Abstract
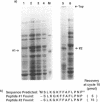
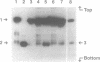
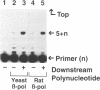
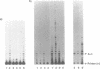
We have shown by activity gel that overexpression in E. coli of a yeast chromosome 3 open reading frame (ORF) designated YCR14C and bearing homology to mammalian DNA polymerases beta results in a new DNA polymerase in the host cells. The molecular mass of this enzyme corresponded to the YCR14C-predicted 67 kDa protein, and NH2-terminal amino acid sequencing confirmed that the expressed protein was encoded by the yeast ORF. This new yeast DNA polymerase was purified to homogeneity from E.coli. In a fashion similar to that of mammalian beta-polymerases, the purified yeast enzyme exhibited distributive DNA synthesis on DNA substrate with a single-stranded template and processive gap-filling synthesis on a short-gapped DNA substrate. Activity of this yeast beta-polymerase-like enzyme was sensitive to the beta-polymerase inhibitor ddNTP and resistant to both 1 mM NEM and neutralizing antibody to E. coli DNA polymerase I. These results, therefore, indicate that YCR14C encodes a DNA beta-polymerase-like enzyme in yeast, and we name it DNA polymerase IV. Yeast strains harboring a deletion mutation of the pol IV gene are viable, they exhibit no increase in sensitivity to ultraviolet light, ionizing radiation or alkylating agents, and sporulation and spore viability are not affected in the mutant.
Full text
PDF




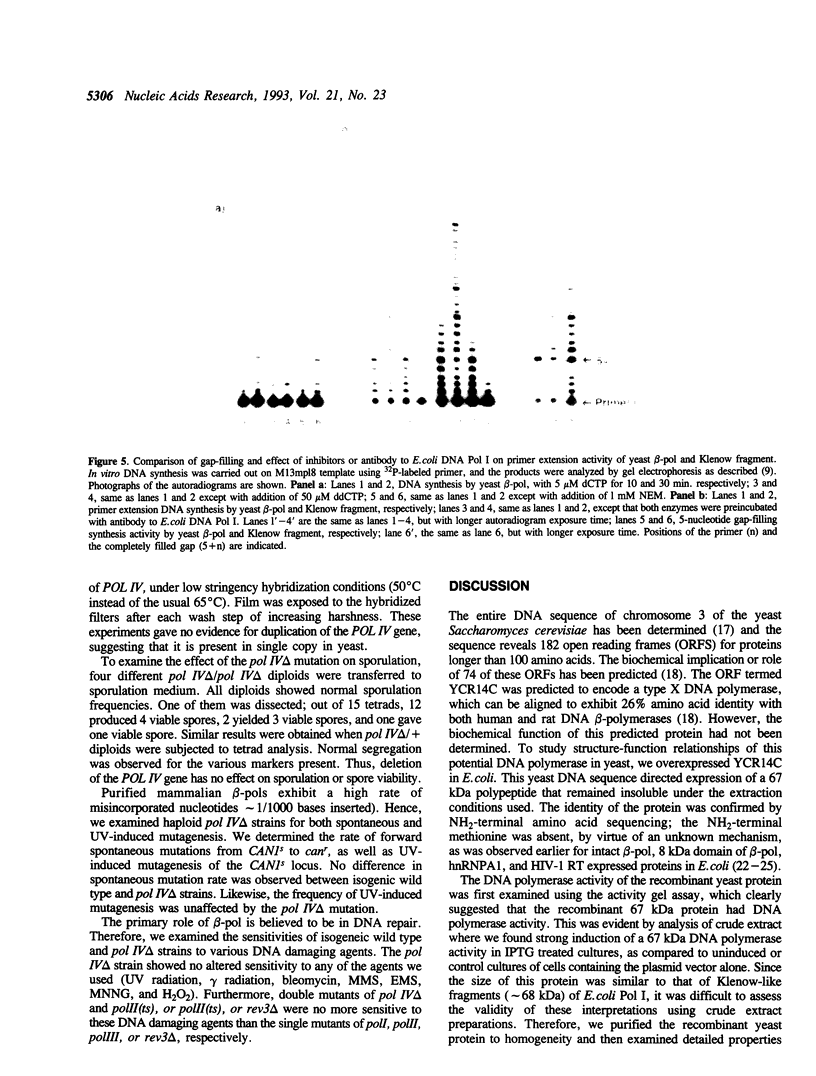

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Badaracco G., Capucci L., Plevani P., Chang L. M. Polypeptide structure of DNA polymerase I from Saccharomyces cerevisiae. J Biol Chem. 1983 Sep 10;258(17):10720–10726. [PubMed] [Google Scholar]
- Baril E. F., Scheiner C., Pederson T. A beta-like DNA polymerase activity in the slime mold Dictyostelium discoideum. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3317–3321. doi: 10.1073/pnas.77.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G. A., Heller H. M., Burgers P. M. DNA polymerase III from Saccharomyces cerevisiae. I. Purification and characterization. J Biol Chem. 1988 Jan 15;263(2):917–924. [PubMed] [Google Scholar]
- Becerra S. P., Kumar A., Lewis M. S., Widen S. G., Abbotts J., Karawya E. M., Hughes S. H., Shiloach J., Wilson S. H., Lewis M. S. Protein-protein interactions of HIV-1 reverse transcriptase: implication of central and C-terminal regions in subunit binding. Biochemistry. 1991 Dec 17;30(50):11707–11719. doi: 10.1021/bi00114a015. [DOI] [PubMed] [Google Scholar]
- Bork P., Ouzounis C., Sander C., Scharf M., Schneider R., Sonnhammer E. Comprehensive sequence analysis of the 182 predicted open reading frames of yeast chromosome III. Protein Sci. 1992 Dec;1(12):1677–1690. doi: 10.1002/pro.5560011216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. M. DNA polymerases from bakers' yeast. J Biol Chem. 1977 Mar 25;252(6):1873–1880. [PubMed] [Google Scholar]
- Cobianchi F., Karpel R. L., Williams K. R., Notario V., Wilson S. H. Mammalian heterogeneous nuclear ribonucleoprotein complex protein A1. Large-scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J Biol Chem. 1988 Jan 15;263(2):1063–1071. [PubMed] [Google Scholar]
- Dianov G., Price A., Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992 Apr;12(4):1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresler S. L., Roberts J. D., Lieberman M. W. Characterization of deoxyribonucleic acid repair synthesis in permeable human fibroblasts. Biochemistry. 1982 May 11;21(10):2557–2564. doi: 10.1021/bi00539a040. [DOI] [PubMed] [Google Scholar]
- Fukasawa H., Yamaguchi M., Chou M. Y., Matsumoto H., Matsukage A. Characterization of two DNA polymerases from cauliflower inflorescence. J Biochem. 1980 Apr;87(4):1167–1175. [PubMed] [Google Scholar]
- Furukawa Y., Yamada R. H., Kohno M. Presence of two DNA polymerases in Tetrahymena pyriformis. Nucleic Acids Res. 1979 Dec 20;7(8):2387–2398. doi: 10.1093/nar/7.8.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Karawya E., Swack J. A., Wilson S. H. Improved conditions for activity gel analysis of DNA polymerase catalytic polypeptides. Anal Biochem. 1983 Dec;135(2):318–325. doi: 10.1016/0003-2697(83)90689-9. [DOI] [PubMed] [Google Scholar]
- Kumar A., Widen S. G., Williams K. R., Kedar P., Karpel R. L., Wilson S. H. Studies of the domain structure of mammalian DNA polymerase beta. Identification of a discrete template binding domain. J Biol Chem. 1990 Feb 5;265(4):2124–2131. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Miller M. R., Chinault D. N. The roles of DNA polymerases alpha, beta, and gamma in DNA repair synthesis induced in hamster and human cells by different DNA damaging agents. J Biol Chem. 1982 Sep 10;257(17):10204–10209. [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Morrison A., Christensen R. B., Alley J., Beck A. K., Bernstine E. G., Lemontt J. F., Lawrence C. W. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989 Oct;171(10):5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbaugh D. W., Linn S. Gap-filling DNA synthesis by HeLa DNA polymerase alpha in an in vitro base excision DNA repair scheme. J Biol Chem. 1984 Aug 25;259(16):10247–10251. [PubMed] [Google Scholar]
- Nowak R., Siedlecki J. A., Kaczmarek L., Zmudzka B. Z., Wilson S. H. Levels and size complexity of DNA polymerase beta mRNA in rat regenerating liver and other organs. Biochim Biophys Acta. 1989 Jul 7;1008(2):203–207. doi: 10.1016/0167-4781(80)90010-x. [DOI] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Boyd J. B. Purification and characterization of a DNA polymerase beta from Drosophila*. J Biol Chem. 1985 Sep 5;260(19):10406–10411. [PubMed] [Google Scholar]
- Sakaguchi K., Lu B. C. Meiosis in Coprinus: characterization and activities of two forms of DNA polymerase during meiotic stages. Mol Cell Biol. 1982 Jul;2(7):752–757. doi: 10.1128/mcb.2.7.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel W., Raffael A. Two groups of deoxyribonucleic acid polymerases from Physarum polycephalum classified by differential sensitivity to N-ethylmaleimide, heparin, cytosine arabinoside triphosphate and ethidium bromide: evidence for a beta-like activity. FEBS Lett. 1980 Nov 17;121(1):81–85. doi: 10.1016/0014-5793(80)81271-3. [DOI] [PubMed] [Google Scholar]
- Seki S., Oda T. DNA repair synthesis in bleomycin-pretreated permeable HeLa cells. Carcinogenesis. 1986 Jan;7(1):77–82. doi: 10.1093/carcin/7.1.77. [DOI] [PubMed] [Google Scholar]
- Singhal R. K., Wilson S. H. Short gap-filling synthesis by DNA polymerase beta is processive. J Biol Chem. 1993 Jul 25;268(21):15906–15911. [PubMed] [Google Scholar]
- Smith C. A., Okumoto D. S. Nature of DNA repair synthesis resistant to inhibitors of polymerase alpha in human cells. Biochemistry. 1984 Mar 27;23(7):1383–1391. doi: 10.1021/bi00302a008. [DOI] [PubMed] [Google Scholar]
- Wang T. S., Korn D. Reactivity of KB cell deoxyribonucleic acid polymerases alpha and beta with nicked and gapped deoxyribonucleic acid. Biochemistry. 1980 Apr 29;19(9):1782–1790. doi: 10.1021/bi00550a009. [DOI] [PubMed] [Google Scholar]
- Wang T. S., Korn D. Specificity of the catalytic interaction of human DNA polymerase beta with nucleic acid substrates. Biochemistry. 1982 Mar 30;21(7):1597–1608. doi: 10.1021/bi00536a021. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wu X., Friedberg E. C. Nucleotide-excision repair of DNA in cell-free extracts of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):4907–4911. doi: 10.1073/pnas.90.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintersberger U., Blutsch H. DNA-dependent DNA polymerase from yeast mitochondria. Dependence of enzyme activity on conditions of cell growth, and properties of the highly purified polymerase. Eur J Biochem. 1976 Sep;68(1):199–207. doi: 10.1111/j.1432-1033.1976.tb10779.x. [DOI] [PubMed] [Google Scholar]