Abstract
iso-Migrastatin (iso-MGS) has been actively pursued recently as an outstanding candidate of antimetastasis agents. Having characterized the iso-MGS biosynthetic gene cluster from its native producer Streptomyces platensis NRRL 18993, we have recently succeeded in producing iso-MGS in five selected heterologous Streptomyces hosts, albeit the low titers failed to meet expectations and cast doubt on the utility of this novel technique for large-scale production. To further explore and capitalize on the production capacity of these hosts, a thorough investigation of these five engineered strains with three fermentation media for iso-MGS production was undertaken. Streptomyces albus J1074 and Streptomyces lividans K4-114 were found to be preferred heterologous hosts, and subsequent analysis of carbon and nitrogen sources revealed that sucrose and yeast extract were ideal for iso-MGS production. After the initial optimization, the titers of iso-MGS in all five hosts were considerably improved by 3–18-fold in the optimized R2YE medium. Furthermore, the iso-MGS titer of S. albus J1074 (pBS11001) was significantly improved to 186.7 mg/L by a hybrid medium strategy. Addition of NaHCO3 to the latter finally afforded an optimized iso-MGS titer of 213.8 mg/L, about 5-fold higher than the originally reported system. With S. albus J1074 (pBS11001) as a model host, the expression of iso-MGS gene cluster in four different media was systematically studied via the quantitative RT–PCR technology. The resultant comparison revealed the correlation of gene expression and iso-MGS production for the first time; synchronous expression of the whole gene cluster was crucial for optimal iso-MGS production. These results reveal new insights into the iso-MGS biosynthetic machinery in heterologous hosts and provide the primary data to realize large-scale production of iso-MGS for further preclinical studies.
Keywords: iso-Migrastatin, Heterologous Streptomyces hosts, Fermentation optimization, Quantitative RT–PCR
Introduction
Natural products play an important role in the treatment of human diseases (Newman 2008). In the past two decades, an explosive growth in the cloning and sequencing of gene clusters encoding natural product biosynthesis and our increasing knowledge of these biosynthetic machineries have greatly accelerated the progress in expressing bio-synthetic gene clusters in heterologous hosts for natural product production (Galm and Shen 2006). This technology can be used to produce natural products derived from slow growing or even uncultured microorganisms, and to activate the biosynthetic potential of microbes from certain “silent” pathways (Wenzel and Muller 2005). Although the unexplored capacity of each heterologous host for the production of a natural product poses a possible bottleneck for its further application, the feasibility of producing complex natural products in model heterologous hosts has clearly been demonstrated (Sosio et al. 2000; Martinez et al. 2004; Zhang et al. 2008).
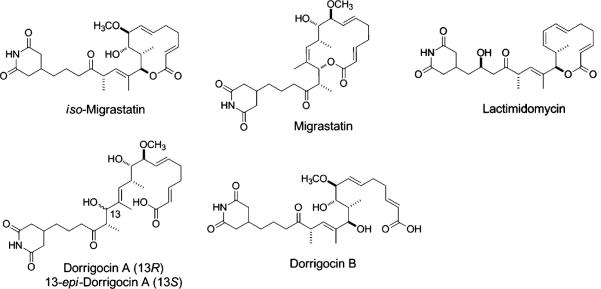
iso-Migrastatin (iso-MGS, Fig. 1) is a 12-membered macrolide belonging to the class of glutarimide-containing polyketides (Woo et al. 2002), which includes lactimidomycin (LTM), migrastatin (MGS), and dorrigocins (DGNs) (Fig. 1). This family has been actively pursued recently because its members represent outstanding candidates for the development of antimetastasis agents (Reymond and Cossy 2008; Ju et al. 2008, 2009). Both MGS and DGNs have been shown to be shunt metabolites of iso-MGS via H2O-mediated rearrangements, whereas iso-MGS is the only bona fide natural product biosynthesized by the native producer Streptomyces platensis NRRL 18993 (Ju et al. 2005). In addition to potently inhibiting human tumor cell migration, iso-MGS is also an antagonist of the muscarinic acetylcholine receptor, is associated with the suppression of multidrug resistance, and is being applied to the study of signal transduction pathways (Nakae et al. 2006; Takemoto et al. 2006; Lim et al. 2009).
Fig. 1.
Structures of glutarimide-containing polyketides including iso-migrastatin, migrastatin, dorrigocin A, B, and 13-epi-dorrigocin A, and lactimidomycin
We have recently cloned and characterized the iso-MGS biosynthetic gene cluster from S. platensis NRRL 18993 (Lim et al. 2009) and succeeded in developing a bacterial artificial chromosome (BAC) vector-based technology to produce iso-MGS in five selected Streptomyces heterologous hosts (Feng et al. 2009). However, the titers of iso-MGS in these engineered strains were still low under the set of media and fermentation conditions tested (Feng et al. 2009), prompting a more thorough examination of fermentation conditions and genetic analyses for these strains. The selection of suitable fermentation conditions may activate or improve the metabolite production, whereas the proper regulation and expression of the exogenously introduced iso-MGS gene cluster are prerequisite for the optimal production of iso-MGS in the heterologous hosts. To quantitatively investigate the relationship between iso-MGS gene expression and iso-MGS titer, as well as the interactions among the 11 genes within the iso-MGS gene cluster, a real-time quantitative reverse-transcription PCR (qRT–PCR) suitable for the accurate measurement of low level mRNA gene expression (Fleige et al. 2006) was applied to quantify the mRNA expression of iso-MGS genes under both original and optimized fermentation conditions.
We report here (1) titer improvement of iso-MGS in five heterologous Streptomyces hosts by rational fermentation optimization and (2) systematic analysis of mRNA expression of the iso-MGS gene cluster in Streptomyces albus J1074 under different culturing conditions by quantitative RT–PCR.
Materials and methods
Streptomyces strains
The five recombinant strains SB11001 [S. albus J1074 (pBS11001)], SB11002 [Streptomyces lividans K4-114 (pBS11001)], SB11003 [Streptomyces coelicolor M512 (pBS11001)], SB11004 [Streptomyces avermitilis SUKA4 (pBS11001)], and SB11005 [S. avermitilis SUKA5 (pBS11001)] have been described previously (Feng et al. 2009) and maintained as spore suspensions in 20% glycerol at −20 °C.
Media and culture condition
ISP-4 (Difco) medium was employed for strain sporulation, with addition of 25 μg/mL apramycin. B2 medium (Nakae et al. 2000; Feng et al. 2009), R2YE medium (Hopwood et al. 2000), and YEME medium (Hopwood et al. 2000) were applied as production medium, respectively. The fermentations of recombinant strains were described previously (Feng et al. 2009).
Isolation and HPLC analysis of iso-MGS produced by the recombinant strains
Isolation of iso-MGS from fermentation broth was previously described (Feng et al. 2009). iso-MGS sample (15 μL) was subjected to HPLC analysis on an Agilent 1100 system equipped with variable wavelength detector. The mobile phase was comprised of buffer A (15% CH3CN in H2O containing 0.1% HOAc pertained to the entire buffer mixture) and buffer B (80% CH3CN in H2O containing 0.1% HOAc pertained to the entire buffer mixture). Analytical HPLC was conducted using an Agilent TC-C18 column eluted with a linear gradient of 100% buffer A and 0% buffer B to 20% buffer A and 80% buffer B over 20 min, followed by 10 min at 20% buffer A and 80% buffer B at a flow rate of 1.0 mL/min with UV detection at 205 nm.
Medium optimization
B2, R2YE, and YEME media were evaluated for the selected recombinant strains, respectively. For each individual strain, the medium most amenable to high production of iso-MGS was chosen as the starting point for fermentation optimization. Major components of the target production medium, such as carbon sources and nitrogen sources, were then examined individually to determine their contribution towards the production of iso-MGS.
Two new hybrid media were designed by exchanging the C sources of R2YE and B2 media. Hybrid R-B (HRB) medium [2% glycerol, 2% dextrin, 0.5% yeast extract, 0.573% N-tris (hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES free acid), 1.012% MgCl2·6H2O, 0.025% K2SO4, 0.01% Difco casamino acids, 2 mL/L trace element solution (FeCl3·6H2O 200 mg/L, ZnCl2 40 mg/L, CuCl2·2H2O 10 mg/L, MnCl2·4H2O10 mg/L, Na2B4O7·10H2O10 mg/L, (NH4)6Mo7O24·4H2O 10 mg/L); after autoclaving, 15 mL/L 20% L-proline, 10 mL/L 0.5% KH2PO4, 7 mL/L 1 M NaOH, and 4 mL/L 5 M CaCl2·2H2O were added] and Hybrid B-R (HBR) medium (10% sucrose, 1% glucose, 1% Bacto soytone, 0.3% yeast extract, 0.2% (NH4)2SO4, 0.2% CaCO3, pH 7.0) were tested as production media for SB11001, respectively.
When NaHCO3 was regarded as possible precursor, after autoclave sterilization, different concentrations of NaHCO3 were added into HBR medium and the optimized HBR medium around 48 h without pH adjustment, respectively.
Real-time quantitative reverse-transcription-PCR in SB11001
SB11001 was cultured in R2YE medium, YEME medium, B2 medium, and optimized HBR medium, respectively. For R2YE medium and optimized HBR medium, cells were sampled from culture broth at 24, 36, 48, 60, 72, 84, 96, and 108 h, respectively. For YEME medium and B2 medium, cells were sampled from culture broth at 24, 48, 72, and 96 h, respectively. Each seed broth used for inoculation was set as the control point (0 h) of the corresponding medium.
Each sample was collected by centrifugation for 10 min at 6,000×g and washed with ddH2O twice at ambient room temperature; the resulting pellet was immediately frozen at −80 °C. After motorized grinding with liquid nitrogen, the powder of mycelia was re-suspended in the TRIzol Reagent (Invitrogen). RNA isolation was performed according to the manufacturer's instruction. Total RNA preparations were treated with DNase I to eliminate possible chromosomal DNA contamination. The absence of DNA contamination was checked by PCR, using primers corresponding to 16S rRNA gene (Table 1). The concentration and purity of total RNA were determined from the 260 nm/280 nm ratio. Integrity of RNA was checked by running 1 μg of RNA on a 1% agarose gel.
Table 1.
Primers for iso-MGS RT–PCR used in this work
| Gene | Primer (5′–3′) |
|---|---|
| mgsA | F: CCTGGAGGTGATCTGGAGCC |
| R: GCATGACGAAGTACGGGTTGC | |
| mgsB | F: CTGGACAGCGTCCGCTACCT |
| R: CGCCCGACAGAGCAGGAAT | |
| mgsC | F: AAGCACATGGAAGAGCGGTTCA |
| R: ACGATGCCGGACAGCGAGAC | |
| mgsD | F: TACGGCATCTTCAAGGACACG |
| R: CGGCAGCACATCGGCATAG | |
| mgsE | F: CGAATGCCAACGGCTATGTG |
| R: GGCGGTCCAGTCGGTGAG | |
| mgsF | F: TCGTCAACTCCCTTGTGCG |
| R: CCCCGTCTGCTGCGTCTC | |
| mgsG | F: CAGGTGTCCCAGTGGCACGAAG |
| R: ATGGAGGAGAAGCCGATGAAGCA | |
| mgsH | F: ACCGCATTCTCGGCTTCCTG |
| R: CGACCGTGATGTCGTGGCTC | |
| mgsI | F: GACGCGGCATTCGACTACAAGG |
| R: TCGGCGATTCTCACGAGCATTTT | |
| mgsJ | F: GCCTCCACCTCGGCTACTG |
| R: GAAGGTCTGACGCACGCTCT | |
| mgsK | F: TCCGAACTTTCCGATGCTGGTC |
| R: GTTTCCGGTCGTGTCGTGCC | |
| 16S rRNA of S. albus | F: CTTCGGTGGTGGATTAGTGG |
| R: CTTCCCTGCTGAAAGAGGTTT |
qRT–PCR reactions were carried out using TOYOBO ReverTra Ace® qPCR RT Kit. The fluorescent quantitative PCR reaction was carried out using ABI PRISM® 7500. Reactions were carried out according to the instructions of TOYOBO SYBR qPCR Mix Kit. The primers used for qRT–PCR are listed in Table 1.
Ninety-six (or fewer) replicated reactions of identical cDNA were performed using real-time PCR SYBR I Green detection. A master mixture containing all of the ingredients was pipetted into individual tubes of a 96-well reaction plate. Relative quantitation method was used to quantify gene expression changes, and 16S rRNA was used as the internal control. The samples were subjected to real-time PCR, and the analysis of individual CT values was completed automatically by the software of ABI PRISM® 7500.
Results
Media and hosts compared and preferred strains selected for future optimization
In addition to the B2 medium and R2YE medium that were successfully used previously for iso-MGS production in the selected heterologous hosts (Feng et al. 2009), YEME medium was also widely employed as a production medium (Hopwood et al. 2000; Galm and Shen 2006). These three commonly used production media were evaluated for iso-MGS production with all five recombinant strains. The titers of iso-MGS in B2, YEME, and R2YE media are shown in Table 2. Comparably, R2YE medium was found to best support iso-MGS production in different strains and was therefore chosen as the starting point for fermentation optimization.
Table 2.
iso-MGS production titers (mg/L) in five host strains
| Strains | Titera (mg/L) | |||
|---|---|---|---|---|
| R2YE medium | B2 medium | YEME medium | Optimized R2YE medium | |
| SB11001 | 43.8 | 2.3 | 8.6 | 128.6 |
| SB11002 | 24.0 | 11.4 | 18.8 | 86.3 |
| SB11003 | 25.0 | 1.9 | 10.1 | 78.2 |
| SB11004 | 2.2 | 0.5 | 0.0 | 42.0 |
| SB11005 | 1.5 | 2.0 | 0.0 | 33.6 |
The iso-MGS titer in the table was the average value of three parallel tests, and all the standard deviations were less than 6%
C-source optimization
The carbon source is essential to various primary or secondary metabolic pathways (Olano et al. 2008). With the invariable 20 g/L glucose in the medium, ten different carbon sources, including sucrose, glucose, dextrin, glycerol, soluble starch, galactose, maltose, fructose, mannitol, and xylose, were tested respectively, each in the concentration range from 50 to 200 g/L. As summarized in Table 3, sucrose, glucose, dextrin, and soluble starch could support cell growth well. Mannitol could only moderately satisfy cell growth, while xylose could not be utilized by the cells. The utilization of L-galactose, maltose, and fructose was distinct in different strains; SB11004 and SB11005 preferred these carbon sources. Cell growth was not synchronous with iso-MGS production. For example, the use of glucose dramatically improved cell growth, yet no iso-MGS could be detected in the broth. The production of iso-MGS was significantly affected by carbon sources (see Table 3). As a result, sucrose was chosen as the candidate and combined with 20 g/L glucose to form a complex main carbon source in the production medium. Comparably, SB11001, SB11002, and SB11003 showed better productivity than SB11004 and SB11005.
Table 3.
Utilization of C sources in five host strains
| C sources | Recombinant strains [iso-MGS titera (mg/L)/growth statusb] | ||||
|---|---|---|---|---|---|
| SB11001 | SB11002 | SB11003 | SB11004 | SB11005 | |
| Sucrose | 43.2/++ | 24.0/++ | 25.0/++ | 2.2/++ | 1.5/++ |
| D-Glucose | 0.0/++ | 0.0/++ | 0.0/++ | 0.0/++ | 0.0/++ |
| Dextrin | 6.8/++ | 10.6/++ | 5.5/++ | 0.4/++ | 1.9/++ |
| Glycerol | 3.4/++ | 3.6/+ | 0.6/+ | 0.0/+ | 0.0/− |
| Starch | 9.1/++ | 11.2/++ | 5.1/++ | 8.9/++ | 1.1/++ |
| L-Galactose | 5.8/+ | 3.5/+ | 0.6/+ | 4.8/++ | 11.6/++ |
| Maltose | 16.9/++ | 20.2/+ | 6.9/+ | 7.9/++ | 8.5/++ |
| D-Fructose | 1.2/+ | 4.0/+ | 4.3/+ | 7.1/++ | 10.7/++ |
| Xylose | 0.0/− | 0.0/− | 0.0/− | 0.0/− | 0.0/− |
| Mannitol | 0.9/+ | 3.1/+ | 2.8/+ | 1.2/+ | 0.8/+ |
The iso-MGS titer in the figure was the average value of three parallel tests, and all the standard deviations were less than 6%
b ++ Good growth, + normal growth, − poor growth
As shown in Fig. 2, various initial sucrose concentrations in the range of 30.0–550 g/L were investigated for the different strains to determine the optimal sucrose concentration. The results indicate that SB11001 and SB11002 may be the better recombinant strains for iso-MGS production of the five heterologous hosts compared. With 100 g/L sucrose, the highest titer of iso-MGS in SB11001 was 43.8 mg/L; whereas with 250 g/L sucrose, the highest titer of iso-MGS in SB11002 was 66.7 mg/L. Thus, these two strains were chosen for further fermentation optimization.
Fig. 2.
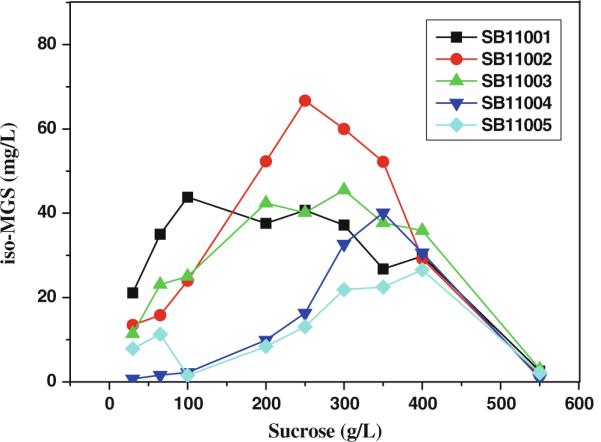
iso-MGS production titers as a function of sucrose concentration in five host strains. All the shake flask experiments were carried out in triplicate. The iso-MGS titer in the figure was the average value of three parallel tests, and all the standard deviations were less than 7%
N-source optimization
N sources play a central role in the growth of microorganisms and the biosynthesis of secondary metabolites. Based on the C-source optimization results (100 g/L sucrose for SB11001 and 250 g/L sucrose for SB11002), seven different nitrogen sources, including corn flour, soybean flour, malt extract, corn gluten meal, fish meal, polypeptone, and yeast extract, were evaluated at a concentration of 10 g/L.
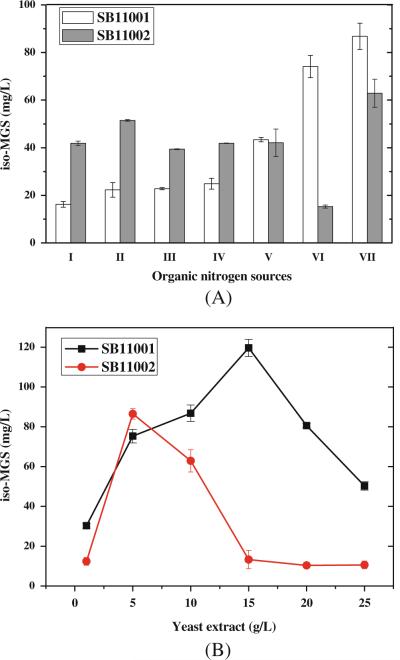
The results clearly indicate that yeast extract was the best organic nitrogen source for high iso-MGS production in both SB11001 and SB11002 (Fig. 3a). Subsequent determination of optimal yeast extract concentration was executed in SB11001 and SB11002, with the range of concentrations from 1.0 to 25 g/L. Results (Fig. 3b) indicate that SB11001 could afford 128.6 mg/L iso-MGS with 15 g/L yeast extract in the fermentation medium, and SB11002 could afford 86.5 mg/L iso-MGS with 5 g/L yeast extract in the fermentation medium. Higher concentrations of yeast extract led to dramatic decreases in iso-MGS production.
Fig. 3.
iso-MGS production titers as a function of different N sources by host strain SB11001 and SB11002. a I Corn flour. II Soybean flour. III Malt extract. IV Corn gluten meal. V Fish meal. VI Polypeptone. VII Yeast extract. b Effects of yeast extract concentration
Hybrid medium evaluation for SB11001
The product profiles of SB11001 in B2 medium were much simpler than in R2YE medium (Feng et al. 2009), prompting us to combine their merits and achieve better and cleaner production of iso-MGS. Thus, HBR medium and HRB medium with exchanged carbon sources were evaluated for SB11001, respectively. The titer of iso-MGS in HBR medium was 66.2 mg/L, much higher than that in HRB medium (11.7 mg/L). Subsequently, a simple orthogonal design had determined the optimal composition of three main components (sucrose 20%, Bacto soytone 2%, and yeast extract 1.5%) within the HBR medium, and the highest titer of iso-MGS was 186.7 mg/L.
Possible precursor addition
It is well known that acetyl-CoA is the accumulated intermediate from the tricarboxylic acid cycle in microorganism carbon source metabolism and acetyl-CoA carboxylase converts acetyl-CoA and HCO3− into malonyl-CoA (Moss and Lane 1972), the major substrate for iso-MGS biosynthesis (Lim et al. 2009). Accordingly, NaHCO3 was regarded as a possible precursor and evaluated in the concentration range of 0 to 3 g/L for iso-MGS production. Different concentrations of NaHCO3 were added into HBR medium and optimized HBR medium around 48 h, respectively. The iso-MGS titers were improved to 104.9 mg/L (about 60% higher) in HBR medium and 213.8 mg/L (about 11% higher) in optimized HBR medium, respectively, with 2 g/L NaHCO3 (Fig. 4).
Fig. 4.
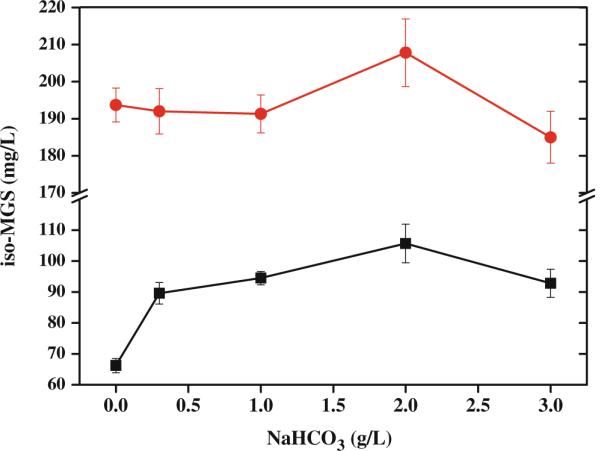
Effect of NaHCO3 addition in two hybrid media on iso-MGS production by host strain SB11001. Filled squares—HBR medium, filled circles—the optimized HBR medium
qRT–PCR results
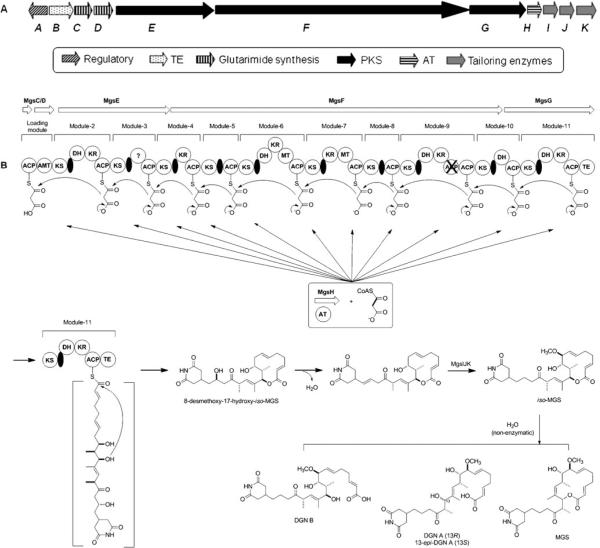
We have previously cloned the iso-MGS gene cluster from S. platensis NRRL 18993, defined the cluster to consist of one regulatory gene (mgsA) and ten biosynthesis genes (mgsBCDEFGHIJK), and proposed a pathway for iso-MGS biosynthesis (Fig. 5) (Lim et al. 2009). To quantitatively investigate the relationship between iso-MGS gene expression and iso-MGS titer, as well as the interactions among the genes within the cluster, qRT–PCR was applied to quantify the mRNA expression of all 11 genes under the original and optimized culture conditions.
Fig. 5.
Organization of the MGS gene cluster (a) and proposed biosynthetic pathway for iso-MGS and its biosynthetic relationship to MGS and DGN A, 13-epi-DGN A, and B (b) (Lim et al. 2009). ACP acyl carrier protein, AMT aminotransferase, AT acyltransferase, DGN dorrigocin, DH dehydratase, iso-MGS iso-migrastatin, KR ketoreductase, KS ketoacyl synthase, MGS migrastatin, MT methyltransferase, PKS polyketide synthase, TE thioesterase, ? domain of unknown function
The 2−ΔΔCT method (Livak and Schmittgen 2001)was used to analyze the relative changes of iso-MGS gene expression from real-time quantitative PCR experiments. The seed broth used for inoculation was set as the control point (0 h), and the 16S rRNA was used as the internal control. The calculation equation was proposed below: ΔΔCT = (CT,Target − CT,16SrRNA)Time x − (CT,Target − CT,16SrRNA)Time 0. The value of 2−ΔΔCT represents the relative fold change of the target gene expression.
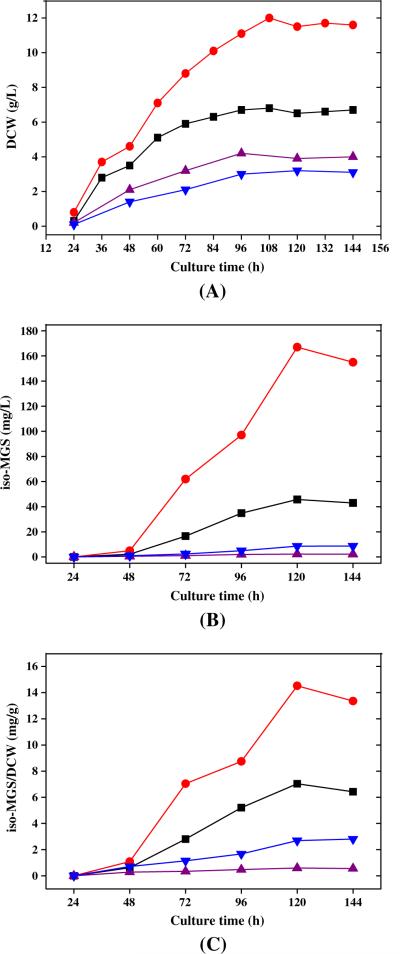
The expressions of all iso-MGS genes were compared in different fermentation media, including optimized HBR, R2YE, YEME, and B2. To emphasize the changes of gene expression during the time course, the value of −ΔΔCT was set as the ordinate to present the relative change of iso-MGS gene expression. The time course of cell growth and iso-MGS production of SB11001 in these four media were also determined, and each medium showed distinct time-course profiles (Fig. 6). The iso-MGS specific productivity (mg iso-MGS/g DCW) was directly correlated with the amount of mRNA derived from the same quantity of cells, in which the relationship between the production of iso-MGS and the expression of iso-MGS genes could be reflected. The comparison indicates that the optimized HBR medium was the best candidate to support both cell growth and high production of iso-MGS. While SB11001 could grow moderately well in the B2 medium, iso-MGS production was largely absent under these growth conditions.
Fig. 6.
Time-course comparisons of cell growth (a), iso-MGS concentration (b), and specific iso-MGS productivity (c) with SB11001 strain among in four different media. All the shake flask experiments were carried out in triplicate. The iso-MGS titer in the figure was the average value of three parallel tests, and all the standard deviations were less than 8%. Filled squares—R2YE medium, filled triangles—B2 medium, filled down-pointing triangles—YEME medium, filled circles—optimized HBR medium
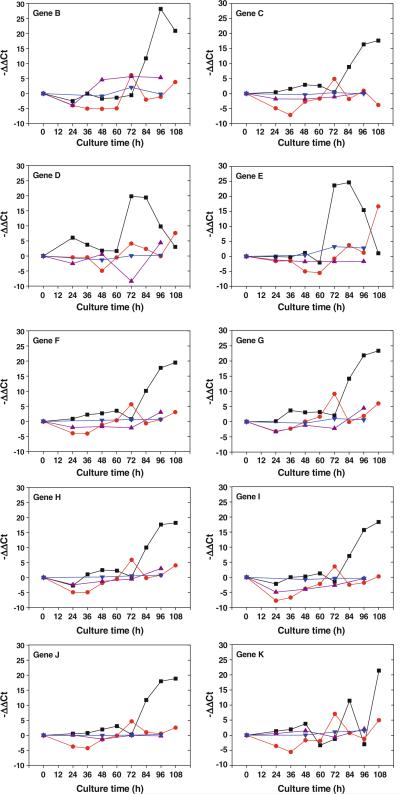
Transcription of ten (mgsBCDEFGHIJK) of the 11 genes within the iso-MGS cluster, with the exception of mgsA, was successfully detected via qRT–PCR analysis. Each transcribed gene presented the characteristic expression change pattern in the respective medium during the fermentation process (Fig. 7). In general, the expression of iso-MGS genes in the B2 medium and YEME medium were comparably stable. The expression of six genes (mgsBDFGHI) in the B2 medium fluctuated slightly during the fermentation, while only two genes (mgsBE) in the YEME medium showed altered expression levels. On the other hand, the gene expressions in either R2YE medium or optimized HBR medium were significantly changed especially during the late phase of fermentation, when the production of secondary metabolites dominated the fermentation process. Most genes showed similar expression change patterns in the optimized HBR medium. The expression started to increase around 60 h and to peak at 72 h. Following a quick relapse, the expression continued to increase until the end of fermentation. The only two exceptions were mgsC and mgsE. The expression of mgsC decreased at the end of fermentation. The expression of mgsE peaked at 84 h instead of 72 h, and showed a distinguishably high expression level at the end of fermentation. The situation in the R2YE medium was more complicated. The expression of seven genes (mgsBCFGHIJ) began to increase at 72 h and reached a significantly high level at the end of fermentation. The expression of mgsD and mgsE were highly enhanced during the 72-h to 84-h interval but quickly relapsed after that until the end of the fermentation. The clear zigzag expression pattern of mgsK was very unique.
Fig. 7.
qRT–PCR results of iso-MGS gene transcription with SB11001 strain among in four different media. The value in the figures was the average value of two parallel tests in individual tubes of a 96-well reaction plate, and the analysis was completed automatically by the instrument with standard deviations less than 5%. Filled squares—R2YE medium, filled triangles—B2 medium, filled down-pointing triangles—YEME medium, filled circles—optimized HBR medium
Discussion
Expression of biosynthetic gene clusters in heterologous hosts for natural product production is playing an increasing role in natural product-based drug discovery and development programs (Galm and Shen 2006). However, the initial titers of natural products in heterologous hosts are often low, likely due to the lack of a native environment that is necessary for the full expression of the target gene cluster. The engineered recombinant strains, therefore, must be complemented with the proper choice of fermentation medium to support cell growth and efficient functional expression of the target biosynthetic machinery.
iso-MGS, isolated originally from S. platensis NRRL 18993, has been previously produced in five Streptomyces heterologous hosts, but the potential of this strategy was not fully realized due to the low iso-MGS titers in these hosts (Feng et al. 2009). To explore the capacity of these engineered recombinant strains, a systematic fermentation optimization approach was taken to optimize iso-MGS titer in each host. S. albus J1074 (as exemplified by SB11001) and S. lividans K4-114 (as exemplified by SB11002) were found to be better hosts for iso-MGS production than the other three. Subsequent evaluation of common carbon and nitrogen sources indicate that sucrose and yeast extract, respectively, are suitable for iso-MGS production by S. albus J1074 (pBS11001) and S. lividans K4-114 (pBS11001). After initial optimization, the titers of iso-MGS in all five hosts were considerably improved by 3- to 18-fold in the optimized R2YE medium (Table 2), the highest of which was 128.6 mg/L by S. albus J1074 (pBS11001).
The product profiles of SB11001 in B2 medium were much simpler than those in the optimized R2YE medium. Thus, two hybrid media, derived from optimized R2YE medium and B2 medium in which carbon sources were exchanged, were subjected to analysis. Iso-MGS titers resulting from the HBR medium were superior to those obtained from the HRB medium. The subsequent three-factor/three-level orthogonal design, which focused on major medium components, including sucrose, Bacto soytone, and yeast extract, revealed the optimal composition of improved HBR medium to be 20% sucrose, 1% glucose, 2% Bacto soytone, 1.5% yeast extract, 0.2% (NH4)2SO4, and 0.2% CaCO3. The application of this medium afforded an iso-MGS titer of 186.7 mg/L. This result not only provided us a simpler medium to support better iso-MGS production but also suggested a new approach for optimization of traditional medium. This strategy may be particularly useful for the improvement of natural product production in heterologous hosts due to the unnatural relationship between the exogenously introduced gene clusters and the selected hosts.
The discovery of the relationship between bicarbonate and iso-MGS production was surprising. NaHCO3 usually is not a common constituent for fermentation media, but our results, derived from both original and optimized HBR media, clearly show that NaHCO3 plays an important role in iso-MGS production. While NaHCO3 may not directly contribute to iso-MGS production, it surely plays a critical role in the biosynthesis of malonyl-CoA, the major precursor for iso-MGS biosynthesis. Thus, improvement of other pathways related to the target metabolite biosynthesis is another alternative way to enhance the metabolite production.
Furthermore, the systematic qRT–PCR analyses of S. albus J1074 (pBS11001) in four different media were carried out to investigate the relationship between gene expression and metabolite production and to better understand the iso-MGS biosynthetic machinery. The RT–PCR analysis revealed that ten of the 11 iso-MGS genes, with the exception of mgsA, were successfully transcribed in the heterologous hosts. This result was further confirmed by quantitative PCR detection. Interestingly, mgsA has been previously characterized as a transcriptional activator essential for MGS and DGN production (Lim et al. 2009), but its function seemed more ambiguous in the heterologous hosts. There are several speculations for this unexpected observation: (1) as a activator gene, very low levels of mgsA expression could satisfy the transcription of the iso-MGS biosynthetic machinery, so the mRNA concentration of mgsA in cells was too low to be detected; or (2) though mgsA may not be transcribed in the heterologous hosts, its biosynthetic function was performed by a homolog endogenous to the heterologous hosts.
Based on the time-course results and qRT–PCR analysis (Figs. 6 and 7), changes to the expression profiles of most genes in B2 medium and YEME medium were minor, explaining the inert activity of iso-MGS biosynthetic machinery and low titers of iso-MGS specific production. On the contrary, though the gene expression levels in the R2YE medium were much higher than those in the optimized HBR medium, iso-MGS specific production in the latter was about 2-fold higher. Thus, the production of iso-MGS is impacted but not directly related to the expression level of its corresponding gene cluster. Moreover, the comparison of gene expression in the B2 medium and the YEME medium revealed that the type-II thioesterase MgsB may not pay a key role in iso-MGS biosynthesis. In contrast, the first polyketide synthase (PKS) MgsE and the amidotransferase MgsD may be extremely important in determining iso-MGS production (Fig. 5) since the expression profile difference of these corresponding genes could significantly affect the production of iso-MGS. The results obtained for the optimized HBR medium and the R2YE medium support this hypothesis (Fig. 7). Furthermore, the gene expression in the optimized HBR medium was the most synchronous one among all four media tested examined. This clearly suggests that the coordinated expression of the whole gene cluster rather than the individual gene is more crucial to the overall titer for the production of the target natural product in heterologous hosts.
Through the application of traditional single factor optimization, hybrid medium strategies, and the addition of a possible substrate, bicarbonate, to enhance precursor biosynthesis, the titer of iso-MGS in the heterologous host S. albus J1074 (as exemplified by recombinant strain SB11001) has been improved to 213.8 mg/L, which is about 4- and 5-fold higher than the titers reported previously for the native producer and the SB11011 recombinant strain, respectively (Feng et al. 2009). This result lends further support to the expression of biosynthetic gene clusters in heterologous hosts as a promising alternative for natural product production and offers a viable route for iso-MGS production by large-scale fermentation. Moreover, subsequent qRT–PCR analysis has provided insight into the relationship between biosynthetic gene expression and metabolite production in the heterologous hosts. An improved understanding of medium constituents and biosynthetic gene expression will greatly facilitate iso-MGS production on scales necessary for the further development of this agent and related analogs as antimetastasis agents and other clinically important therapeutics.
Acknowledgments
This work was supported in part by NIH of US (grants CA106150 and CA113297 to B.S.) and The Nature Science Foundation of China (Grant No. 20736008), The Ministry of Science and Technology (Grant No. 2007AA021702), and The Natural Science Foundation of Zhejiang, China, (R4090041), all to Z.X.
References
- Feng ZY, Wang LY, Rajski SR, Xu ZN, Coeffet-LeGal MF, Shen B. Engineered production of iso-migrastatin in heterologous Streptomyces hosts. Bioorgan Med Chem. 2009;17(6):2147–2153. doi: 10.1016/j.bmc.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT–PCR. Biotechnol Lett. 2006;28:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- Galm U, Shen B. Expression of biosynthetic gene cluster in heterologous hosts for natural product production and combinatorial biosynthesis. Expert Opin Drug Discov. 2006;1(5):409–437. doi: 10.1517/17460441.1.5.409. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation; Norwich: 2000. [Google Scholar]
- Ju JH, Lim SK, Jiang H, Shen B. Migrastatin and dorrigocins are shunt metabolites of iso-migrastatin. J Am Chem Soc. 2005;127:1622–1623. doi: 10.1021/ja043808i. [DOI] [PubMed] [Google Scholar]
- Ju JH, Rajski SR, Lim SK, Seo JW, Peters NR, Hoffmann FM, Shen B. Evaluation of new migrastatin and dorrigocin congeners unveils cell migration inhibitors with dramatically improved potency. Bioorg Med Chem Lett. 2008;18:5951–5954. doi: 10.1016/j.bmcl.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JH, Rajski SR, Lim SK, Seo JW, Peters NR, Hoffmann FM, Shen B. Lactimidomycin, iso-migrastatin and related glutarimide-containing 12-membered macrolides are extremely potent inhibitors of cell migration. J Am Chem Soc. 2009;131(4):1370–1371. doi: 10.1021/ja808462p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Ju JH, Zazopoulos E, Jiang H, Seo JW, Chen YH, Feng ZY, Rajski SR, Farnet CM, Shen B. iso-Migrastatin, migrastatin, and dorrigocin production in Streptomyces platensis NRRL 18993 is governed by a single biosynthetic machinery featuring an acyltransferase-less type I polyketide synthase. J Biol Chem. 2009;284(43):29746–29756. doi: 10.1074/jbc.M109.046805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen KJ. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martinez A, Kolvek SJ, Yip CLT, Hopke J, Brown KA, MacNeil IA, Osburne MS. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microb. 2004;70(4):2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J, Lane MD. Acetyl coenzyme A carboxylase III: further studies on the relation of catalytic activity to polymeric state. J Biol Chem. 1972;247(16):4944–4951. [PubMed] [Google Scholar]
- Nakae K, Yoshimoto Y, Sawa T, Homma Y, Hamada M, Takeuchi T, Imoto M. Migrastatin, a new inhibitor of tumor cell migration from Streptomyces sp. MK929-43F1. Taxonomy, fermentation, isolation and biological activities. J Antibiot. 2000;53(10):1130–1136. doi: 10.7164/antibiotics.53.1130. [DOI] [PubMed] [Google Scholar]
- Nakae K, Nishimura Y, Ohba S, Akamatsu Y. Migrastatin acts as a muscarinic acetylcholine receptor antagonist. J Antibiot. 2006;59:685–692. doi: 10.1038/ja.2006.91. [DOI] [PubMed] [Google Scholar]
- Newman DJ. Natural products as leads to potential drugs: an old process or the new hope for drug discovery? J Med Chem. 2008;51(9):2589–2599. doi: 10.1021/jm0704090. [DOI] [PubMed] [Google Scholar]
- Olano C, Lombó F, Méndez C, Salas JA. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng. 2008;10(5):281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Reymond S, Cossy J. Migrastatin and analogues: new anti-metastatic agents. CR Chim. 2008;11:1447–1462. [Google Scholar]
- Sosio M, Giusino F, Cappellano C, Bossi E, Puglia AM, Donadio S. Artificial chromosomes for antibiotic-producing actinomycetes. Nat Biotechnol. 2000;18:343–345. doi: 10.1038/73810. [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Tashiro E, Imoto M. Suppression of multidrug resistance by migrastatin. J Antibiot. 2006;59:435–438. doi: 10.1038/ja.2006.62. [DOI] [PubMed] [Google Scholar]
- Wenzel SC, Muller R. Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr Opin Biotech. 2005;16(6):594–606. doi: 10.1016/j.copbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Woo EJ, Starks CM, Carney JR, Arslanin R, Cadapan L, Zavala S, Licari P. Migrastatin and a new compound, isomigrastatin, from Streptomyces platensis. J Antibiot. 2002;55(2):141–146. doi: 10.7164/antibiotics.55.141. [DOI] [PubMed] [Google Scholar]
- Zhang HR, Wang Y, Pfeifer BA. Bacterial hosts for natural product production. Mol Pharmaceut. 2008;5(2):212–225. doi: 10.1021/mp7001329. [DOI] [PubMed] [Google Scholar]