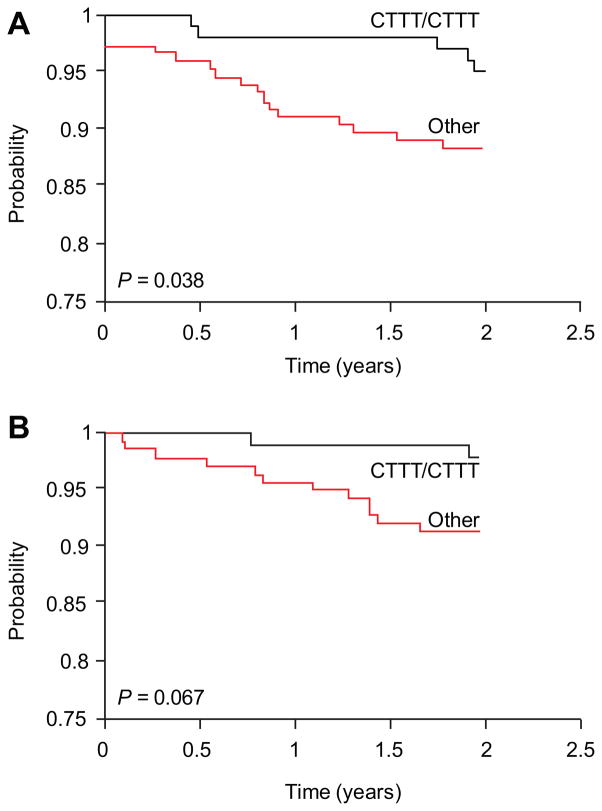
First, we estimated the association between haplotypes and Overall Survival (OS) and Event-Free Survival (EFS). In the entire cohort of 369 patients from all three clinical protocols, there was not a significant difference among the ADRB2 haplotypes. Because the more recent protocols (Total 14 and 15) used significantly more intensive chemotherapy, we analyzed the 245 patients who were uniformly treated in the initial clinical protocol (Total 13B). The median follow-up time is 11.9 years. Among the 245 patients, 47 had adverse events (e.g., disease relapse and no response) by the end of follow-up. OS and EFS data were stratified for risk factors using established criteria (16), and high-risk and low-risk patients were analyzed separately. This analysis revealed that there was no difference in OS or EFS when patients with the CTTT/CTTT genotype were compared to patients with either the GCCC/CTTT or GCCC/GCCC genotypes, stratified by ALL risk groups (overall P-value is 0.75 [OS] and 0.61 [EFS]), and no difference was found for the ADRB2 haplotype when the entire period of follow-up was analyzed. However, the heterozygous GCCC/CTTT or the homozygous GCCC/GCCC promoter diplotype was associated with significantly worse 2-year OS when compared to the homozygous CTTT/CTTT diplotype (P = 0.038, Figure 3A), a similar association was found for the 2-year EFS (P = 0.067, Figure 3B).
Figure 3. Overall and event-free survival in ALL patients with and without the homozygous ADRB2 CTTT promoter haplotype.
Kaplan-Meier analysis of (A) Overall and (B) event-free survival in ALL patients with and without the homozygous ADRB2 CTTT promoter haplotype based on the first two years after diagnosis.
Discussion
Despite steady improvement in the treatment of ALL over the last four decades, treatment failure remains a major problem (2). A barrier to further progress is a lack of understanding why some patients with the same molecular subtype of ALL are not cured by treatment that is effective in 80% of patients. Previously, we identified specific gene expression patterns in ALL cells prior to treatment that were related to chemotherapy sensitivity and treatment outcome (17;18). To date few studies have investigated the relationship between post-treatment leukemia cell characteristics and treatment outcome (19). Indeed, it is likely that the response of cancer cells to drug treatment may provide additional information about the underlying causes of treatment failure.
In this study, we investigated treatment-specific changes in gene expression in leukemia cells after treatment with MTX and MP, and identified genes that exhibited significantly different changes between relapsed patients and those who remained in continuous complete remission. Of the six genes that best discriminated treatment response, five are annotated with known biological functions. Three genes exhibited higher expression in the relapse samples after treatment (CD97, insulin-like growth factor binding protein IGFBP4, transcription elongation factor B TCEB3), and three genes exhibited lower expression after treatment (beta 2-adrenergic receptor ADRB2, carbohydrate sulfotransferase 1 CHST1, family with sequence similarity 168 FAM168B), with the latter being a hypothetical protein of which the biological function is unknown. CD97 is a G protein-coupled receptor, the expression of which is rapidly up-regulated in activated lymphocytes (20). A high level of CD97 expression has been associated with a more aggressive stage and lymph node metastases of thyroid carcinoma (21). Insulin-like growth factor binding proteins (IGFBP1-6) can either inhibit or enhance cellular responses to IGF-I and IGF-II (22), with IGFBP4 being predominantly an inhibitor of IGF-induced mitogenesis (23). CHST1 is involved in keratan sulfate biosynthesis. Carbohydrate sulfotransferases are transmembrane enzymes in the golgi network, that play a critical role in extracellular signaling and adhesion (24). Significantly lower sulfotransferase activity has been reported in human breast cancer cell lines (25) and has been associated with progression and metastasis of colon cancer (26;27).
The beta-2 adrenergic receptor (ADRB2) has been reported to regulate apoptosis, leading us to focus on potential mechanisms underlying differences in its up-regulation in leukemia cells after treatment (8;9). The beta-2 adrenergic receptor is a plasma membrane bound G-protein coupled receptor. This gene has been of particular interest in pulmonary and cardiovascular diseases and several SNPs have been identified (11). In peripheral mononuclear blood cells, there was a minimal effect of the −367, −47, 46, 79 SNPs on receptor expression and on cAMP generation after agonist stimulation (28;29). However, currently no studies have been done in acute leukemia. We observed a 9-fold lower level of ADRB2 expression after in vivo treatment in leukemia cells of patients who ultimately relapsed. Consistent with this finding, a lower density of beta 2-adrenergic receptors in chronic lymphocytic leukemia cells has been associated with disease progression (30) and ADRB2 inhibition leads to cell invasion and transformation of benign prostate cells (31). Furthermore, ADRB2 has been linked to apoptosis in lymphoid tissue, via two mechanisms. The first involves activation of adenylate cyclase which elevates intracellular cyclic adenosine monophosphate (cAMP) (8;32) and the second mechanism involves activation of Src-family tyrosine kinases (32). cAMP has been shown to induce apoptosis in human leukemia (33–36), lymphoma (37) and ovarian cancer cells (38) by mechanisms that have not fully been elucidated. Putative mechanisms include the activation of protein kinase A (PKA) and the phosphorylation of downstream targets such as Bcl-2 (37;39). This raises the possibility that therapeutic agents that elevate cAMP may augment the efficacy of current treatment for childhood ALL, particularly the subset of patients who respond poorly due to ADRB2 down-regulation. In this regard, novel pharmacological agents that can increase cAMP (i.e., phosphodiesterase inhibitors) have been shown to synergize with conventional antileukemic agents in human leukemia cell lines in vitro (34). The pro-apoptotic effects mediated via the beta 2-adrenergic receptor signaling pathway are consistent with our findings of significantly higher ADRB2 expression in ALL blasts from patients who remained in remission compared to those who relapsed, as the latter patients avidly down-regulated ADRB2 expression after methotrexate and mercaptopurine treatment.
The current findings indicate that treatment-specific changes in gene expression can provide new insights into how individual anticancer agents evoke their cytotoxic effects and how cancer cells respond to avoid cytotoxicity or trigger apoptosis. Furthermore, differences in changes of gene expression after MTX plus MP were able to identify patients more likely to have an early relapse of their leukemia. MTX and MP constituted the majority of therapy given to these patients over 2.5 to 3 years of treatment, rendering the current study particularly well suited to assess the relationship between genomic responses to these agents and treatment outcome.
We next investigated whether genetic regulatory SNPs (rSNP) in the promoter of ADRB2 could influence treatment-induced changes observed in ADRB2 expression and potentially response to chemotherapy. Our analyses revealed four previously described rSNPs within ADRB2 promoter region grouped into two haplotypes. Although no significant association was found between ADRB2 basal expression level and genotype, patients harboring at least one allele with the CTTT haplotype had significantly greater up-regulation of ADRB2 expression after MTX treatment (P = 0.0076, Figure 2). This suggests that these promoter polymorphisms either introduce a new trans-regulator binding site or alter the binding affinity for a regulatory protein that is activated in response to drug treatment. The current findings should stimulate additional research to define the precise mechanism by which these SNPs alter treatment-induced changes in ADRB2 expression.
In a cohort of 245 uniformly treated children with long follow-up information, we assessed whether the ADRB2 CTTT haplotype was related to treatment outcome. The GCCC haplotype was significantly associated with worse OS and a similar trend was observed for EFS. This is consistent with our initial finding that the large majority of patients who remain in complete remission increased expression of ADRB2 after treatment. Of note, the haplotype is not significantly related to survival using all 369 patients, but treatment administered in the protocol Total 14 and 15 is significantly more intense and is thus not comparable. We conclude that the haplotype has an effect in patients treated on a less-intense ALL treatment protocols (not uncommon in efforts to reduce toxicity). The greater risk of relapse during the first 2 years of treatment largely coincides with the period of time when methotrexate and mercaptopurine are being given as continuation therapy. Significance was not maintained for the 5-year period of long-term follow-up, possibly due to more direct effects of treatment (intrinsic resistance), versus late relapse or of development of resistant subclones.
In conclusion, our study has shown that inherited differences in the ADRB2 promoter haplotype influence treatment-induced changes in ADRB2 expression in leukemia cells and that this is associated with a greater risk of disease relapse during the first two years of treatment. This illustrates the potential of treatment-induced changes in gene expression to identify genomic determinants of treatment response and to potentially reveal novel therapeutic targets.
Material an Methods
Primary leukemia cells
Leukemia cells were isolated from 369 patients with newly diagnosed ALL who were enrolled on St. Jude Children’s Research Hospital Total Therapy Studies 13B, 14 and 15 (40;41). The St. Jude Institutional Review Board approved all protocols and informed consent was obtained from parents or guardians, and consent or assent was also obtained from patients, when appropriate. Cells were obtained from bone marrow aspirates performed at diagnosis (pre-treatment) (42) and one day post-treatment with mercaptopurine (MP) [1 g/m2 I.V.] or high-dose methotrexate (HDMTX) [1 g/m2 I.V.] given alone, or mercaptopurine in combination with either low-dose MTX (LDMTX/MP) [180 mg/m2 orally] or high-dose MTX (HDMTX/MP) [1.0 g/m2 I.V.]). A stratified randomization (by immunophenotype and DNA ploidy) was used to assign the initial treatment. Immunophenotype, DNA ploidy and chromosomal translocations were determined for each diagnostic sample, as previously described (4;42;43). Leukemic blast cells were isolated by Ficoll-Hypaque gradient and the percentage of leukemic blasts was similar in the diagnostic and post-treatment bone marrow samples (median 97% and 95%, respectively). All patients had sufficient blasts in their post-treatment bone marrow aspirates to permit RNA and DNA isolation from 5–10 million leukemia cells.
Trial registration: Total 15, Therapy for Newly Diagnosed Patients With Acute Lymphoblastic Leukemia, http://www.ClinicalTrials.gov (NCT00137111); Total 14, Total Therapy Study 14 for Newly Diagnosed Patients With Acute Lymphoblastic Leukemia (NCT00187005); Total 13BH, Phase III Randomized Study of Antimetabolite-Based Induction plus High-Dose MTX Consolidation for Newly Diagnosed Pediatric Acute Lymphocytic Leukemia at Intermediate or High Risk of Treatment Failure (NCI-T93-0101D); Total 13BL, Phase III Randomized Study of Antimetabolite- Based Induction plus High-Dose MTX Consolidation for Newly Diagnosed Pediatric Acute Lymphocytic Leukemia at Lower Risk of Treatment Failure (NCI-T93-0103D).
Gene expression profiling
Total RNA was extracted with TriReagent (MRC, Cincinnati, OH) from cryopreserved mononuclear cell suspensions from bone marrows at diagnosis and one day post-treatment (n = 95). High quality RNA was hybridized to the HG-U95Av2 (n = 53) or HG-U133Av 2.0 (n = 57) oligonucleotide microarray according to the manufacturer’s protocol (Affymetrix, Santa Clara, CA). These microarrays contain 12,599 or 22,284 gene probe sets, representing approximately 9,670 or 14,500 human genes, plus approximately 1495 or 3900 EST clones with unknown function (44). Reproducibility and independent validation of gene expression levels and fold-change in gene expression, were established as previously described (7). Scaled gene expression values for pre-treatment, post-treatment and fold-change (post-treatment versus pre-treatment ratio) were calculated using the default settings of Affymetrix Microarray Suite software version 5 (MAS 5.0) and GeneChip Operating System (Affymetrix, Santa Clara, CA). The HG-U95Av2 data is available via the accession number GSE412 in MIAME-compliant format at the Gene Expression Omnibus hosted by the NCBI (http://www.ncbi.nlm.nih.gov/geo/). Seven patients were excluded from that data set because they had no germline DNA available for genotyping (Supplemental Table 1).
Genotyping
Germline genomic DNA was isolated from normal blood cells obtained from 369 ALL patients after they achieved complete remission. Six SNPs (single nucleotide polymorphism, Supplemental Table 2) in the ADRB2 gene were detected by polymerase chain reaction (PCR), using primers to amplify a 693 bp fragment containing the polymorphic sites (forward primer: 5′-TTGAGACCTCAAGCCGCGCAGGCGCCCA-3′; reverse primer: 5′-CGATGGCCAGGACGATGAGAGACATGACGA-3′). The primers were designed based on NCBI accession number M15169. Aliquots of 20 ng genomic DNA were mixed in a 50 μl reaction volume containing 15 pmol of each primer, 0.2 mM of each dNTPs, 1X PCR reaction buffer with DMSO, 1.5 mM MgCl2 (Roche, Alameda, CA). After heating at 95 C for 10 minutes, 1μl of GC-RICH PCR system enzyme mix (Roche, Alameda, CA) to each tube of the reaction were added with holding the thermal cycler at 95°C. PCR amplification was done with 30 cycles: 95°C for 30 seconds, 65°C for 30 seconds, 72°C for 1 minutes and followed by a final extension step at 72°C for 7 minutes. PCR fragments were purified before sequencing by Shrimp Alkaline Phosphatases and Exonuclease I (USB Corporation, Cleveland, OH). Sequencing analyses were done by ABI Big Dye terminator reagents 3.0 on an ABI PRISM 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA).
Statistical analysis
Analysis was performed in R 2.9.0 software (www.r-project.org) if not otherwise specified. Analyses were performed on both fold-changes in gene expression after treatment and on post-treatment expression levels alone. The raw expression signals were log-transformed and probe sets were filtered out if assigned by MAS 5.0 as “absent”, or if assigned as “no change” in all patients. Principal component analysis and hierarchical clustering were performed using GeneMaths XT software (AppliedMaths, Belgium). Several supervised learning methods were used to select the most discriminating gene probe sets, including linear discriminant analysis with variance (LDA) (GeneMaths XT) and t-statistics. Probe sets were ranked according to their discriminating power, i.e., by P-values in t-statistics and by contribution to the discriminating components in LDA. To establish that these probe sets could discriminate treatment outcome, leave-one-out cross-validation using support vector machines was performed with the top-ranked probe sets.
Cox proportional hazard regression model was used to identify gene expression changes related to treatment outcome, with ALL lineage included as the second covariate. Genes related to treatment outcome were rank ordered by P-values based on their contribution to treatment outcome. Permutation test and LDA were performed as described elsewhere (17).
A total of 369 patients from St. Jude Total 13B (n = 245), Total 14 (n = 52) and Total 15 (n = 72) protocols were genotyped. Supplemental Figure 3 illustrates which analyses were available for which patients. OS was defined as the time interval from date of diagnosis to death from any cause or to last follow up. For EFS, an event was defined as all types of relapse, death due to any cause, second malignancy and no remission. The date of first event was used in calculating event-free survival. EFS time of zero were assigned to patients who did not achieve a complete remission by day 43 of induction therapy. Time was censored at the last follow-up date if no failure was observed. Kaplan-Meier estimator of the survival function was used and the comparisons between survival distributions were made using log-rank test. Cumulative incidences of second malignancies observed in the cohort were analyzed using Gray’s estimator with incorporation of competing events.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the outstanding technical support of Nancy Anderson, Michael Shipman, Mark Wilkinson and the staff in the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital. The authors also thank Drs. Paul Insel, Julie Johnson and John Lima for helpful discussions regarding the beta-2 adrenergic receptor.
Supported in part by the following NIH grants; R37 CA36401 (WEE, MVR, CHP), R01 CA78224 (WEE, MVR, CHP), RO1 CA51001 (MVR, CHP), RO1 CA71907 (JRD), U01 GM61393 (MVR, WEE), U01 GM61394, Cancer Center Support Grant CA21765, by a F.M. Kirby Clinical Research Professorship from the American Cancer Society (CHP), and by the American Lebanese Syrian Associated Charities (ALSAC), F32 1F32CA141762-01 (SWP), and a grant “Avenir” by the Institut National du Cancer (INCa), Boulogne Bilancourt, France (MHC.)
Footnotes
Authors disclosure of potential conflicts of interest
The authors indicated no potential conflicts of interest.
References
- 1.Pui CH, Campana D, Evans WE. Childhood acute lymphoblastic leukaemia -current status and future perspectives. The Lancet Oncology. 2001;2(10):597–607. doi: 10.1016/S1470-2045(01)00516-2. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–78. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 3.Simone JV. Childhood leukemia as a model for cancer research: the Richard and Hinda Rosenthal Foundation Award Lecture. Cancer Res. 1979;39(11):4301–7. [PubMed] [Google Scholar]
- 4.Pui CH, Evans WE. Acute lymphoblastic leukemia. N Engl J Med. 1998;339(9):605–15. doi: 10.1056/NEJM199808273390907. [DOI] [PubMed] [Google Scholar]
- 5.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 6.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990):464–8. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 7.Cheok MH, Yang W, Pui CH, Downing JR, Cheng C, Naeve CW, et al. Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat Genet. 2003;34(1):85–90. doi: 10.1038/ng1151. [DOI] [PubMed] [Google Scholar]
- 8.Gu C, Ma YC, Benjamin J, Littman D, Chao MV, Huang XY. Apoptotic signaling through the beta -adrenergic receptor. A new Gs effector pathway. J Biol Chem. 2000;275(27):20726–33. doi: 10.1074/jbc.M000152200. [DOI] [PubMed] [Google Scholar]
- 9.Ma YC, Huang XY. Novel signaling pathway through the beta-adrenergic receptor. Trends Cardiovasc Med. 2002;12(1):46–9. doi: 10.1016/s1050-1738(01)00138-4. [DOI] [PubMed] [Google Scholar]
- 10.Scott MG, Swan C, Wheatley AP, Hall IP. Identification of novel polymorphisms within the promoter region of the human beta2 adrenergic receptor gene. Br J Pharmacol. 1999;126(4):841–4. doi: 10.1038/sj.bjp.0702385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liggett SB. beta(2)-adrenergic receptor pharmacogenetics. Am J Respir Crit Care Med. 2000;161(3 Pt 2):S197–S201. doi: 10.1164/ajrccm.161.supplement_2.a1q4-10. [DOI] [PubMed] [Google Scholar]
- 12.Liggett SB. Polymorphisms of the beta2-adrenergic receptor. N Engl J Med. 2002;346(7):536–8. doi: 10.1056/NEJM200202143460718. [DOI] [PubMed] [Google Scholar]
- 13.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000;97(19):10483–8. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnatty SE, Abdellatif M, Shimmin L, Clark RB, Boerwinkle E. Beta 2 adrenergic receptor 5′ haplotypes influence promoter activity. Br J Pharmacol. 2002;137(8):1213–6. doi: 10.1038/sj.bjp.0704935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pui CH, Sandlund JT, Pei D, Campana D, Rivera GK, Ribeiro RC, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children’s Research Hospital. Blood. 2004;104(9):2690–6. doi: 10.1182/blood-2004-04-1616. [DOI] [PubMed] [Google Scholar]
- 17.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–42. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 18.Lugthart S, Cheok MH, den Boer ML, Yang W, Holleman A, Cheng C, et al. Identification of genes associated with chemotherapy crossresistance and treatment response in childhood acute lymphoblastic leukemia. Cancer Cell. 2005;7(4):375–86. doi: 10.1016/j.ccr.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Lee SC, Xu X, Chng WJ, Watson M, Lim YW, Wong CI, et al. Post-treatment tumor gene expression signatures are more predictive of treatment outcomes than baseline signatures in breast cancer. Pharmacogenetics and Genomics. 2009 doi: 10.1097/FPC.0b013e328330a39f. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 20.Hamann J, Eichler W, Hamann D, Kerstens HM, Poddighe PJ, Hoovers JM, et al. Expression cloning and chromosomal mapping of the leukocyte activation antigen CD97, a new seven-span transmembrane molecule of the secretion receptor superfamily with an unusual extracellular domain. J Immunol. 1995;155(4):1942–50. [PubMed] [Google Scholar]
- 21.Hoang-Vu C, Bull K, Schwarz I, Krause G, Schmutzler C, Aust G, et al. Regulation of CD97 protein in thyroid carcinoma. J Clin Endocrinol Metab. 1999;84(3):1104–9. doi: 10.1210/jcem.84.3.5557. [DOI] [PubMed] [Google Scholar]
- 22.Clemmons DR, Busby WH, Arai T, Nam TJ, Clarke JB, Jones JI, et al. Role of insulin-like growth factor binding proteins in the control of IGF actions. Prog Growth Factor Res. 1995;6(2–4):357–66. doi: 10.1016/0955-2235(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer MC, Schmid C, Waldvogel M, Schlapfer I, Futo E, Masiarz FR, et al. Characterization of recombinant human insulin-like growth factor binding proteins 4, 5, and 6 produced in yeast. J Biol Chem. 1992;267(18):12692–9. [PubMed] [Google Scholar]
- 24.Bowman KG, Bertozzi CR. Carbohydrate sulfotransferases: mediators of extracellular communication. Chem Biol. 1999;6(1):R9–R22. doi: 10.1016/S1074-5521(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 25.Brockhausen I, Yang JM, Burchell J, Whitehouse C, Taylor-Papadimitriou J. Mechanisms underlying aberrant glycosylation of MUC1 mucin in breast cancer cells. Eur J Biochem. 1995;233(2):607–17. doi: 10.1111/j.1432-1033.1995.607_2.x. [DOI] [PubMed] [Google Scholar]
- 26.Vavasseur F, Dole K, Yang J, Matta KL, Myerscough N, Corfield A, et al. O-glycan biosynthesis in human colorectal adenoma cells during progression to cancer. Eur J Biochem. 1994;222(2):415–24. doi: 10.1111/j.1432-1033.1994.tb18880.x. [DOI] [PubMed] [Google Scholar]
- 27.Yamori T, Kimura H, Stewart K, Ota DM, Cleary KR, Irimura T. Differential production of high molecular weight sulfated glycoproteins in normal colonic mucosa, primary colon carcinoma, and metastases. Cancer Res. 1987;47(10):2741–7. [PubMed] [Google Scholar]
- 28.Lipworth B, Koppelman GH, Wheatley AP, Le JI, Coutie W, Meurs H, et al. Beta2 adrenoceptor promoter polymorphisms: extended haplotypes and functional effects in peripheral blood mononuclear cells. Thorax. 2002;57(1):61–6. doi: 10.1136/thorax.57.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panebra A, Schwarb MR, Glinka CB, Liggett SB. Allele-specific binding of airway nuclear extracts to polymorphic beta2-adrenergic receptor 5′ sequence. Am J Respir Cell Mol Biol. 2007;36(6):654–60. doi: 10.1165/rcmb.2006-0394OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamp T, Liebl B, Haen E, Emmerich B, Hallek M. Defects of beta 2-adrenergic signal transduction in chronic lymphocytic leukaemia: relationship to disease progression. Eur J Clin Invest. 1997;27(2):121–7. doi: 10.1046/j.1365-2362.1997.700623.x. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Cao Q, Mehra R, Laxman B, Yu J, Tomlins SA, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12(5):419–31. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Ma YC, Huang J, Ali S, Lowry W, Huang XY. Src tyrosine kinase is a novel direct effector of G proteins. Cell. 2000;102(5):635–46. doi: 10.1016/s0092-8674(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 33.Myklebust JH, Josefsen D, Blomhoff HK, Levy FO, Naderi S, Reed JC, et al. Activation of the cAMP signaling pathway increases apoptosis in human B-precursor cells and is associated with downregulation of Mcl-1 expression. J Cell Physiol. 1999;180(1):71–80. doi: 10.1002/(SICI)1097-4652(199907)180:1<71::AID-JCP8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa R, Streiff MB, Bugayenko A, Kato GJ. Inhibition of PDE4 phosphodiesterase activity induces growth suppression, apoptosis, glucocorticoid sensitivity, p53, and p21(WAF1/CIP1) proteins in human acute lymphoblastic leukemia cells. Blood. 2002;99(9):3390–7. doi: 10.1182/blood.v99.9.3390. [DOI] [PubMed] [Google Scholar]
- 35.Siegmund B, Welsch J, Loher F, Meinhardt G, Emmerich B, Endres S, et al. Phosphodiesterase type 4 inhibitor suppresses expression of anti-apoptotic members of the Bcl-2 family in B-CLL cells and induces caspase-dependent apoptosis. Leukemia. 2001;15(10):1564–71. doi: 10.1038/sj.leu.2402232. [DOI] [PubMed] [Google Scholar]
- 36.Thompson EB, Medh RD, Zhou F, Ayala-Torres S, Ansari N, Zhang W, et al. Glucocorticoids, oxysterols, and cAMP with glucocorticoids each cause apoptosis of CEM cells and suppress c-myc. J Steroid Biochem Mol Biol. 1999;69(1–6):453–61. doi: 10.1016/s0960-0760(99)00063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan L, Herrmann V, Hofer JK, Insel PA. beta-adrenergic receptor/cAMP-mediated signaling and apoptosis of S49 lymphoma cells. Am J Physiol Cell Physiol. 2000;279(5):C1665–C1674. doi: 10.1152/ajpcell.2000.279.5.C1665. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava RK, Srivastava AR, Cho-Chung YS, Longo DL. Synergistic effects of retinoic acid and 8-Cl-cAMP on apoptosis require caspase-3 activation in human ovarian cancer cells. Oncogene. 1999;18(9):1755–63. doi: 10.1038/sj.onc.1202464. [DOI] [PubMed] [Google Scholar]
- 39.Steinberg RA, Coffino P. Two-dimensional gel analysis of cyclic AMP effects in cultured S49 mouse lymphoma cells: protein modifications, inductions and repressions. Cell. 1979;18(3):719–33. doi: 10.1016/0092-8674(79)90126-0. [DOI] [PubMed] [Google Scholar]
- 40.Pui CH, Pei D, Sandlund JT, Ribeiro RC, Rubnitz JE, Raimondi SC, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia. 2009 doi: 10.1038/leu.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360(26):2730–41. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeoh E-J, Ross ME, Surtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 43.Silverman LB, Gelber RD, Dalton VK, Asselin BL, Barr RD, Clavell LA, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Dana-Farber Consortium Protocol 91-01. Blood. 2001;97(5):1211–8. doi: 10.1182/blood.v97.5.1211. [DOI] [PubMed] [Google Scholar]
- 44.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21(1 Suppl):20–4. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.