Abstract
Messenger RNA (mRNA) stability is a critical determinant that affects gene expression. Many pathways have evolved to modulate mRNA stability in response to developmental, physiological and/or environmental stimuli. Eukaryotic mRNAs have a considerable range of half-lives, from as short as a few minutes to as long as several days. Human globin mRNAs constitute an example of highly stable mRNAs. However, a wide variety of naturally occurring mutations that result in the clinical syndrome of thalassemia can trigger accelerated mRNA decay thus controlling mRNA quality prior to translation. Distinct surveillance mechanisms have been described as being targeted for specific defective globin mRNAs. Here, we review mRNA stability mechanisms implicated in the control of β-globin gene expression and the surveillance pathways that prevent translation of aberrant β-globin mRNAs. In addition, we emphasize the importance of these pathways in modulating the severity of the β-thalassemia phenotype.
Keywords: mRNA stability, mRNA quality control, nonsense-mediated mRNA decay (NMD), beta-globin, beta-thalassemia, clinical phenotype
Introduction
Post-transcriptional controls play an important role in gene expression regulation. A major component of these controls is exerted at the level of mRNA stability, which is determined by a combination of interactions between cis-acting mRNA sequences and trans-acting mRNA-binding proteins. The cis-acting determinants include the 5’ m7Gppp cap structure, the 5’ and 3’ untranslated regions (UTRs), the 3’ polyadenylated [poly(A)] tail and sequences in the coding region.1–4 The closed loop structure shared by almost all eukaryotic mRNAs that link the 5’ cap structure to the 3’ poly(A) tail through the interaction of the cap binding protein complex with the poly(A) binding protein (PABP) contributes to mRNA stability as it protects the termini from exonuclease activity.1–3 Examples of other determinants that regulate mRNA stability include AU-rich elements in the 3’UTR of some interleukin mRNAs and the iron-responsive element in the transferrin receptor mRNA, or the JNK-response element in the 5’UTR of the interleukin-2 transcript.4 Otherwise, mRNA degradation typically initiates with deadenylation of the 3’ poly(A) tail followed by decapping and degradation of the mRNA body in either the 5’ to 3’ or the 3’ to 5’ direction. In the 5' to 3’ decay pathway, the cap structure is cleaved by the Dcp2 decapping enzyme and the monophosphate RNA is degraded by proteins from the Xrn family. Degradation from the 3’ end is carried out by the exosome complex5 after deadenylation by PARN, PAN2 and CCR4-POP2-NOT protein complexes.4,5
Intrinsic stabilities of eukaryotic mRNA can differ by over a 100-fold, from several minutes to several days.6 Short-lived mRNAs usually encode proteins that must undergo dramatic changes in expression levels within a brief time frame. Such proteins include cell cycle control factors, proto-oncogenes, and cytokines.7 In contrast, mRNAs that encode highly abundant functional proteins tend to be highly stable. Examples of such highly stable mRNAs include crystallins,6 collagens8 and globins.8,9 Globin mRNAs are unusually stable. The importance of globin mRNA stability in red blood cell differentiation and function is related to the fact that the final reticulocyte stage of erythroid development lacks the nucleus. However, translation needs to continue for up to three days after enucleation and cessation of transcription sustaining the synthesis of the globin proteins. In most studies, globin mRNA half-lives vary from 10 to 24 hours.3
Several naturally occurring mutations affecting the expression of human globin genes, resulting in the clinical syndrome of thalassemia, have been described. The outcome of many of these thalassemic mutations is the inhibition of efficient mRNA transcription and/or processing, resulting in decreased mRNA levels. Destabilization of abnormally processed mRNAs may result from mutations that affect normal splicing or block polyadenylation.3 Mutations that introduce premature translation-termination codons (PTCs; non-sense codons) or affect the termination codon, triggering accelerated mRNA decay, have also been described.3,10 Nonsense mutations are amongst the most prevalent β-globin mutations that cause β-thalassemia whereas the antitermination mutation (TAA→CAA) at the major fetal/adult α2-globin gene, known as hemoglobin Constant Spring (αCS), accounts for the majority of non-deletional forms of α-thalassemia worldwide. In this review, we present the major cellular mechanisms that specify the role of mRNA stability in the control of β-globin gene expression during terminal differentiation of erythroid cells. We also provide an overview of the surveillance pathways that have been described to prevent translation of aberrant β-globin mRNAs. Furthermore, we emphasize the importance of these mechanisms in the control of gene expression, as well as in modulating the severity of the β-thalassemia phenotype.
Human globin mutations and mRNA stability determinants
Human α- and β-globin genes descend from a common ancestral gene, share general and specific structural features, and both encode extraordinarily stable mRNAs in erythroid cells, which allow co-expression of α- and β-globin proteins to reach high levels. Human globin gene expression is a highly regulated process requiring transcriptional and post-transcriptional controls. Although the mechanisms involved in regulation of α-globin mRNA stability have been characterized in detail, neither the cis-elements nor the trans-acting factors that specify the high stability of β-globin mRNA are well known. Different studies have suggested a β-globin mRNA half-life in the range of 10 to 20 hours.8,9,11–13
Even though various naturally occurring mutations are known to affect globin gene expression, few provide information about the mechanism underlying the high mRNA stability. An example studied in detail is the αCS mutation. First described in 1971 by Clegg et al.,14 αCS mutation allows the ribosome to translate an additional 31 codons into the 3’UTR, originating the hemoglobin Constant Spring. Since heterozygotes for the mutation express the C-terminally extended globin protein at only 3% of the wild-type α-globin level and the αCS mRNA is transcribed at normal rates while the αCS protein is stable,14 the loss of expression reflects a defect in the cytoplasmic αCS mRNA accumulation.15–17 Studies in the human α-globin mRNA showed that the loss of stability is related to the ribosome readthrough beyond the native stop codon that interferes with a determinant within the 3’UTR. This determinant comprises three non-contiguous C-rich sites (pyrimidine-rich element, PRE) and provides the binding site for the α-globin poly(C)-binding protein (αCP).15,18–20 Three mechanisms have been shown to contribute to mRNA stability via αCP. First, this α-complex appears to inhibit poly(A) shortening,17, 21 probably via a direct interaction of αCP with the poly(A) binding protein (PABP).21 Second, the binding of αCP protects mRNA from an erythroid endoribonuclease attack.21 Third, the mechanism by which the α-complex can confer RNA stability in vitro was also identified and shown to involve inhibition of 3’ to 5’ exonucleolytic degradation.22
A frameshift mutation between codons 146 and 147 of the human β-globin gene that consists of a dinucleotide CA insertion, which abolishes the normal stop codon at position 147, was also described.23 This insertion results in the elongation of the β-chain by 11 aminoacid residues at its C-terminus generating hemoglobin Tak.23 This hemoglobin has a high oxygen affinity24,25 causing secondary polycythemia. Most individuals with this condition have asymptomatic erythrocytosis, but a homozygote for the Tak mutation has already been reported with hypoxemia and aggravated respiratory distress.25 These data indicate that, unlike the αCS mRNA, the Tak mutation in the β-globin mRNA, which permits ribosomes to read 11 codons into the 3’UTR before they encounter an in-frame termination codon, have little impact on the overall β-globin mRNA stability, suggesting that crucial stability elements are located downstream of this region. It has also been suggested that α- and β-globin mRNAs might be stabilized through potentially divergent mechanisms.
Naturally occurring mutations have also been described within the β-globin 3’UTR.26 However, the possibility that these naturally occurring mutations affect β-globin mRNA stability has not been clarified.26–28 Bilenoglu et al.27 have shown that a 13 nucleotide (nt) deletion, located 17 nt upstream of the poly(A) signal, seems to reduce the accumulation of cytoplasmic mRNA without significantly impairing its stability. On the other hand, Cai et al.28 described a T→C substitution in the same sequence, 12 nt upstream of the poly(A) signal. These authors hypothesize that it might lead to destabilization of the encoded β-globin mRNA.28 Besides, mutations in the polyadenylation signal of β-globin have been identified in β+-thalassemic patients resulting in inefficient cleavage and absence of polyadenylation of mRNA at the normal poly(A) site.29–32 The importance of these mutations in gene expression relies on the critical role of polyadenylation on mRNA stability and export to the cytoplasm. Polyadenylation also affects the efficiency of translation in the cytoplasm: PABP1 bound to the poly(A) tail seems to help recruiting ribosomes to the cap site through mRNA circularization.33
Human β-globin mRNA stability elements
Some experimental results suggested a role for the β-globin 3’UTR in the mRNA stability mechanism. Indeed, the replacement of the 3’ end of the c-fos mRNA (an unstable mRNA) with the 3’ end of the β-globin mRNA increases the half-life of the resultant hybrid mRNA, whereas the reciprocal hybrid transcript shows a decreased half-life compared with the normal β-globin mRNA.34 Thus, after PRE was first described in the α-globin mRNA,35 the 3’UTR of the β-globin mRNA was also screened for the presence of a stability element comparable to the one found in α-3’UTR. The destabilizing effect of ribosomal entry into the β-globin 3’UTR was assessed by introducing two tandem antitermination mutations into the wild-type human β-globin gene that permit ribosomes to translate 37 codons into the 3’UTR of the encoded β-globin mRNA.36 The mutant β-globin mRNA was less stable in cultured erythroid cells, indicating that, as in human α-globin mRNA, an unperturbed 3’UTR is crucial to maintain mRNA stability, and that both α and β elements are sensitive to disruption by readthrough ribosomes.36 However, the extent of readthrough necessary for mRNA destabilization differs between both mRNAs. While the α-globin mRNA is destabilized by ribosomes that readthrough for 4 codons into the 3’UTR, destabilization of the β-globin mRNA needs translation for more than 10 codons.36,37 Also, β-globin mRNA stability is destroyed only when two mutations separated by 35 nt are located in cis, suggesting that β-globin mRNA stability motif might be composed of two independent and redundant elements. Consistent with these data is the evidence that the naturally occurring C→G mutation 6 nt downstream of the normal termination codon fails to destabilize the corresponding β-globin mRNA.38 Actually, the available data suggest that the α- and β-globin mRNA stability elements are structurally and functionally distinct.3, 36
An mRNP complex mediating the high stability of normal human β-globin mRNAs
With the aim of characterizing trans-acting factors involved in the mechanism underlying the high stability of the human β-globin mRNA, Yu and Russell39 described an mRNP complex that assembles on the 3’UTR of the β-globin mRNA and exhibits some of the properties of the α-complex. The β-globin mRNP complex was shown to contain one or more factors homologous to the αCP. Sequence analysis implicated a specific 14-nt pyrimidine-rich track within the β-globin 3’UTR at 34 nt downstream of the normal UAA codon, as the site of the mRNP assembly.39 In agreement with these data is the observation that a naturally occurring 13 nt deletion within the β-globin 3’UTR, 90 nt downstream the native termination codon, does not alter mRNA stability.40 The existence of a β-globin mRNP complex involved in the mechanism by which mRNA is highly stable had already been suggested previously. This study showed that a HeLa whole-cell extract contains a factor that protects β-globin mRNA from attack by RNases present in an MEL cell cytoplasmic extract.41 It is possible that this factor may be a constituent of the αCP-homologous β-globin mRNA complex. Moreover, Jiang et al.42 identified a new determinant in the β-globin 3’UTR, positioned on an mRNA half-stem opposite PRE that functions as a binding site for the RNA-binding factor nucleolin. The results from this study lead the authors to hypothesize a model for β-globin mRNA stability that is related to, but distinct from the mechanism that stabilizes α-globin mRNA. The proposed model relies on the potential of β-globin 3’UTR to assume a highly stable stem-loop structure that incorporates the β-PRE and nucleolin-binding sites into half-stems. This native structure would be remodeled by nucleolin binding, which would facilitate αCP access to β-PRE and, therefore, enhance β-globin mRNA stability.42
mRNA surveillance mechanisms
Gene expression regulation at the post-transcriptional level allows cells to rapidly adapt to changes in their environment by altering the patterns of gene expression.43 The detection and degradation of defective and improperly-processed transcripts is essential to ensure that information flowing from the nucleus into the cytoplasm is of high quality prior to translation. Thus, cells have evolved surveillance mechanisms to guarantee a tight mRNA quality control, acting at several steps of mRNA biogenesis.44 Mammalian mRNA surveillance pathways include the nonstop decay pathway (NSD), the nonsense-mediated mRNA decay (NMD) and the ribosome extension-mediated decay (REMD) (Table 1).
Table 1.
Mammalian mRNA surveillance pathways.
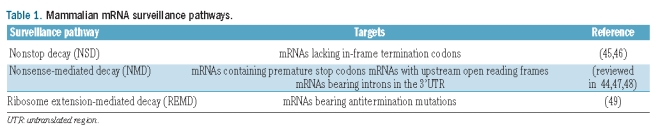
The NSD targets mRNAs lacking in-frame termination codons. This mechanism is translation-dependent and results in rapid decay of the aberrant transcripts.45 In the absence of a stop codon, translation proceeds along the poly(A) tail, the ribosome stalls at the 3’ end and the empty ribosomal A site is recognized by Ski7 protein. Then, Ski7 protein recruits Ski2-Ski3-Ski8 protein complex, which facilitates ribosome release and results in degradation of the target transcripts by the exosome.46
Recently, Kong and Liebhaber49 described the REMD, a novel surveillance mechanism. This mechanism accounts for marked repression of αCS globin synthesis. Transcripts harboring the αCS mutation are translated for an additional 31 codons into the 3’UTR before terminating at the poly(A) signal (see above). This ribosome extension triggers accelerated deadenylation of the αCS transcript resulting in reduced stability of the αCS mRNA.50 REMD has a different mechanistic profile to other surveillance pathways by (i) its functional linkage to accelerated deadenylation; (ii) its independence from the NMD factor UPF1; and (iii) cell-type restriction. This unusual pathway of mRNA surveillance is likely to act as a modifier of additional genetic defects and may reflect post-transcriptional controls peculiar to erythroid and other differentiated cell lineages.49
The best characterized surveillance mechanism is NMD. The physiological importance of NMD is to limit the synthesis of C-terminally truncated proteins, thus protecting the cell from their deleterious dominant-negative or gain-of-function effects. The biological and medical significance of the NMD pathway is suggested by the fact that approximately one-third of all inherited genetic disorders are due to PTCs, and in many of these cases, NMD influences the severity of the clinical phenotype.47,51 The discovery of the NMD pathway has provided an explanation for the long-lasting observation that the cells degrade PTC-containing mRNAs. PTCs can be generated by a series of events like naturally occurring frameshift and nonsense mutations, splicing errors, leaky 40S scanning or utilization of minor AUG initiation sites.52,53
Overview of the NMD mechanism
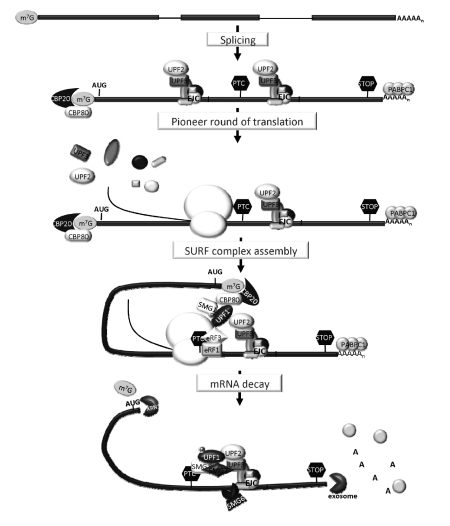
NMD is an evolutionary conserved mechanism found to be implicated in the surveillance and regulation of gene expression in all eukaryotes examined so far. Although the core process is conserved among species, important differences concerning the mechanism of PTC definition and the specific degradation pathways of the target transcripts have been identified. In mammalian systems, both splicing and translation are crucial for recognition of the translation termination event as premature.44 Generally, in mammalian cells, PTCs located more than 50–55 nt upstream of the last exon-exon junction are able to target mRNA for NMD, whereas, PTCs located downstream of this boundary are not,54 suggesting that the process of splicing is implicated in mammalian NMD. This process involves the assembly of a “mark”, during splicing, 20–24 nt upstream of exon junctions, therefore called EJCs (exon-junction complexes), that communicates the location of the translation termination event relative to the splice junctions to the NMD machinery44,48 (Figure 1). The activation of NMD in the presence of a premature stop codon and a competent downstream EJC requires a link between the factors involved in translation termination and the EJC complex. That link is provided by the UPF (up-frameshift) proteins. According to current models, recognition of a nonsense codon as premature involves a direct interaction of EJC-bound UPF2 with the UPF1, a RNA-dependent ATPase and 5’ to 3’ helicase protein, whose activation upon phosphorylation by SMG1 protein is required for NMD55–57 (Figure 1). Under these circumstances, UPF1 also interacts with eRF1-eRF3 complex at the PTC, providing the molecular link between the terminating ribosome and the EJC. An interaction between UPF1 and UPF3 may also be involved in the NMD process.57 These conserved NMD factors, the SMG proteins and the eRF1-eRF3 complex constitute the SURF complex.58 Recently, Hwang et al.59 showed that inhibition of binding of UPF1 to CBP80 results in inhibition of SMG1 and UPF1 association with eRF1 and eRF3 during SURF complex assembly at a PTC, and the subsequent association of SMG1 and UPF1 with the EJC. These results suggest that CBP80 at the mRNA 5’ end can bridge SMG1-UPF1 to eRF1-eRF3 positioned at a PTC59 (Figure 1).
Figure 1.
Simplified model of the nonsense-mediated mRNA decay (NMD) pathway. During splicing, exon-junction complexes (EJCs) assemble at 20–24 nucleotides (nt) upstream of exon-exon junctions. The translating ribosome displaces the EJCs during the pioneer round of translation. If a PTC is located more than 50–54 nt upstream from the last EJC, the terminating complex is able to interact with EJC-associated NMD factors. The CBP80 protein at the mRNA 5’ end can bridge UPF1-SMG-1 to eRF1-eRF3 at the PTC, leading to the formation of the SURF complex. The interaction of UPF1 with EJC-bound UPF2 induces the SMG1-mediated phosphorylation of UPF1 and dissociation of eRF1-eRF3. Phosphorylated UPF1 recruits SMG-7 and SMG-5 proteins and EJC recruits SMG-6. The decay of the PTC-containing transcript is triggered by SMG-6 which possesses endonuclease activity. The degradation pathways of PTC-bearing mRNAs involve decapping (by DCP1/DCP2) followed by 5’ to 3’ exonucleolytic activity (by XRN1) as well as accelerated dead-enylation and subsequent 3’ to 5’ exonu-cleolytic degradation (catalysed by the exosome complex).
The termination event is considered critical in the process of distinguishing a premature termination codon from a normal one. The eRF1 protein has a structure mimicking that of a tRNA molecule. It recognizes the stop codon in the A site of the ribosome and catalyzes the hydrolysis of the peptidyl-tRNA bond.60 The eRF3 protein is a ribosome- and eRF1-dependent GTPase, and stimulates eRF1 activity in a GTP-dependent manner.61 Emerging data showing that NMD can occur in the absence of splicing or downstream EJCs, and that the presence of a 3’UTR intron is not sufficient to define a stop codon as premature, challenged the requirement for splicing and EJCs in mammalian NMD.44 Moreover, recent data suggesting that PTC definition of EJC-independent NMD relies on the 3’UTR length, depending on the distance between the termination codon and PABPC1, supports a role for PABPC1 in PTC definition.62,63 Accordingly, tethering studies performed by us and others indicate that PABPC1 is able to superimpose its inhibitory effect over EJCs provided that PABPC1 is favorably located relatively to the PTC.62–65 In fact, if PABPC1 is in close proximity to the PTC, it can impair the interaction between UPF1 and eRF3, and so inhibits NMD.63,65
The major decay pathways associated with regular mRNA turnover are rate-limiting steps involved in the disruption of the protective 5’ cap and 3’ poly(A) terminal structures. However, in mammalian cells, the decay step of NMD can be initiated by decapping and deadenylation. On the other hand, it was recently shown that SMG6, recruited by the EJC to the NMD target,66 possesses the endonuclease activity responsible for initiating NMD in human cells67 (Figure 1). In either case, the next step is the exonucleolytic decay of the resulting RNA fragments. The determinants which direct the NMD targets for each decay pathway have yet to be clarified.
When general rules for NMD are not obeyed: example of the nonsense β-globin transcripts
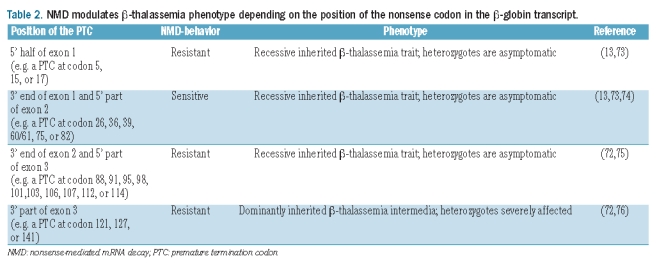
The human β-globin gene has two intervening non-coding sequences (introns) and three coding regions (exons) that are flanked by 5’ and 3’ non-coding sequences, the untranslated regions.68 Thus, after the two introns are spliced out, two EJCs might be assembled on the transcript.69,70 A β-globin NMD-behavior corroborating the “50–55 nt boundary rule” has been described. Indeed, the first indications regarding whether a β-globin PTC elicits NMD, concerning its position relatively to the downstream intron, were obtained in the laboratories of Kulozik and Maquat71,72 (Table 2). These authors showed that β-globin nonsense mutations located in the 3’ region of exon 1 (for example, at codon 26) and within the 5’ two-thirds of exon 2 (at codons 36, 60/61, 75, and 82) elicit NMD. In contrast, mRNAs bearing PTCs towards the 3’ end of exon 2 (at codons 88, 91, 95, 98, 101 and 103) and those with PTCs in exon 3 (at codons 106, 107, 114, 121 and 141) are all NMD-resistant. This illustrates a clear boundary between 48 and 66 nt upstream of the last exon-exon junction. Although the onset of the NMD process usually requires a minimum distance between the nonsense mutation and the final exon-exon junction, on β-globin transcripts the requirement for a maximum distance appears to be considerably less important. Indeed, a β-globin mRNA with a nonsense mutation at codon 3974 remains NMD competent, even when the distance between the PTC and the 3’ exon-exon junction is increased from the normal 180 nt up to 654 nt.77 This is a much longer distance than the one required for NMD in the triosephosphate isomerase (TPI) transcripts.78–80 On the other hand, an intronless human β-globin gene, carrying a nonsense mutation at codon 39, generates an mRNA that lacks the EJCs and, consequently, is immune to NMD.77 This is supported by the fact that naturally intronless mRNAs containing PTCs are not usually degraded by NMD.81 Furthermore, the insertion of an intron more than 50 nt downstream of the native stop codon, redefines this codon as premature and targets this mRNA to NMD.72 However, exceptions to the “50–55 nt boundary rule” have also been reported for the human β-globin mRNAs. In fact, we have shown that β-globin transcripts containing naturally occurring nonsense mutations in the 5’ region of exon 1 accumulate to levels similar to those of normal β-globin transcripts, being, unexpectedly according to their position, NMD-resistant.73 Furthermore, a functional analysis of these mRNAs with 5’ proximal nonsense mutations demonstrated that their resistance to NMD does not reflect abnormal RNA splicing or translation reinitiation and is independent of promoter identity and erythroid specificity.13 Instead, the proximity of the nonsense codon to the translation initiation AUG is the basis of their NMD resistance. Therefore, the AUG-proximity effect can override the “50–55 nt boundary rule” in establishing the overall efficiency of NMD.13 This NMD-resistance can be explained by the inherent nature of an extremely premature termination event, that could be sufficient to maintain PABPC1, bound simultaneously to the poly(A) tail and to the cap-associated eIF4G, in close proximity to the PTC. PABPC1 would then be able to superimpose its inhibitory effect on NMD activation.44 In addition to the AUG-proximity effect, translation re-initiation 3’ to the nonsense codon may contribute to the alleviation of the NMD effect and, as an outcome, modulates disease phenotypes.82,83 If re-initiation occurs in frame downstream of the PTC, it produces an N-terminally truncated protein that may or may not be functional. These two parameters can independently contribute to the NMD-resistance of a nonsense-mutated mRNA with implications for genotype-phenotype correlations in genetic disorders.84
Table 2.
NMD modulates β-thalassemia phenotype depending on the position of the nonsense codon in the β-globin transcript.
The NMD was also found to be triggered in hybrid mouse-human β-globin transcripts (carrying nonsense mutations at codons 21/22, 39 or 60/61) in the absence of the last intron71 and, therefore, in the absence of the downstream EJC. Thus, the existence of a fail-safe mechanism was suggested where, in the absence of intron 2, a cis-acting sequence located within the coding region could function in NMD; this effect was also described for TPI mRNA.78 Nevertheless, these results are in contrast with those described by Neu-Yilik et al.,77 who have shown that splicing is an indispensable component of the β-globin NMD pathway. In addition, a study using the IVS1 +5 G→A thalassemic β-globin allele as a model system showed that the splicing of the corresponding pre-mRNAs at a criptic site generates a mature transcript carrying a PTC at codon 30 (located more than 55 nt 5’ of the final exon-exon junction) that is immune to NMD.85 Although the mechanism underlying this NMD-behavior was not clarified, it was demonstrated that neither abnormalities of splicing nor translation re-initiation at downstream AUG codons is the cause of the NMD resistance of this transcript.85
Impact of the NMD mechanism on the β-thalassemia phenotype
The β-globin mRNA has been widely used as a model system for the study of the NMD mechanism and how it modulates the clinical outcome of genetic disorders. In fact, the deleterious effects that can be caused by the presence of C-terminally truncated proteins, and the benefits that can be achieved by the elimination of the mRNAs that encode these proteins, are clearly illustrated in β-thalassemia. This disorder exemplifies the phenotypic impact of the polar effect of PTCs at different positions within the same β-globin gene.51 If a PTC is located at a position that activates NMD, the disease results in a recessive mode of inheritance and heterozygotes are asymptomatic (Figure 2 and Table 2). In this case, the defective β-globin transcript is degraded by NMD resulting in limited synthesis of the truncated product. The excess of free α-globin, as well as the limited amount of truncated β-globin protein (if translated) are proteolytically degraded. Indeed, it has been shown that these defects result in very low amounts of β-globin chain production from the affected allele causing a reduction of about 50% of total β-globin chain synthesis in the heterozygote. This produces the clinically asymptomatic phenotype of β-thalassemia trait (also known as thalassemia minor), as heterozygotes usually synthesize enough β-globin from the remaining normal allele to support near-normal hemoglobin levels51 (Figure 2 and Table 2). An example of such a defect is the nonsense mutation CAG→TAG at codon 39 of the β-globin gene which accounts for a high percentage of β-thalassemia alleles in the Mediterranean region.86 If the PTC is located at a position that does not induce NMD, at less than 55 nt upstream of the last exon-exon junction or at the 5’-part of exon 3, the amount of truncated protein that is synthesized might be small enough to be efficiently degraded by the proteolytic system of the red blood cell, together with the excess of free α-globin chains. In this case, the heterozygotes are also asymptomatic and the disease still results in a recessive mode of inheritance (Figure 2 and Table 2). Such an example is the naturally-occurring nonsense mutation at codon 112 (TGT→TGA) of the β-globin gene. In fact, carriers for this mutation present hematologic data compatible with typical β-thalassemia recessive trait, being essentially healthy and only showing mostly insignificant abnormalities in hematologic indices.75 In contrast, if the PTC is located further downstream at exon 3 of the β-globin mRNA, for example at codon 121 (GAA→TAA) or 127 (CAG→TAG),88 NMD is also not triggered. However, instead, substantial amounts of mutant β-globin mRNA are present in the patients, encoding for truncated translation products that are long enough to overburden the cellular proteolytic system (Figure 2 and Table 2). Under these conditions, accumulation of the truncated products, as well as free α-globin chains in excess, act in a dominant negative manner, leading to deleterious effects on the cell, as the nonfunctional globin chains cause toxic precipitation of insoluble globins.51 This condition is related to a symptomatic form of the disease in heterozygotes called “thalassemia intermedia” and a dominant mode of inheritance.76 It is mostly associated with mutations located in the final two-thirds of the third exon of the β-globin gene (Figure 2 and Table 2). We have shown that naturally occurring PTCs in the 5’ region of exon 1, such as at codon 15 (TGG→TGA), do not elicit NMD (see above).73 Nevertheless, these mRNAs with short open reading frames codify for very short peptides that, if translated, are degraded. Thus, carriers of these extremely premature nonsense mutations are also asymptomatic for β-thalassemia (Figure 2 and Table 2).
Figure 2.
Nonsense-mediated mRNA decay (NMD) modulates disease phenotype. Map of the NMD-resistant and NMD-sensitive regions in the human β-globin mRNA. AUG-proximal premature termination codons (PTCs) do not trigger NMD but heterozygotes are asymptomatic, as the translated short β-globin peptides, along with the α-globin chains in excess, are effectively degraded. If the PTC is located downstream of codon 23 and more than 55 nucleotides (nt) upstream of the last exon-exon junction, the corresponding transcript is targeted for NMD and heterozygotes are also protected from thalassemia. In contrast, transcripts bearing PTCs located less than 55 nt upstream of the last exon-exon junction, or in the 5’-part of exon 3, escape NMD, resulting in the production of truncated proteins that are small enough to be efficiently degraded, along with the α-globin chains in excess, and heterozygotes are still asymptomatic for β-thalassemia. However, if the PTC is located further downstream, the encoded truncated nonfunctional β-globin proteins overwhelm the cellular proteolytic system and cause toxic precipitation of insoluble globin chains. Heterozygotes with these mutations are affected with dominantly inherited β-tha-lassemia intermedia.
From these studies, it becomes clear that NMD can represent a crucial modulator of the clinical phenotype not only of β-thalassemia, but also of many other genetic disorders. In most conditions, this mechanism is beneficial and helps in the elimination of transcripts which might encode C-terminally truncated proteins with dominant-negative or gain-of-function effects. However, it is important to note that there are some examples in which NMD aggravates the disease phenotype because truncated proteins that would otherwise retain some normal function are not produced, leading to haploinsufficiency.87
Conclusions
The understanding that mRNA stability can impact on gene expression regulation and, consequently, on the genotype-phenotype correlation in many genetic disorders has led to a number of highly useful studies regarding mRNA decay and quality control pathways. The human β-globin mRNA as the product of a normal or a naturally-occurring mutated allele has proved to be an excellent experimental model system, not only to study the outcome of specific mRNA defects, but also to clarify many general features of mRNA stability control and surveillance as well as their role in the regulation of gene expression. Further studies will be important to reveal novel aspects of these pathways, which might provide perspectives for the development of new strategies to treat diseases associated to mutations that target the corresponding transcripts for rapid decay. As the severity of disease in patients with β-thalassemia is directly related to the relative excess in β-globin synthesis, therapies that decrease this chain synthesis disequilibrium are predicted to reduce the clinical severity of β-thalassemia. PTCs are a major cause of β-thalassemia worldwide. They may induce rapid mRNA decay through the NMD mechanism or generate non-functional truncated β-globin chains. Therefore, therapies that increase β-globin mRNA stability and/or suppress the nonsense codon recognition would increase the level of full-length protein and could, therefore, be of great value in the treatment of β-thalassemia patients. In fact, development and testing of therapies to correct the effect of PTC-containing transcripts are ongoing88 for treatment of several PTC-associated diseases.
Supplementary Material
Acknowledgments
The authors would like to thank João Lavinha for comments on the manuscript.
Footnotes
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
Funding: this work was partially supported by Fundação Luso-Americana para o Desenvolvimento and Center for Biodiversity, Functional and Integrative Genomics from Fundação para a Ciência e a Tecnologia. I Peixeiro and AL Silva were supported by Fellowships from Fundação para a Ciência e a Tecnologia.
References
- 1.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59(3):423–50. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89(6):831–8. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 3.Russell JE, Morales J, Liebhaber SA. The role of mRNA stability in the control of globin gene expression. Prog Nucleic Acid Res Mol Biol. 1997;57:249–87. doi: 10.1016/s0079-6603(08)60283-4. [DOI] [PubMed] [Google Scholar]
- 4.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265(1–2):11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 5.Tomecki R, Drazkowska K, Dziembowski A. Mechanisms of RNA degradation by the eukaryotic exosome. Chembiochem. 2010;11(7):938–45. doi: 10.1002/cbic.201000025. [DOI] [PubMed] [Google Scholar]
- 6.Li XA, Beebe DC. Messenger RNA stabilization in chicken lens development: a reexamination. Dev Biol. 1991;146(1):239–41. doi: 10.1016/0012-1606(91)90463-d. [DOI] [PubMed] [Google Scholar]
- 7.Hamalainen L, Oikarinen J, Kivirikko KI. Synthesis and degradation of type I procollagen mRNAs in cultured human skin fibroblasts and the effect of cortisol. J Biol Chem. 1985;260(2):720–5. [PubMed] [Google Scholar]
- 8.Aviv H, Voloch Z, Bastos R, Levy S. Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell. 1976;8(4):495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- 9.Ross J, Sullivan TD. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66(5):1149–54. [PubMed] [Google Scholar]
- 10.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 11.Bastos RN, Aviv H. Theoretical analysis of a model for globin messenger RNA accumulation during erythropoiesis. J Mol Biol. 1977;110(2):205–18. doi: 10.1016/s0022-2836(77)80069-7. [DOI] [PubMed] [Google Scholar]
- 12.Bastos RN, Volloch Z, Aviv H. Messenger RNA population analysis during erythroid differentiation: a kinetical approach. J Mol Biol. 1977;110(2):191–203. doi: 10.1016/s0022-2836(77)80068-5. [DOI] [PubMed] [Google Scholar]
- 13.Inacio A, Silva AL, Pinto J, Ji X, Morgado A, Almeida F, et al. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J Biol Chem. 2004;279(31):32170–80. doi: 10.1074/jbc.M405024200. [DOI] [PubMed] [Google Scholar]
- 14.Clegg JB, Weatherall DJ, Milner PF. Haemoglobin Constant Spring--a chain termination mutant? Nature. 1971;234(5328):337–40. doi: 10.1038/234337a0. [DOI] [PubMed] [Google Scholar]
- 15.Waggoner SA, Liebhaber SA. Regulation of alpha-globin mRNA stability. Exp Biol Med (Maywood) 2003;228(4):387–95. doi: 10.1177/153537020322800409. [DOI] [PubMed] [Google Scholar]
- 16.Liebhaber SA, Kan YW. alpha-Thalassemia caused by an unstable alpha-globin mutant. J Clin Invest. 1983;71(3):461–6. doi: 10.1172/JCI110790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales J, Russell JE, Liebhaber SA. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J Biol Chem. 1997;272(10):6607–13. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Kiledjian M, Weiss IM, Liebhaber SA. Detection and characterization of a 3' untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol Cell Biol. 1995;15(3):1769–77. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiledjian M, Wang X, Liebhaber SA. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14(17):4357–64. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong J, Ji X, Liebhaber SA. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol Cell Biol. 2003;23(4):1125–34. doi: 10.1128/MCB.23.4.1125-1134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Kiledjian M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol. 2000;20(17):6334–41. doi: 10.1128/mcb.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodgers ND, Wang Z, Kiledjian M. Regulated alpha-globin mRNA decay is a cytoplasmic event proceeding through 3'-to-5' exosome-dependent decapping. RNA. 2002;8(12):1526–37. [PMC free article] [PubMed] [Google Scholar]
- 23.Flatz G, Kinderlerer JL, Kilmartin JV, Lehmann H. Haemoglobin Tak: a variant with additional residues at the end of the beta-chains. Lancet. 1971;1(7702):732–3. doi: 10.1016/s0140-6736(71)91994-5. [DOI] [PubMed] [Google Scholar]
- 24.Imai K, Lehmann H. The oxygen affinity of haemoglobin Tak, a variant with an elongated beta chain. Biochim Biophys Acta. 1975;412(2):288–94. doi: 10.1016/0005-2795(75)90043-4. [DOI] [PubMed] [Google Scholar]
- 25.Tanphaichitr VS, Viprakasit V, Veerakul G, Sanpakit K, Tientadakul P. Homozygous hemoglobin Tak causes symptomatic secondary polycythemia in a Thai boy. J Pediatr Hematol Oncol. 2003;25(3):261–5. doi: 10.1097/00043426-200303000-00016. [DOI] [PubMed] [Google Scholar]
- 26.Jankovic L, Dimovski AJ, Kollia P, Karageorga M, Loukopoulos D, Huisman TH. A C----G mutation at nt position 6 3' to the terminating codon may be the cause of a silent beta-thalassemia. Int J Hematol. 1991;54(4):289–93. [PubMed] [Google Scholar]
- 27.Bilenoglu O, Basak AN, Russell JE. A 3'UTR mutation affects beta-globin expression without altering the stability of its fully processed mRNA. Br J Haematol. 2002;119 (4):1106–14. doi: 10.1046/j.1365-2141.2002.03989.x. [DOI] [PubMed] [Google Scholar]
- 28.Cai SP, Eng B, Francombe WH, Olivieri NF, Kendall AG, Waye JS, et al. Two novel beta-thalassemia mutations in the 5' and 3' non-coding regions of the beta-globin gene. Blood. 1992;79(5):1342–6. [PubMed] [Google Scholar]
- 29.Rund D, Filon D, Dowling C, Kazazian HH, Jr, Rachmilewitz EA, Oppenheim A. Molecular studies of beta-thalassemia in Israel. Mutational analysis and expression studies. Ann NY Acad Sci. 1990;612:98–105. doi: 10.1111/j.1749-6632.1990.tb24295.x. [DOI] [PubMed] [Google Scholar]
- 30.Ghanem N, Girodon E, Vidaud M, Martin J, Fanen P, Plassa F, et al. A comprehensive scanning method for rapid detection of beta-globin gene mutations and polymorphisms. Hum Mutat. 1992;1(3):229–39. doi: 10.1002/humu.1380010310. [DOI] [PubMed] [Google Scholar]
- 31.Jankovic L, Efremov GD, Petkov G, Kattamis C, George E, Yang KG, et al. Two novel polyadenylation mutations leading to beta(+)-thalassemia. Br J Haematol. 1990;75 (1):122–6. doi: 10.1111/j.1365-2141.1990.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 32.Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH., Jr Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985;4(2):453–6. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proudfoot NJ. Genetic dangers in poly(A) signals. EMBO Rep. 2001;2(10):891–2. doi: 10.1093/embo-reports/kve207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabnick KS, Housman DE. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988;8(8):3244–50. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss IM, Liebhaber SA. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3' nontranslated region. Mol Cell Biol. 1995;15(5):2457–65. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell JE, Liebhaber SA. The stability of human beta-globin mRNA is dependent on structural determinants positioned within its 3' untranslated region. Blood. 1996;87(12):5314–23. [PubMed] [Google Scholar]
- 37.Weiss IM, Liebhaber SA. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol Cell Biol. 1994;14(12):8123–32. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sgourou A, Papachatzopoulou A, Psiouri L, Antoniou M, Zoumbos N, Gibbs R, et al. The beta-globin C-->G mutation at 6 bp 3' to the termination codon causes beta-thalassaemia by decreasing the mRNA level. Br J Haematol. 2002;118(2):671–6. doi: 10.1046/j.1365-2141.2002.03627.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Russell JE. Structural and functional analysis of an mRNP complex that mediates the high stability of human beta-globin mRNA. Mol Cell Biol. 2001;21(17):5879–88. doi: 10.1128/MCB.21.17.5879-5888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23(14):4805–13. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolle CA, Benz EJ., Jr Cellular factor affecting the stability of beta-globin mRNA. Gene. 1988;62(1):65–74. doi: 10.1016/0378-1119(88)90580-x. [DOI] [PubMed] [Google Scholar]
- 42.Jiang Y, Xu XC, Russell JE. A Nucleolin-Binding 3' Untranslated Region Element Stabilizes β-Globin mRNA In Vivo. Mol Cell Biol. 2006;26(6):2419–29. doi: 10.1128/MCB.26.6.2419-2429.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–8. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 44.Silva AL, Romao L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett. 2009;583(3):499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 45.Vasudevan S, Peltz SW, Wilusz CJ. Non-stop decay--a new mRNA surveillance pathway. Bioessays. 2002;24(9):785–8. doi: 10.1002/bies.10153. [DOI] [PubMed] [Google Scholar]
- 46.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 47.Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18(7):315–21. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J. 2010;430(3):365–77. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 49.Kong J, Liebhaber SA. A cell type-restricted mRNA surveillance pathway triggered by ribosome extension into the 3' untranslated region. Nat Struct Mol Biol. 2007;14(7):670–6. doi: 10.1038/nsmb1256. [DOI] [PubMed] [Google Scholar]
- 50.Voon HP, Vadolas J. Controlling alpha-globin: a review of alpha-globin expression and its impact on beta-thalassemia. Haematologica. 2008;93(12):1868–76. doi: 10.3324/haematol.13490. [DOI] [PubMed] [Google Scholar]
- 51.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36 (8):801–8. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 52.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299(1–2):1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5(2):89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 54.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17(3):316–25. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 55.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6 (9):1226–35. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12(5):1187–200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 57.Singh G, Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem Sci. 2003;28(9):464–6. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- 58.Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581(15):2845–53. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 59.Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39(3):396–409. doi: 10.1016/j.molcel.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, et al. The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100(3):311–21. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 61.Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2(4):334–41. [PMC free article] [PubMed] [Google Scholar]
- 62.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3' untranslated region. PLoS Biol. 2008;6(4):e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6(4):e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20(3):355–67. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14(3):563–76. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, et al. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev. 2010;24(21):2440–50. doi: 10.1101/gad.604610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16(1):49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 68.Huisman TH. The structure and function of normal and abnormal haemoglobins. Baillieres Clin Haematol. 1993;6(1):1–30. doi: 10.1016/s0950-3536(05)80064-2. [DOI] [PubMed] [Google Scholar]
- 69.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19(24):6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14(9):1098–108. [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4(7):801–15. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17 (12):3484–94. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romao L, Inacio A, Santos S, Avila M, Faustino P, Pacheco P, et al. Nonsense mutations in the human beta-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation. Blood. 2000;96(8):2895–901. [PubMed] [Google Scholar]
- 74.Baserga SJ, Benz EJ., Jr Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci USA. 1988;85(7):2056–60. doi: 10.1073/pnas.85.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Divoky V, Gu LH, Indrak K, Mocikova K, Zarnovicanova M, Huisman TH. A new beta zero-thalassaemia nonsense mutation (codon 112, T-->A) not associated with a dominant type of thalassaemia in the heterozygote. Br J Haematol. 1993;83(3):523–4. doi: 10.1111/j.1365-2141.1993.tb04682.x. [DOI] [PubMed] [Google Scholar]
- 76.Hall GW, Thein S. Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood. 1994;83(8):2031–7. [PubMed] [Google Scholar]
- 77.Neu-Yilik G, Gehring NH, Thermann R, Frede U, Hentze MW, Kulozik AE. Splicing and 3' end formation in the definition of nonsense-mediated decay-competent human beta-globin mRNPs. EMBO J. 2001;20(3):532–40. doi: 10.1093/emboj/20.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18(9):5272–83. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun X, Maquat LE. mRNA surveillance in mammalian cells: the relationship between introns and translation termination. RNA. 2000;6(1):1–8. doi: 10.1017/s1355838200991660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruiz-Echevarria MJ, Gonzalez CI, Peltz SW. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17(2):575–89. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104(2):173–6. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- 82.Silva AL, Pereira FJ, Morgado A, Kong J, Martins R, Faustino P, et al. The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context. RNA. 2006;12(12):2160–70. doi: 10.1261/rna.201406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang J, Maquat LE. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16(4):826–33. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Inacio A, Silva AL, Morgado A, Pereira FJ, Lavinha J, Romao L. Comment on 'Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease'. Eur J Hum Genet. 2007 May;15(5):533–4. doi: 10.1038/sj.ejhg.5201808. author reply 534. [DOI] [PubMed] [Google Scholar]
- 85.Danckwardt S, Neu-Yilik G, Thermann R, Frede U, Hentze MW, Kulozik AE. Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood. 2002;99(5):1811–6. doi: 10.1182/blood.v99.5.1811. [DOI] [PubMed] [Google Scholar]
- 86.Cao A, Gossens M, Pirastu M. Beta thalassaemia mutations in Mediterranean populations. Br J Haematol. 1989;71(3):309–12. doi: 10.1111/j.1365-2141.1989.tb04285.x. [DOI] [PubMed] [Google Scholar]
- 87.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14(10):1074–81. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 88.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12(7):306–16. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.