Abstract
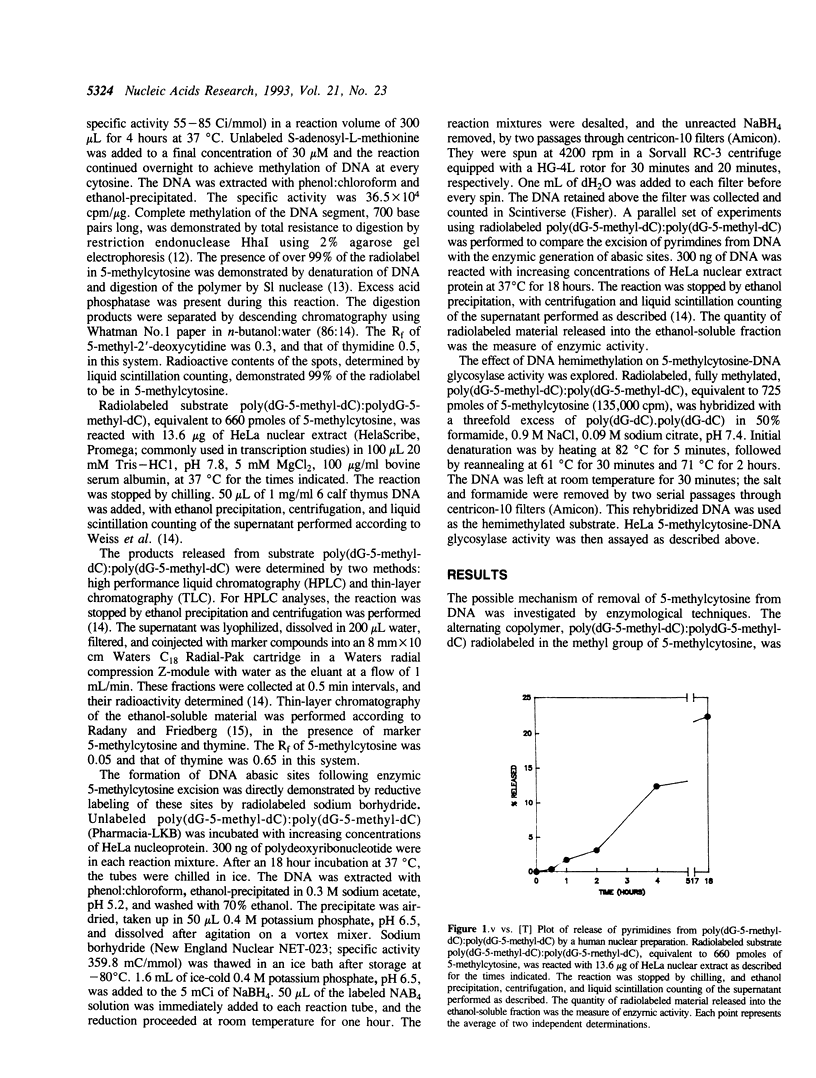
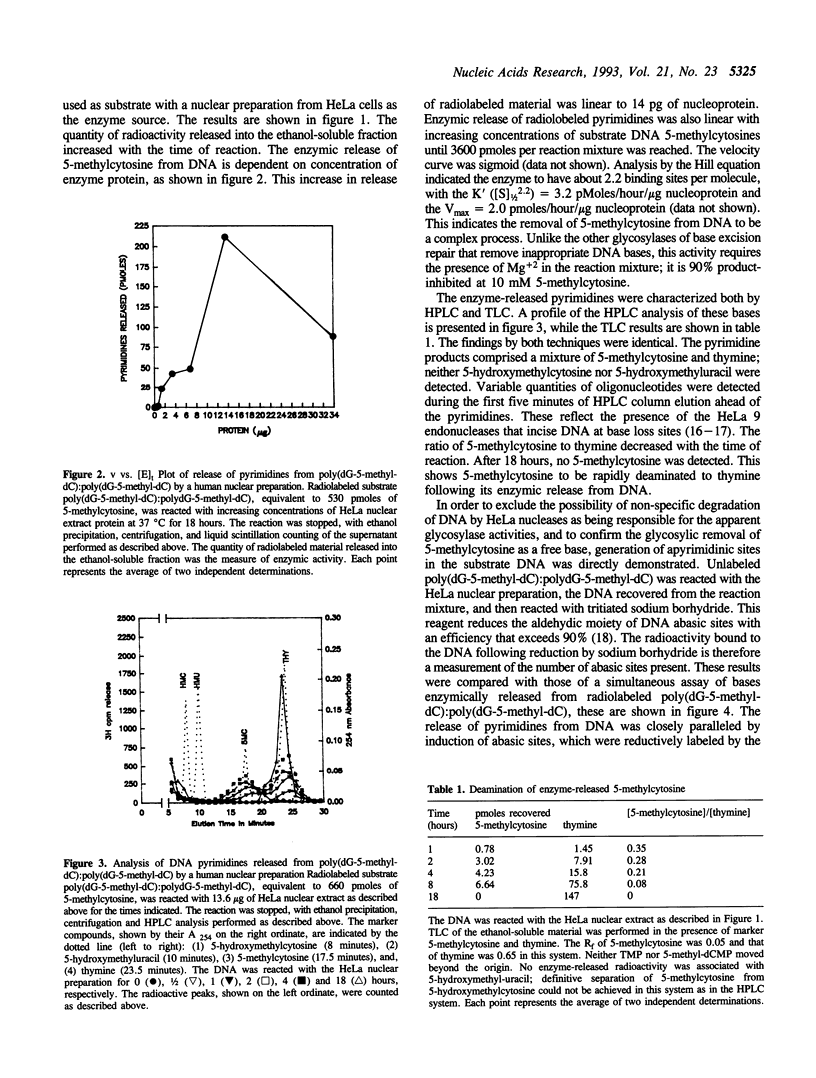
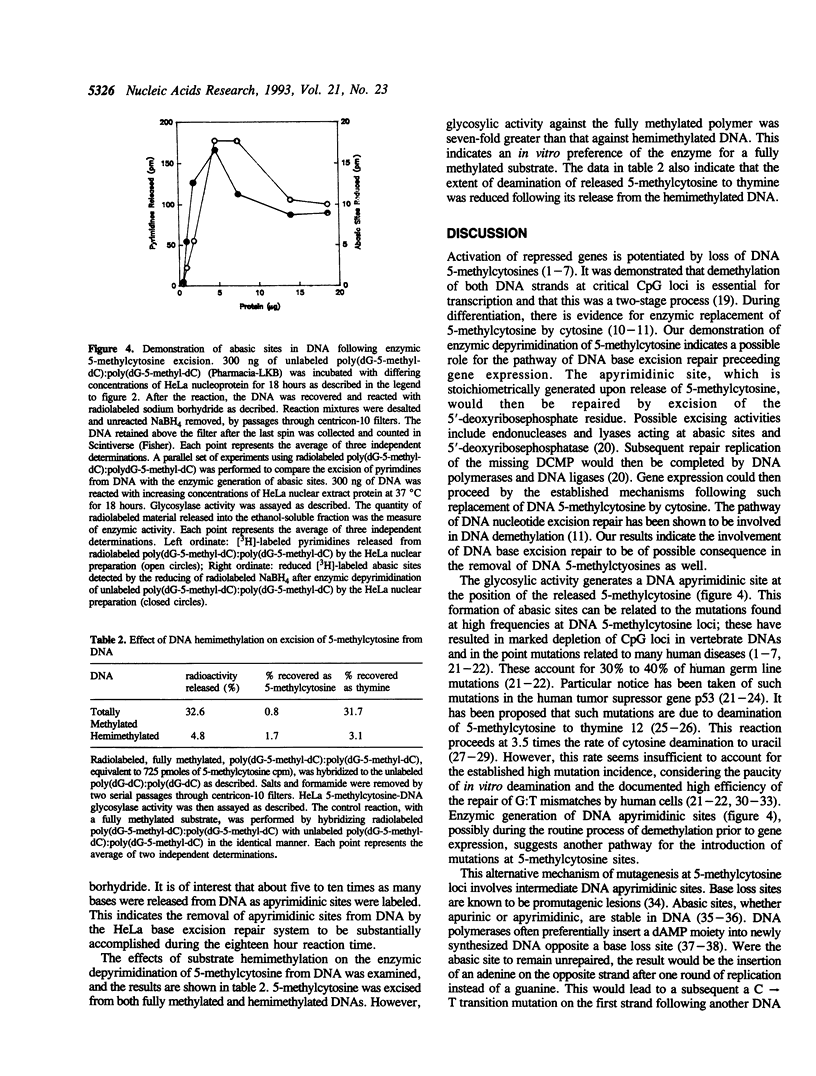
DNA 5-methylcytosine is a major factor in the silencing of mammalian genes; it is involved in gene expression, differentiation, embryogenesis and neoplastic transformation. A decrease in DNA 5-methylcytosine content is associated with activation of specific genes. There is much evidence indicating this to be an enzymic process, with replacement of 5-methylcytosine by cytosine. We demonstrate here enzymic release of 5-methylcytosines from DNA by a human 5-methylcytosine-DNA glycosylase activity, which affords a possible mechanism for such replacement. This activity generates promutagenic apyrimidinic sites, which can be related to the high frequency of mutations found at DNA 5-methylcytosine loci. The recovery of most released pyrimidines as thymines indicates subsequent deamination of free 5-methylcytosines by a 5-methylcytosine deaminase activity. This prevents possible recycling of 5-methylcytosine into replicative DNA synthesis via a possible 5-methyl-dCTP intermediate synthesized through the pyrimidine salvage pathway. Taken together, these findings indicate mechanisms for removal of 5-methylcytosines from DNA, hypermutability of DNA 5-methylcytosine sites, and exclusion of 5-methylcytosines from DNA during replication.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L. DNA methylation. The effect of minor bases on DNA-protein interactions. Biochem J. 1990 Jan 15;265(2):309–320. doi: 10.1042/bj2650309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. H., Hellewell S. B., Ingram V. M. Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol Cell Biol. 1984 Sep;4(9):1800–1806. doi: 10.1128/mcb.4.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982 Dec 21;21(26):6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987 Sep 11;50(6):945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- Castaing B., Boiteux S., Zelwer C. DNA containing a chemically reduced apurinic site is a high affinity ligand for the E. coli formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1992 Feb 11;20(3):389–394. doi: 10.1093/nar/20.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Duker N. J., Hart D. M., Grant C. L. Stability of the DNA apyrimidinic site. Mutat Res. 1982 Feb;103(2):101–106. doi: 10.1016/0165-7992(82)90012-4. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Norris K. F., Wang R. Y., Kuo K. C., Gehrke C. W. DNA cytosine methylation and heat-induced deamination. Biosci Rep. 1986 Apr;6(4):387–393. doi: 10.1007/BF01116426. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Zhang X. Y., Inamdar N. M. Spontaneous deamination of cytosine and 5-methylcytosine residues in DNA and replacement of 5-methylcytosine residues with cytosine residues. Mutat Res. 1990 May;238(3):277–286. doi: 10.1016/0165-1110(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Harris C. C. Chemical and physical carcinogenesis: advances and perspectives for the 1990s. Cancer Res. 1991 Sep 15;51(18 Suppl):5023s–5044s. [PubMed] [Google Scholar]
- Hergersberg M. Biological aspects of cytosine methylation in eukaryotic cells. Experientia. 1991 Dec 1;47(11-12):1171–1185. doi: 10.1007/BF01918381. [DOI] [PubMed] [Google Scholar]
- Holliday R., Grigg G. W. DNA methylation and mutation. Mutat Res. 1993 Jan;285(1):61–67. doi: 10.1016/0027-5107(93)90052-h. [DOI] [PubMed] [Google Scholar]
- Holliday R., Ho T. Gene silencing in mammalian cells by uptake of 5-methyl deoxycytidine-5'-triphosphate. Somat Cell Mol Genet. 1991 Nov;17(6):537–542. doi: 10.1007/BF01233618. [DOI] [PubMed] [Google Scholar]
- Holliday R. Mutations and epimutations in mammalian cells. Mutat Res. 1991 Sep-Oct;250(1-2):351–363. doi: 10.1016/0027-5107(91)90192-q. [DOI] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Jekunen A., Vilpo J. A. 5-Methyl-2'-deoxycytidine. Metabolism and effects on cell lethality studied with human leukemic cells in vitro. Mol Pharmacol. 1984 May;25(3):431–435. [PubMed] [Google Scholar]
- Jones P. A., Rideout W. M., 3rd, Shen J. C., Spruck C. H., Tsai Y. C. Methylation, mutation and cancer. Bioessays. 1992 Jan;14(1):33–36. doi: 10.1002/bies.950140107. [DOI] [PubMed] [Google Scholar]
- Jost J. P. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane C. M., Linn S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J Biol Chem. 1981 Apr 10;256(7):3405–3414. [PubMed] [Google Scholar]
- Lewis J., Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991 Jul 22;285(2):155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Repair of intrinsic DNA lesions. Mutat Res. 1990 May;238(3):305–311. doi: 10.1016/0165-1110(90)90022-4. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Preston B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Nyce J. Gene silencing in mammalian cells by direct incorporation of electroporated 5-methyl-2'-deoxycytidine 5'-triphosphate. Somat Cell Mol Genet. 1991 Nov;17(6):543–550. doi: 10.1007/BF01233619. [DOI] [PubMed] [Google Scholar]
- Paroush Z., Keshet I., Yisraeli J., Cedar H. Dynamics of demethylation and activation of the alpha-actin gene in myoblasts. Cell. 1990 Dec 21;63(6):1229–1237. doi: 10.1016/0092-8674(90)90418-e. [DOI] [PubMed] [Google Scholar]
- Radany E. H., Friedberg E. C. A pyrimidine dimer-DNA glycosylase activity associated with the v gene product of bacterophage T4. Nature. 1980 Jul 10;286(5769):182–185. doi: 10.1038/286182a0. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Szyf M., Kafri T., Roll M., Giloh H., Scarpa S., Carotti D., Cantoni G. L. Replacement of 5-methylcytosine by cytosine: a possible mechanism for transient DNA demethylation during differentiation. Proc Natl Acad Sci U S A. 1986 May;83(9):2827–2831. doi: 10.1073/pnas.83.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout W. M., 3rd, Coetzee G. A., Olumi A. F., Jones P. A. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990 Sep 14;249(4974):1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983 Sep 13;22(19):4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Scarano E., Iaccarino M., Grippo P., Parisi E. The heterogeneity of thymine methyl group origin in DNA pyrimidine isostichs of developing sea urchin embryos. Proc Natl Acad Sci U S A. 1967 May;57(5):1394–1400. doi: 10.1073/pnas.57.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teebor G. W., Duker N. J. Human endonuclease activity for DNA apurinic sites. Nature. 1975 Dec 11;258(5535):544–547. doi: 10.1038/258544a0. [DOI] [PubMed] [Google Scholar]
- Vilpo J. A., Vilpo L. M. Biochemical mechanisms by which reutilization of DNA 5-methylcytosine is prevented in human cells. Mutat Res. 1991 Jan;256(1):29–35. doi: 10.1016/0921-8734(91)90030-f. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Kuo K. C., Gehrke C. W., Huang L. H., Ehrlich M. Heat- and alkali-induced deamination of 5-methylcytosine and cytosine residues in DNA. Biochim Biophys Acta. 1982 Jun 30;697(3):371–377. doi: 10.1016/0167-4781(82)90101-4. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Gallagher P. E., Brent T. P., Duker N. J. Cytosine photoproduct-DNA glycosylase in Escherichia coli and cultured human cells. Biochemistry. 1989 Feb 21;28(4):1488–1492. doi: 10.1021/bi00430a010. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. In vitro correction of G.T mispairs to G.C pairs in nuclear extracts from human cells. Nature. 1989 May 18;339(6221):234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks A., Seldran M., Jost J. P. An estrogen-dependent demethylation at the 5' end of the chicken vitellogenin gene is independent of DNA synthesis. Nucleic Acids Res. 1984 Jan 25;12(2):1163–1177. doi: 10.1093/nar/12.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]



