Abstract
We analyzed the metaproteome of the bacterial community resident in the hindgut paunch of the wood-feeding ‘higher' termite (Nasutitermes) and identified 886 proteins, 197 of which have known enzymatic function. Using these enzymes, we reconstructed complete metabolic pathways revealing carbohydrate transport and metabolism, nitrogen fixation and assimilation, energy production, amino-acid synthesis and significant pyruvate ferredoxin/flavodoxin oxidoreductase protein redundancy. Our results suggest that the activity associated with these enzymes may have more of a role in the symbiotic relationship between the hindgut microbial community and its termite host than activities related to cellulose degradation.
Keywords: Nasutitermes, mass spectrometry, microbial communities
Within the family Termitidae (higher termites), the P3 segment (hindgut paunch) microbiome is capable of fixing nitrogen and potentially degrading lignocellulose (Breznak and Brune, 1994). In contrast to the families of ‘lower termites' (for example, Kalotermitidae, Rhinotermitidae), where flagellated protists and the termite are sources of cellulases (Cleveland, 1923; Watanabe et al., 2002; Tokuda et al., 2004), the contribution and sources of hydrolytic enzymes in the higher termite are not well defined. For example, a recent metagenomic study of the Nasutitermes P3 microbiome (Warnecke et al., 2007) identified domains representative of several families of carbohydrate active enzymes (CAZy families) (Cantarel et al., 2009) and then used liquid chromatography-tandem mass spectrometry-based proteomics to look for evidence of CAZy expression.
We report results from a liquid chromatography-tandem mass spectrometry-based global proteome characterization (Supplementary Materials and Methods) of the Nasutitermes (laboratory maintained colony collected in Dania Beach, FL) P3 microbiome, in which we searched for evidence supporting CAZy family expression. Metaproteomic and metagenomic data sets were obtained from different termite nests. We observed 886 proteins (Supplementary Information) identified using ⩾2 unique peptide sequences as a requirement for confident protein identification. The confidently identified proteins represent 1.2% of the predicted proteome (based on the metagenomic sequence); however, these proteins make up 235 protein families (Pfams) or 11.5% of the 2050 Pfams predicted by the metagenome. The limited coverage may reflect the small volume of P3 hindgut fluid available for analysis (100 μl equates to ∼100 P3 hindguts), but suggests that the identified proteins are the most abundant and/or easily detected using liquid chromatography-tandem mass spectrometry. Among the 886 proteins, 70 are unannotated and 58 are characterized as hypothetical, predicted or putative in the annotation. The most represented proteins are flagellin-related hook-associated proteins and methyl-accepting chemotaxis proteins that contain 50 and 43 copies (redundancy), respectively. Chemotaxis is hypothesized to have an important role in compartmentalizing bacteria to specific regions of the termite gut according to the functional roles of the bacteria (Warnecke et al., 2007).
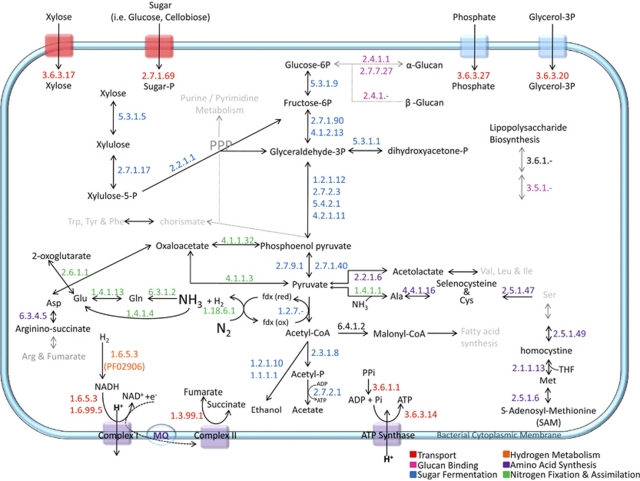
Of the 886 proteins identified in this study, 197 had Enzyme Commission (EC) numbers, which allowed us to construct enzymatic pathways and gain insight into the molecular mechanisms defining the symbiotic relationship between the P3 hindgut microbiome and its termite host. Figure 1 shows expressed genes for transport, glucan binding, sugar fermentation, hydrogen metabolism, amino-acid synthesis, and nitrogen fixation and assimilation pathways. The transportation and fermentation pathways are of particular interest with regard to possible energy substrates available for the microbiome. For example, xylose (wood sugar) is transported into the cytoplasm by ATPases (EC3.6.3.17), and glucose, monosaccharide and disaccharide substrates are transported into the bacterial cells through the phosphoenolpyruvate-dependent sugar phosphotransferase system (EC2.7.1.69). Phosphotransferase catalyzes phosphorylation of these incoming sugar substrates simultaneously with their translocation across the cell membrane, after which phosphorylated glucose (glucose-6P) can be metabolized and phosphorylated disaccharide can be broken down by β-glucan hydrolases (EC2.4.1.-). The collective actions of many bacterial enzymes transport and metabolize reducing sugars to alcohol, energy (ATP) and acetate (Supplementary Table S1). Bacterial acetate is a dominant end product for energy and biosynthesis in the termite; however, molecular hydrogen (H2) is an alternative energy source and is abundantly produced during both lignocellulose fermentation and N2 fixation. H2 is metabolized by iron-only hydrogenase enzymes (EC1.6.5.3) (Vignais and Colbeau, 2004).
Figure 1.
Molecular snapshot of the metabolic pathways within the termite gut microbial community. The enzymatic pathways of multiple enzymes are illustrated. Black arrows represent complete pathways; every enzyme was identified. Gray arrows and gray text represent incomplete pathways. The legend is color coded to the EC numbers. Ala, alanine; Arg, arginine; Asp, aspartate; Cys, cysteine; Gln, glutamine; Glu, glutamate; Ile, isoleucine; Leu, leucine; Met, methionine; Phe, phenylalanine; PPi, pyrophosphate; PPP, pentose phosphate pathway; Ser, serine; THF, tetrahydrofolate; Trp, tryptophan; Tyr, tyrosine; Val, valine.
Importantly, the sugar fermentation and nitrogen fixation pathways are coupled by the activity of pyruvate ferredoxin/flavodoxin oxidoreductase (EC1.2.7.-) through oxidative decarboxylation of pyruvate to acetyl-CoA under anaerobic conditions (Figure 1). The electrons generated from this reaction are transferred to ferredoxin (or some other electron carrier) and are used by nitrogenase in the reduction of N2. Pyruvate ferredoxin/flavodoxin oxidoreductase was observed with the highest degree of redundancy among the 197 enzymes, that is, 24 observed copies (Supplementary Table S2). Within this group, 4 copies (out of a possible 6) are annotated as EC1.2.7.- in the metagenome (Supplementary Figure S1, Supplementary Table 1). The role of this enzyme in acetate and nitrogen fixation and its high degree of redundancy suggest this enzyme's catalytic function is critical to the microbiome and the termite. The reported absence of pyruvate dehydrogenase activity (aerobic conversion of pyruvate to acetyl-CoA) in Nasutitermes walkeri tissues (Slaytor et al., 1997) and other termites (O'Brien and Breznak, 1984) has led to a long-standing hypothesis that acetate production by the microbiome is critical for producing energy in the termite (reviewed by Breznak and Brune (1994)). More recently, activity of the pyruvate dehydrogenase complex was reported for mitochondria extracted from N. walkeri and Coptotermes formosanus; however, pyruvate dehydrogenase activity in whole tissue homogenates remained elusive (Itakura et al., 2003).
After global characterization, we looked for supporting evidence of CAZy family expression. To improve the likelihood of observing these Pfams, we prepared samples from several fractionations of the hindgut fluid (Supplementary Materials and Methods and Supplementary Table S3) based on cellulase activity recently reported in similar fractions for Nasutitermes species (Tokuda and Watanabe, 2007). We searched our data for proteins having domains previously identified by Warnecke et al. (2007), using Pfam hidden Markov models (Krogh et al., 1994) (Supplementary Materials and Methods) and associated with glycoside hydrolysis and carbohydrate binding (Table 1). We identified 48 proteins within 22 CAZy and Pfams, of which 13 were reported for Nasutitermes (Warnecke et al., 2007). However, of these 48 proteins, only 8 within 6 families passed the ⩾2 unique peptide filtering criterion. The absence of confidently observed enzymes involved in cellulose degradation maybe because of these enzymes not being readily detected using liquid chromatography-tandem mass spectrometry (for example, secreted CAZy enzymes were insoluble and/or remained firmly bound to lignocellulose), the need for a more representative genome, or that native cellulose degradation may occur elsewhere in the termite. A recent report of an abundance of endo-β-1,4-glucanase in the midgut of Nasutitermes suggests that early stages of native cellulose degradation may depend on enzymes secreted by the termite host before complete degradation by the microbiome (Tokuda and Watanabe, 2007). In conclusion, the presented proteomic data shed further light on which genes are expressed to support the symbiotic relationship between the hindgut microbiome and its host.
Table 1. Glycoside hydrolases and carbohydrate-binding modules.
| CAZy family | Pfam HMM name | Pfam accessionb | Previously reported proteomics dataa |
Our analysis |
|
|---|---|---|---|---|---|
| ⩾1 peptides | 1 peptide | ⩾2 peptides | |||
| GH1 | Glyo_hydro_1 | Pfam00232 | 1 | 1 | |
| GH3 | Glyco_hydro_3 | Pfam00933 | 1 | 3 | |
| GH4 | Glyco_hydro_4 | Pfam02056 | 2 | 1 | |
| GH5 | Cellulase | Pfam00150 | 2 | 1 | |
| GH8 | Glyco_hydro_8 | Pfam01270 | 1 | ||
| GH9 | Glyco_hydro_9 | Pfam00759 | 1 | ||
| GH10 | Glyco_hydro_10 | Pfam00331 | 1 | 1 | |
| GH13 | Alpha-amylase | Pfam00128 | 2 | 5 | |
| GH23 | SLT | Pfam01464 | 2 | ||
| GH42 | Glyco_hydro_42 | Pfam02449 | 1 | ||
| GH43 | Glyco_hydro_43 | Pfam04616 | 5 | ||
| GH57 | Glyco_hydro_57 | Pfam03065 | 2 | ||
| GH77 | Glyco_hydro_77 | Pfam02446 | 4 | 3 | |
| GH88 | Glyco_hydro_88 | Pfam07470 | 1 | 1 | |
| GH94 | Glyco_transf_36 | Pfam06165 | 8 | 8 | 2 |
| CBM4 | CBM_4_9 | Pfam02018 | 1 | ||
| CBM6 | CBM_6 | Pfam03422 | 1 | ||
| CBM11 | CBM_11 | Pfam03425 | 2 | 2 | |
| Big_2 | Pfam02368 | 4 | |||
| Big_3 | Pfam07523 | 1 | |||
| CBM_X | Pfam06204 | 8 | 8 | 2 | |
| Glyco_hydro_3_C | Pfam01915 | 3 | 2 | 1 | |
| GT36_AF | Pfam06205 | 8 | 9 | 2 | |
| CBM_48 (Isoamylase_N) | Pfam02922 | 1 | |||
Many proteins contain more than one Pfam domain (Supplementary Tables 3 and 4 corresponding Pfams denoted in red).
Acknowledgments
The research described in this paper was funded by the Genomes to Life program sponsored by the US Department of Energy's Office of Biological and Environmental Research and performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract DE-ACO5-76RLO 1830. We also thank Penny Colton for technical editing. The data used in the analysis can requested at http://ober-proteomics.pnl.gov/.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Breznak JA, Brune A. Role of microorganisms in the digestion of lignocellulose by termites. Annu Rev Entomol. 1994;39:453–487. [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland LR. Symbiosis between termites and their intestinal protozoa. Proc Natl Acad Sci USA. 1923;9:424–428. doi: 10.1073/pnas.9.12.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura S, Tanaka H, Enoki A, Chappell DJ, Slaytor M. Pyruvate and acetate metabolism in termite mitochondria. J Insect Physiol. 2003;49:917–926. doi: 10.1016/s0022-1910(03)00150-1. [DOI] [PubMed] [Google Scholar]
- Krogh A, Brown M, Mian IS, Sjölander K, Haussler D. Hidden Markov models in computational biology: applications to protein modeling. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- O'Brien RW, Breznak JA. Enzymes of acetate and glucose metabolism in termites. Insect Biochem. 1984;14:639–643. [Google Scholar]
- Slaytor M, Veivers PC, Lo N. Aerobic and anaerobic metabolism in the higher termite Nasutitermes walkeri (Hill) Insect Biochem Mol Biol. 1997;27:291–303. [Google Scholar]
- Tokuda G, Lo N, Watanabe H, Arakawa G, Matsumoto T, Noda H. Major alteration of the expression site of endogenous cellulases in members of an apical termite lineage. Mol Ecol. 2004;13:3219–3228. doi: 10.1111/j.1365-294X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- Tokuda G, Watanabe H. Hidden cellulases in termites: revision of an old hypothesis. Biol Lett. 2007;3:336–339. doi: 10.1098/rsbl.2007.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais PM, Colbeau A. Molecular biology of microbial hydrogenases. Curr Issues Mol Biol. 2004;6:159–188. [PubMed] [Google Scholar]
- Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Nakashima K, Saito H, Slaytor M. New endo-beta-1,4-glucanases from the parabasalian symbionts, Pseudotrichonympha grassii and Holomastigotoides mirabile of Coptotermes termites. Cell Mol Life Sci. 2002;59:1983–1992. doi: 10.1007/PL00012520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.