Abstract
The earthworm gut provides ideal in situ conditions for ingested heterotrophic soil bacteria capable of anaerobiosis. High amounts of mucus- and plant-derived saccharides such as glucose are abundant in the earthworm alimentary canal, and high concentrations of molecular hydrogen (H2) and organic acids in the alimentary canal are indicative of ongoing fermentations. Thus, the central objective of this study was to resolve potential links between fermentations and active fermenters in gut content of the anecic earthworm Lumbricus terrestris by 16S ribosomal RNA (rRNA)-based stable isotope probing, with [13C]glucose as a model substrate. Glucose consumption in anoxic gut content microcosms was rapid and yielded soluble organic compounds (acetate, butyrate, formate, lactate, propionate, succinate and ethanol) and gases (carbon dioxide and H2), products indicative of diverse fermentations in the alimentary canal. Clostridiaceae and Enterobacteriaceae were users of glucose-derived carbon. On the basis of the detection of 16S rRNA, active phyla in gut contents included Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes, Gemmatimonadetes, Nitrospirae, Planctomycetes, Proteobacteria, Tenericutes and Verrucomicrobia, taxa common to soils. On the basis of a 16S rRNA gene similarity cutoff of 87.5%, 82 families were detected, 17 of which were novel family-level groups. These findings (a) show the large diversity of soil taxa that might be active during gut passage, (b) show that Clostridiaceae and Enterobacteriaceae (fermentative subsets of these taxa) are selectively stimulated by glucose and might therefore be capable of consuming mucus- and plant-derived saccharides during gut passage and (c) indicate that ingested obligate anaerobes and facultative aerobes from soil can concomitantly metabolize the same source of carbon.
Keywords: anaerobic metabolism, gut bacteria, invertebrate microbiology, soil microbiology, 16S rRNA SIP, Lumbricus terrestris
Introduction
Earthworms can be the major macrofauna of soil and can have profound effects on the structure and fertility of soils (Lee, 1985; Edwards and Bohlen, 1996; Lavelle et al., 1997; Makeschin, 1997). The earthworm gut is oxygen (O2) limited and provides ideal in situ conditions for ingested heterotrophic soil bacteria capable of anaerobiosis (Horn et al., 2003; Drake and Horn, 2007). Exoenzymes produced by ingested microorganisms stimulate the degradation of complex organic matter and thus enhance the ability of the worm to assimilate nutrients through a mutualistic digestive system (Brown et al., 2000).
The earthworm alimentary canal contains high amounts of mucus- and plant-derived saccharides (Martin et al., 1987; Trigo et al., 1999). Glucosamine, galactosamine, glucose, galactose, mannose and fucose are components of mucopolysaccharides from Lumbricus sp. (Rahemtulla and Lovtrup, 1975) and occur in hydrolyzed alimentary canal contents (Wüst et al., 2009a). The total concentration of monosaccharide equivalents in the aqueous phase of alimentary canal contents can be >100 m, with many monosaccharides such as glucose approximating 10 m or more (Horn et al., 2003; Drake and Horn, 2007; Wüst et al., 2009a). The occurrence of such high amounts of saccharides and the anoxia of the worm gut should favor fermentation in situ. Indeed, high concentrations of organic acids and molecular hydrogen (H2) occur in the alimentary canal (Horn et al., 2003; Wüst et al., 2009a). For example, over 30 m of organic acids (for example, acetate, lactate, succinate, butyrate, propionate and formate) occur in the aqueous phase of the midgut of Lumbricus terrestris (Wüst et al., 2009a), indicating that the midgut harbors a high diversity of fermentative processes and associated taxa, which might include both facultative aerobes and obligate anaerobes. Furthermore, living earthworms emit denitrification-derived nitrous oxide (N2O), which can be concomitant to the emission of H2 (Karsten and Drake, 1997; Matthies et al., 1999; Wüst et al., 2009a, 2009b). Although the occurrence of H2 and organic acids is evidence for in situ fermentation, the fermentative microorganisms in the alimentary canal are unknown. Indeed, although diverse bacteria are present in the earthworm gut (Karsten and Drake, 1997; Furlong et al., 2002; Ihssen et al., 2003; Singleton et al., 2003; Horn et al., 2005; Byzov et al., 2009), active bacterial taxa remain unresolved.
On the basis of these collective findings, we hypothesized that diverse obligate anaerobes and facultative aerobes ferment saccharides in the earthworm alimentary canal. The central objective of this study was to resolve potential links between fermentations and active fermenters in gut content of the anecic earthworm L. terrestris by 16S ribosomal RNA (rRNA)-based stable isotope probing, with [13C]glucose as a model substrate.
Materials and methods
Earthworms and soil
Adult L. terrestris L. were purchased from ANZO (Bayreuth, Germany) and maintained in soil at 15 °C for ∼5 days. Soil was obtained from the meadow Trafo Wiese in Bayreuth (Germany), which is described elsewhere (Horn et al., 2003). Worms had an average weight of 4.2±0.7 g (n=45). Gut transit times (Hartenstein et al., 1981) were evaluated to determine whether the fitness and feeding activities of purchased earthworms were similar to that of earthworms collected in the field. In brief, the procedure determines the length of time required for Coomassie Brilliant Blue R-stained soil to pass through the earthworm (Hartenstein et al., 1981). Average gut transit times approximated 11 h, which is similar to that of earthworms collected in the field (Hartenstein and Amico, 1983).
Anoxic gut content microcosms
Earthworms were washed with sterile water, killed by brief immersion in 70 °C water and dissected under anoxic conditions in an O2-free chamber (Mecaplex, Grenchen, Switzerland) containing a gas phase of 100% dinitrogen (N2). Midgut and hindgut contents were pooled. A dilution of gut contents was necessary for obtaining adequate samples for chemical and molecular analyses. Gut content was homogenized (1:10 w/v) with sterile anoxic sodium phosphate buffer (pH 7) that contained (in grams per liter) 1.90 NaH2PO4·H2O and 3.36 Na2HPO4·2H2O. Tubes (27 ml) containing ∼8 ml gut homogenates were sealed with rubber stoppers and aluminum caps and flushed with sterile N2 (100%). Either [12C]glucose or [13C]glucose was added from sterile anoxic stock solutions (100 m) to a final concentration of ∼10 m. Tubes lacking supplemental substrates served as controls. Tubes were incubated upright in the dark at 15 °C. Headspace and liquid phase were sampled with sterile syringes. Samples for nucleic acid extraction were stored at −80 °C. Values are the mean values of triplicate experiments.
Analytical techniques
Moisture content was determined by weighing gut content before and after drying at 60 °C for 72 h. pH was measured with a U457-S7/110 combination pH electrode (Ingold, Steinbach, Germany). Carbon dioxide (CO2) and H2 were measured with a 5890 series II gas chromatograph equipped with a thermal conductivity detector (Hewlett-Packard, Palo Alto, CA, USA; Küsel and Drake, 1995). The concentrations of gases represent the total in both the gas and liquid phases, and are shown per unit volume of the liquid phase (as are the soluble compounds). Soluble organic compounds were analyzed with a 1090 series II high performance liquid chromatograph (Hewlett-Packard; Wüst et al., 2009a).
Nucleic acid extraction
RNA and DNA were coextracted by bead-beating lysis, organic solvent extraction and precipitation (Griffiths et al., 2000). The purification of RNA from DNA was performed with a Qiagen RNA/DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
Separation of 13C- and 12C-labeled RNA
A gradient solution (buoyant density 1.785 g ml−1) of cesium trifluoroacetate (buoyant density 2.0 g ml−1; 83.8%), gradient buffer (pH 8; 100 m Tris; 100 m KCl; 1 m EDTA; 13.5%) and formamide (3.2%) was added to 500 ng RNA and filled into OptiSeal Tubes (Beckmann, Fullerton, CA, USA). All gradients were set up with the same gradient solution to minimize potential differences that might otherwise occur in the labeling patterns. 13C- and 12C-labeled RNA were separated by isopycnic centrifugation (130 000 × g at 20 °C for 67 h; vertical rotor, VTi 65.2, Beckmann; Lüders et al., 2004) and fractions (450 μl each) were collected (Figure 1). The densities of the gradient solutions were determined by weighing at 25 °C. RNA was precipitated as described (Degelmann et al., 2009). RNA concentrations were determined using a Quant-iT RiboGreen RNA Assay Kit (Invitrogen, Karlsruhe, Germany) according to the manufacturer's protocol, and RNA was stored at −80 °C.
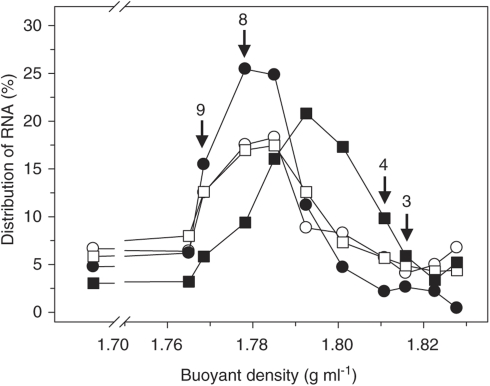
Figure 1.
Distribution of RNA in gradient fractions of glucose treatments. Arrows with fraction numbers indicate ‘heavy' (3 and 4) and ‘light' (8 and 9) fractions, which were chosen for further analyses. RNA was measured in all gradient fractions of one replicate. Symbols: empty symbols, RNA extracted at the start of incubation; filled symbols, RNA extracted at 51 h of incubation; circles, [12C]glucose treatment; squares, [13C]glucose treatment.
Reverse transcription PCR
Reverse transcription of RNA was performed with random hexamers and Superscript III reverse transcriptase (SuperScript VILO cDNA Synthesis Kit; Invitrogen) according to the manufacturer's protocol.
PCR and cloning
Complementary DNA was amplified by the bacterial 16S rRNA gene-specific primer set 27F/907RM (Lane, 1991) using a 5 Prime mastermix (5 Prime, Hamburg, Germany). PCR conditions were as follows: initial denaturation at 95 °C for 5 min, 4 cycles at 95 °C for 60 s, at 40 °C for 60 s and at 72 °C for 90 s, and 30 subsequent cycles at 95 °C for 60 s, 50 °C for 30 s, and 72 °C for 90 s. The final elongation was at 72 °C for 5 min. Purified PCR products from fractions three and four that represented labeled RNA (‘heavy' fractions; buoyant density 1.809–1.817 g ml−1) were pooled and fractions eight and nine that represented unlabeled RNA (‘light' fractions; buoyant density 1.767–1.779 g ml−1) were also pooled (Figure 1). PCR products were ligated into pGEM-T vector plasmids (Promega, Mannheim, Germany), and competent cells of Escherichia coli JM109 were transformed (Promega; protocol as per manufacturer's instructions). Clones with the correct insert were determined with M13 PCR (primer set M13F/M13R) according to published protocol (Messing, 1983) and selected for sequencing at Macrogen (Seoul, South Korea).
Sequence analyses
Analysis of 16S rRNA complementary DNA sequences (∼880 bp) was performed with MEGA (http://www.megasoftware.net; release Beta 4.1.; Tamura et al., 2007) and ARB (http://www.arb-home.de; version 2005; Ludwig et al., 2004). Chimeric sequences were identified by RDP Chimera Check 2.7 (http://rdp8.cme.msu.edu; Cole et al., 2003) and excluded from further analyses. The number of sequences per clone library ranged between 81 and 96. BLASTn 2.2.21 was applied for comparing sequences to those in public databases (Zhang et al., 2000). Sequences were assigned to taxonomic groups using the RDP Classifier with a confidence threshold of 80% (Wang et al., 2007). Sequences were aligned using the SINA Webaligner (http://www.arb-silva.de) and imported into the latest 16S rRNA gene-based database obtained from the SILVA homepage (Pruesse et al., 2007). Sequences were assigned to a novel family-level group on the basis of a 16S rRNA gene similarity <87.5% to the next cultivated species (Yarza et al., 2008; Schellenberger et al., 2010). Coverages based on family level were calculated as described (Good, 1958; Schloss et al., 2004). The detected bacterial diversity based on family level was analyzed by rarefaction analysis (Hurlbert, 1971). Phylogenetic trees were calculated using AxML and the neighbor-joining method (Saitou and Nei, 1987), which included a bootstrap test with 10 000 replicates (Felsenstein, 1985). A taxon was considered to be labeled when its abundance in the clone library from the heavy fractions of the [13C]glucose treatment was higher than its abundance in the clone library from the heavy fractions of the [12C]glucose treatment.
Nucleotide sequence accession numbers
The sequences obtained in this study are available from the EMBL nucleotide sequence database under accession numbers FN659069–FN659775.
Results
Effect of supplemental glucose on fermentation
Initial concentrations of glucose approximated 1 m in unsupplemented anoxic gut content microcosms (Figure 2a). The slight initial increase in glucose concentrations that occurred was indicative of the hydrolysis of polysaccharides in gut contents (Wüst et al., 2009a). The detection of glucose in unsupplemented controls and the detection of cellobiose in all microcosms (Table 1) confirm the occurrence of endogenous saccharides in midgut and hindgut contents (Wüst et al., 2009a). In this regard, as gut contents were diluted 1–10 for the preparation of microcosms (see Materials and methods), the initial concentration of 1 m glucose in unsupplemented microcosms indicated that the in situ concentration of glucose in gut contents approximated 10 m.
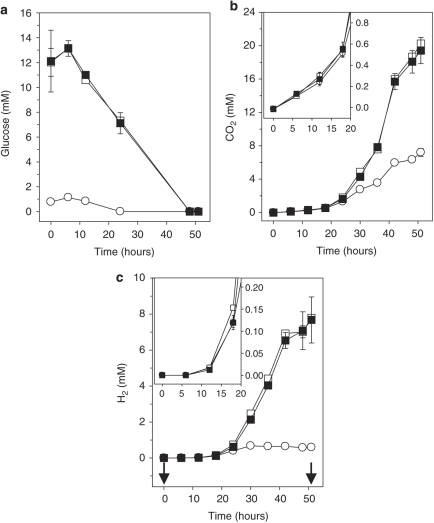
Figure 2.
Effect of supplemental glucose (a) on the production of CO2 (b) and H2 (c). The symbols indicate the means of triplicates; error bars indicate s.d.. Arrows indicate the time when samples were taken for RNA stable isotope probing analyses. Insets in (b) and (c) are enlargements of the initial incubation period. Symbols: ▪, results from [13C]glucose treatment; □, results from [12C]glucose treatment; ○, results from unsupplemented control.
Table 1. Effect of supplemental glucose on the formation of soluble organic compounds in gut content microcosmsa.
| Supplement | Time (h) |
Soluble organic compound (m) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Acetate | Lactate | Butyrate | Formate | Succinate | Propionate | Ethanol | Cellobiose | ||
| None | 0 | 0.1±0.0 | 0.2±0.0 | 0±0 | 0±0 | 0±0 | 0±0 | 0±0 | 0±0 |
| 6 | 0.2±0.0 | 0.2±0.0 | 0±0 | 0±0 | 0±0 | 0±0 | 0±0 | 0.1±0.0 | |
| 12 | 0.2±0.0 | 0.2±0.0 | 0±0 | 0±0 | 0±0 | 0±0 | 0±0 | 0±0 | |
| 24 | 1.3±0.4 | 0.4±0.2 | 0±0 | 0.3±0.1 | 0.1±0.0 | 0±0 | 0.5±0.5 | 0±0 | |
| 48 | 2.6±0.4 | 0.1±0.0 | 0±0 | 0±0 | 0.1±0.0 | 0±0 | 0.3±0.1 | 0±0 | |
| 51 | 4.0±0.2 | 0±0 | 0±0 | 0±0 | 0±0 | 0.5±0.0 | 0.5±0.0 | 0±0 | |
| [12C]glucose | 0 | 0.2b | 0.2b | 0b | 0b | 0b | 0b | 0b | 0.2b |
| 6 | 0.2±0.0 | 0.2±0.0 | 0±0 | 0.1±0.1 | 0±0 | 0±0 | 0±0 | 0.2±0.0 | |
| 12 | 0.3±0.1 | 0.2±0.0 | 0±0 | 0.1±0.1 | 0±0 | 0±0 | 0±0 | 0.1±0.0 | |
| 24 | 1.9±0.2 | 0.6±0.2 | 0±0 | 1.0±0.3 | 0.1±0.0 | 0±0.1 | 0.2±0.4 | 0.1±0.0 | |
| 48 | 5.9±0.6 | 3.7±0.1 | 1.0±0.1 | 0.4±0.1 | 0.4±0.1 | 0±0 | 2.1±0.1 | 0±0 | |
| 51 | 8.9±0.4 | 4.3±0.0 | 1.5±0.1 | 0±0 | 0.6±0.0 | 0.5±0.0 | 4.2±1.1 | 0±0 | |
| [13C]glucose | 0 | 0.2b | 0.2b | 0b | 0b | 0b | 0b | 0b | 0.2b |
| 6 | 0.2±0.0 | 0.1±0.0 | 0±0 | 0±0 | 0±0 | 0±0 | 0±0 | 0.1±0.0 | |
| 12 | 0.3±0.0 | 0.2±0.0 | 0±0 | 0.1±0.1 | 0±0 | 0±0 | 0±0 | 0.1±0.0 | |
| 24 | 1.6±0.4 | 0.5±0.2 | 0±0 | 1.1±0.3 | 0±0 | 0±0 | 0.4±0.2 | 0.1±0.0 | |
| 48 | 5.5±1.5 | 3.4±1.0 | 0.8±0.3 | 0.5±0.2 | 0.4±0.2 | 0±0 | 1.7±0.6 | 0±0 | |
| 51 | 8.0±1.9 | 3.9±0.8 | 1.5±0.6 | 0.1±0.1 | 0.5±0.2 | 0.3±0.0 | 3.6±1.0 | 0±0 | |
Unless otherwise stated, values are the means of triplicates (± s.d.).
No s.d. is given because two replicates were analyzed, and the value is the average for the replicates.
Glucose was completely consumed within 48 h in glucose-supplemented microcosms and within 24 h in unsupplemented microcosms. Consumption of supplemental glucose was relatively linear and rapid. Product profiles of [12C]- and [13C]glucose incubations were nearly identical (Figure 2, Table 1), indicating that microbial activities were similar in both treatments. Approximately 20 m CO2 and 8 m H2 accumulated in incubations with supplemental glucose, whereas ∼7 m CO2 and 0.6 m H2 accumulated in unsupplemented microcosms (Figures 2b and c), suggesting that supplemental glucose decreased the ratios between CO2 and H2 production. However, it cannot be excluded that use of alternative electron acceptors consumed a portion of the H2 formed from either endogenous matter or supplemental glucose. CO2 was produced without apparent delay, whereas the detection of H2 was minimal during the initial incubation period.
All incubations started at a pH of ∼7.3. pH values in glucose-supplemented microcosms at 51 h of incubation were lower than those in unsupplemented microcosms (6.9 and 7.5, respectively), which was consistent with the enhanced production of organic acids (Table 1). Acetate, butyrate, lactate, propionate, succinate and ethanol were end products in glucose-supplemented microcosms. Up to 1 m of formate was transiently produced in glucose-supplemented microcosms, indicating that formate was an intermediate product during the degradation of glucose. Lactate, formate and succinate occurred as transient products in unsupplemented microcosms, whereas acetate, propionate and ethanol were end products. Acetate was the dominant organic acid detected in all treatments in terms of concentration. Minor concentrations (that is, <0.06 m) of products indicative of amino-acid fermentation (that is, isobutyrate and -methylbutyrate; Nanninga and Gottschal, 1985; Gottschalk, 1988) were only detected in unsupplemented incubations (data not shown).
At 51 h of incubation, 72% and 60% of supplemental carbon were recovered in fermentation products of [12C]- and [13C]glucose incubations, respectively (based on amounts of CO2 and soluble organic compounds detected; Figure2 and Table 1; percentages were calculated after subtracting concentrations in unsupplemented controls). The unrecovered carbon could have been dissimilated to undetected products or assimilated and perhaps converted to intracellular storage polymers. Highest percentages of supplemental 12C- and 13C-carbon were recovered in CO2 (19% and 16%, respectively), lactate (18% and 16%, respectively) and acetate (14% and 9%, respectively). Approximately 74% and 63% of supplemental electrons were recovered in fermentation products of [12C]- and [13C]glucose incubations, respectively. The highest percentages of electrons derived from supplemental [12C]- and [13C]glucose were recovered in lactate (18% and 15%, respectively), ethanol (18% and 15%, respectively) and butyrate (both 17%).
Active bacteria in gut contents
A total of 707 16S rRNA complementary DNA sequences were analyzed from ‘heavy' and ‘light' fractions of [12C]- and [13C]glucose incubations (Table 2). Family-level coverage approximated 93% for sequences obtained at the beginning of incubation and 97% for sequences obtained after 51 h of incubation. Sequences were assigned to the phyla Proteobacteria (34.5%), Firmicutes (32.4%), Actinobacteria (20.9%), Planctomycetes (4.0%), Bacteroidetes (3.5%), Acidobacteria (1.6%), Verrucomicrobia (1.0%), Chloroflexi (0.3%), Gemmatimonadetes (0.3%), Nitrospirae (0.3%), Tenericutes (0.1%) and Cyanobacteria (0.1%), or were defined as unclassified Bacteria (0.8% Table 2; percentages reflect the relative amount of a given taxon). In total, 82 families were detected, 17 of which were defined as novel family-level groups on the basis of a 16S rRNA gene similarity cutoff of 87.5% (Yarza et al., 2008; Schellenberger et al., 2010).
Table 2. Phylogenetic distribution of 16S rRNA cDNA sequences obtained from [12C]- and [13C]glucose treatmentsa.
|
Phylogenetic affiliation (total relative abundance of phylum) |
Relative abundance of sequences (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phylum | Order, family |
[12C]glucose |
[13C]glucose |
||||||
|
t0 |
t51 |
t0 |
t51 |
||||||
| L | H | L | H | L | H | L | H | ||
| Acidobacteria (1.6) | Acidobacteriales, | ||||||||
| Acidobacteriaceae | 3.7 | 1.1 | — | — | 6.3 | 1.2 | — | — | |
| Actinobacteria (20.9) | Novel group 4b | — | — | — | — | 1.0 | 1.2 | — | — |
| Novel group 6b | 4.9 | 1.1 | — | 1.1 | 2.1 | 1.2 | — | — | |
| Novel group 7b | — | — | — | — | 1.0 | — | — | — | |
| Novel group 8b | — | — | — | — | — | 2.3 | — | — | |
| Novel group 17b | 1.2 | — | — | — | — | — | — | — | |
| Acidimicrobiales, | |||||||||
| Acidimicrobiaceae | 1.2 | — | — | — | — | 2.3 | — | — | |
| Iamiaceae | — | — | — | — | 2.1 | 1.2 | — | — | |
| Actinomycetales, | |||||||||
| Novel group 5b | — | — | — | — | — | 1.2 | — | — | |
| Actinosynnemataceae | — | 1.1 | — | — | — | — | — | — | |
| Beutenbergiaceae | — | — | — | — | 1.0 | — | — | — | |
| Cellulomonadaceae | 1.2 | 1.1 | — | — | 1.0 | — | — | — | |
| Cryptosporangiaceae | — | 1.1 | — | — | 1.0 | — | 1.1 | — | |
| Frankiaceae | — | 2.2 | — | — | — | 1.2 | — | — | |
| Intrasporangiaceae | — | 2.2 | — | — | 1.0 | — | — | — | |
| Microbacteriaceae | 2.5 | 9.0 | — | — | 5.2 | 4.7 | — | — | |
| Micrococcaceae | 1.2 | 1.1 | — | — | — | — | — | — | |
| Micromonosporaceae | — | 2.2 | — | — | — | 2.3 | 1.1 | — | |
| Mycobacteriaceae | — | 3.4 | — | — | — | 2.3 | — | — | |
| Nakamurellaceae | — | 2.2 | — | — | 2.1 | 4.7 | — | — | |
| Nocardiaceae | 1.2 | 2.2 | — | 1.1 | — | 1.2 | 1.1 | — | |
| Nocardioidaceae | 6.2 | 4.5 | — | — | 8.3 | 8.1 | — | — | |
| Propionibacteriaceae | — | — | — | — | 2.1 | 1.2 | 1.1 | — | |
| Pseudonocardiaceae | 2.5 | 4.5 | — | — | — | 4.7 | — | — | |
| Sporichthyaceae | 2.5 | 3.4 | — | — | 1.0 | — | — | — | |
| Streptomycetaceae | 1.2 | — | — | — | — | — | — | — | |
| Rubrobacterales, | |||||||||
| Novel group 1b | 2.5 | 5.6 | — | — | 1.0 | 2.3 | — | — | |
| Rubrobacteraceae | 2.5 | 3.4 | — | — | 3.1 | 2.3 | — | — | |
| Solirubrobacterales, | |||||||||
| Patulibacteraceae | — | — | — | 1.1 | — | 1.2 | — | — | |
| Bacteroidetes (3.5) | Flavobacteriales, | ||||||||
| Flavobacteriaceae | 11.1 | 2.2 | — | — | 5.2 | 4.7 | — | — | |
| Sphingobacteriales, | |||||||||
| Chitinophagaceae | — | — | — | — | — | 1.2 | — | — | |
| Flexibacteraceae | — | — | — | — | 2.1 | 1.2 | — | — | |
| Novel group 16b | — | — | — | — | 1.0 | — | — | — | |
| Chloroflexi (0.3) | Novel group 14b | — | 1.1 | — | — | — | 1.2 | — | — |
| Cyanobacteria (0.1) | Family I | — | — | — | — | — | 1.2 | — | — |
| Firmicutes (32.4) | Bacillales, | ||||||||
| Bacillaceae | 1.2 | 1.1 | 6.6 | 5.7 | 1.0 | — | 5.7 | 3.4 | |
| Incertae Sedis XII | — | 1.1 | — | — | 1.0 | — | — | — | |
| ‘Paenibacillaceae' | — | — | 6.6 | 4.6 | 2.1 | — | 5.7 | — | |
| Clostridiales, | |||||||||
| Clostridiaceae | 1.2 | 1.1 | 27.5 | 25.3 | — | — | 9.1 | 50.6 | |
| ‘Lachnospiraceae' | — | — | 1.1 | 2.3 | — | — | — | 2.2 | |
| ‘Peptostreptococcaceae' | — | — | 16.5 | 17.2 | 1.0 | — | 51.1 | 4.5 | |
| Veillonellaceae | — | — | — | 1.1 | — | — | — | — | |
| Gemmatimonadetes (0.3) | Gemmatimonadales, | ||||||||
| Gemmatimonadaceae | — | 1.1 | — | — | 1.0 | — | — | — | |
| Nitrospirae (0.3) | Nitrospirales, | ||||||||
| Nitrospiraceae | — | — | — | — | 1.0 | 1.2 | — | — | |
| Planctomycetes (4.0) | Novel group 15b | — | — | — | — | 1.0 | — | — | — |
| Planctomycetales, | |||||||||
| Planctomycetaceae | 7.4 | 7.9 | — | — | 4.2 | 10.5 | 1.1 | — | |
| Proteobacteria (34.5) | |||||||||
| Alphaproteobacteria | Caulobacterales, | ||||||||
| Caulobacteraceae | — | — | — | — | 1.0 | — | — | — | |
| Rhizobiales, | |||||||||
| Beijerinckiaceae | 1.2 | — | — | — | — | — | — | — | |
| Bradyrhizobiaceae | 4.9 | 3.4 | — | — | 6.3 | 5.8 | 2.3 | — | |
| Hyphomicrobiaceae | 2.5 | 1.1 | — | — | 4.2 | 2.3 | — | — | |
| Methylobacteriaceae | 1.2 | — | — | — | — | — | — | — | |
| Methylocystaceae | 1.2 | 1.1 | — | — | — | — | — | — | |
| Phyllobacteriaceae | — | 3.4 | — | — | — | — | — | — | |
| Xanthobacteraceae | 1.2 | 1.1 | — | — | 1.0 | — | — | — | |
| Rhodobacterales, | |||||||||
| Rhodobacteraceae | 2.5 | 3.4 | — | — | — | — | — | — | |
| Rhodospirillales, | |||||||||
| Acetobacteraceae | 1.2 | 2.2 | — | — | — | — | — | — | |
| Rhodospirillaceae | 1.2 | 5.6 | — | — | 4.2 | 4.7 | 1.1 | — | |
| Betaproteobacteria | Burkholderiales, | ||||||||
| Alcaligenaceae | — | 1.1 | — | — | — | — | — | — | |
| Incertae sedis 5 | — | — | — | — | 2.1 | 1.2 | — | — | |
| Oxalobacteraceae | — | — | — | — | — | — | 1.1 | — | |
| Gammaproteobacteria | Aeromonadales, | ||||||||
| Aeromonadaceae | 1.2 | 2.2 | 14.3 | 20.7 | 1.0 | 2.3 | 9.1 | 4.5 | |
| Alteromonadales, | |||||||||
| Shewanellaceae | — | — | — | — | 1.0 | — | — | — | |
| Enterobacteriales, | |||||||||
| Enterobacteriaceae | 2.5 | — | 26.4 | 18.4 | 1.0 | — | 9.1 | 34.8 | |
| Legionellales, | |||||||||
| Legionellaceae | — | — | — | — | — | 1.2 | — | — | |
| Pseudomonadales, | |||||||||
| Pseudomonadaceae | — | — | — | — | — | 2.3 | — | — | |
| Xanthomonadales, | |||||||||
| Sinobacteraceae | — | — | — | — | 2.1 | — | — | — | |
| Xanthomonadaceae | 1.2 | — | — | — | 1.0 | — | — | — | |
| Deltaproteobacteria | Novel group 9b | 3.7 | 1.1 | — | 1.1 | — | 1.2 | — | — |
| Novel group 10b | — | 1.1 | — | — | — | — | — | — | |
| Unclassified group 1c | 6.2 | 1.1 | — | — | 2.1 | 3.5 | — | — | |
| Myxococcales, | |||||||||
| Cystobacteraceae | — | 1.1 | 1.1 | — | 1.0 | — | — | — | |
| Polyangiaceae | 6.2 | — | — | — | 5.2 | 1.2 | — | — | |
| Novel group 2b | — | — | — | — | 1.0 | 2.3 | — | — | |
| Novel group 3b | 2.5 | — | — | — | — | — | — | — | |
| Tenericutes (0.1) | Unclassified group 2d | — | — | — | — | — | 1.2 | — | — |
| Verrucomicrobia (1.0) | Unclassified group 3e | — | 2.2 | — | — | 1.0 | 1.2 | — | — |
| Opitutales, | |||||||||
| Opitutaceae | 1.2 | — | — | — | — | — | — | — | |
| Verrucomicrobiales, | |||||||||
| Novel group 11b | — | — | — | — | — | 1.2 | — | — | |
| Subdivision 3 | — | — | — | — | 1.0 | — | — | — | |
| WS3 (0.1) | Incertae sedis | — | — | — | — | 1.0 | — | — | — |
| Unclassified Bacteria (0.8) | Novel group 12b | — | 1.1 | — | — | 1.0 | — | — | — |
| Novel group 13b | 2.5 | — | — | — | 1.0 | 1.2 | — | — | |
Abbreviations: cDNA, complementary DNA; H, ‘heavy' fractions (fractions 3 and 4; Figure 1); L, ‘light' fractions (fractions 8 and 9; Figure 1); rRNA, ribosomal RNA; t0, at the start of incubation; t51, at 51 h of incubation.
Sequences were assigned to families using the RDP Classifier (Wang et al., 2007) and Blast (Zhang et al., 2000). Number of sequences in the eight clone libraries ranged between 81 and 96 per library. Quotation marks indicate nonvalidated taxa (Euzéby, 2010).
Novel family-level group based on a 16S rRNA gene similarity <87.5% (Yarza et al., 2008).
Next cultivated species: Candidatus ‘Entotheonella palauensis' (AF130847; 86.2–90.9% 16S rRNA gene similarity).
Next cultivated species: Candidatus ‘Lumbricincola sp. Lr-C2' (FM165584; 90.2% 16S rRNA gene similarity).
Next cultivated species: Candidatus ‘Xiphinematobacter rivesi' (AF217461; 89.5–90.3% 16S rRNA gene similarity).
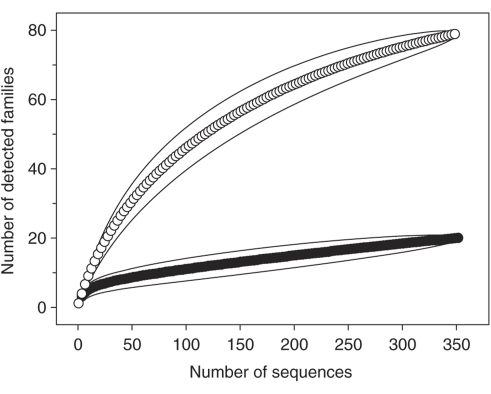
The rarefaction curve for sequences obtained at the start of incubation was significantly higher than the rarefaction curve for sequences obtained after 51 h of incubation (Figure 3). This result indicates that a subset of the total taxa was enriched during incubation, thus yielding a lower detected relative bacterial diversity. Sequences obtained at the start of incubation were assigned to 79 families, 17 of which were novel family-level groups, indicating a high and partially unknown diversity of microorganisms in the earthworm gut. Approximately one-third of these 79 families were affiliated with Actinobacteria, a phylum from which diverse family-level taxa have been isolated from soil (Joseph et al., 2003). The detection of 20 families (which included two novel family-level groups) at 51 h of incubation under anoxic conditions suggests that organisms of these families were capable of anaerobiosis.
Figure 3.
Rarefaction curves of 16S rRNA complementaryDNA sequences were obtained at the start of incubation (empty circles) and after 51 h of incubation (filled circles). Each curve is based on the total sequences obtained from [12C]glucose- and [13C]glucose-supplemented incubations. Solid lines enclosing the symbols indicate the 95% confidence intervals.
Dominant users of glucose-derived carbon
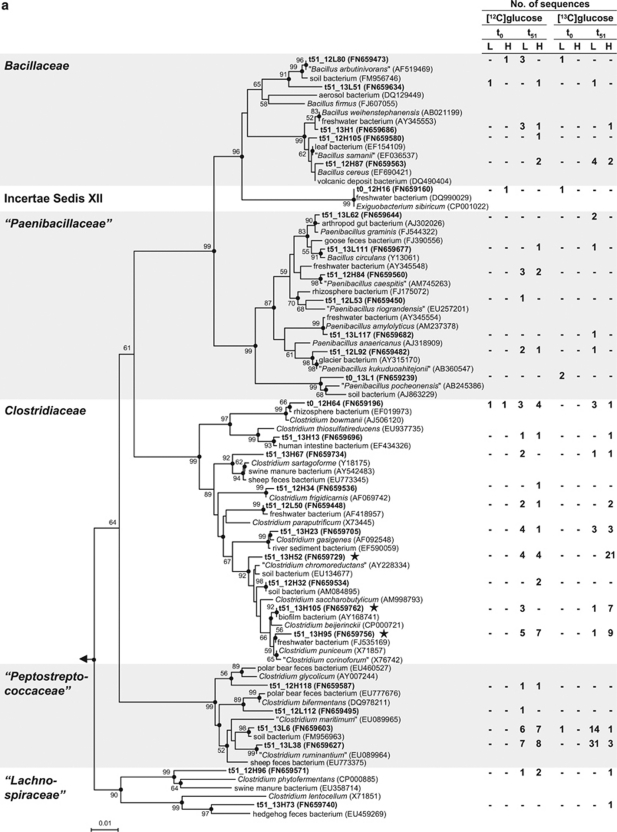
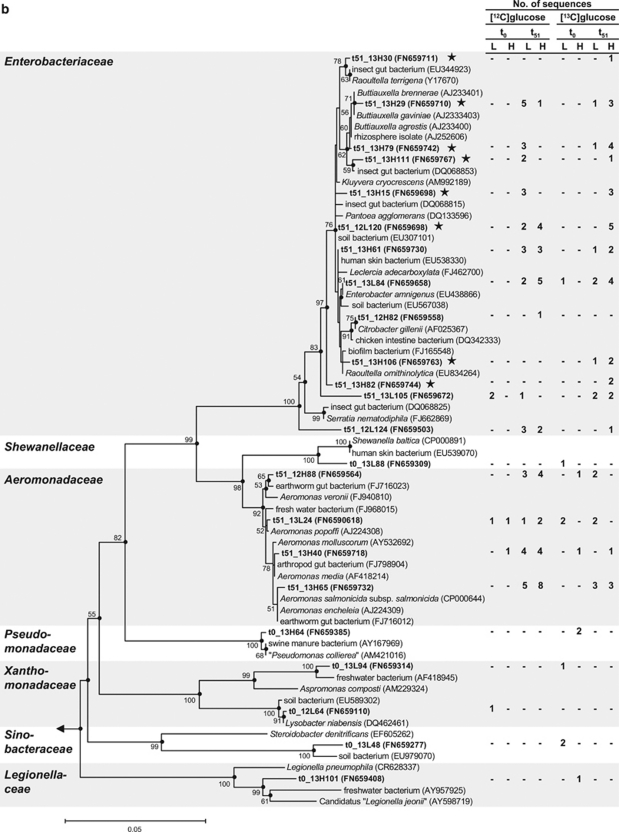
RNA obtained from [13C]glucose treatments became enriched in 13C on the basis of the shift of RNA toward the heavy fractions at the end of the 51 h incubation, a shift not observed with [12C]glucose treatments (Figure 1). Sequences obtained from the ‘heavy' fractions of 51 h samples from [13C]glucose treatments were assigned to families Aeromonadaceae, Bacillaceae, Clostridiaceae, Enterobacteriaceae, ‘Lachnospiraceae' and ‘Peptostreptococcaceae' (Table 2; quotation marks indicate nonvalidated taxa; Euzéby, 2010). Bacillaceae and Clostridiaceae are spore forming, and it is thus noteworthy that gut passage can enhance the germination of spores (Fischer et al., 1997). In [13C]glucose treatments, the relative abundances of sequences affiliated with Clostridiaceae and Enterobacteriaceae were higher in the ‘heavy' fractions than in the ‘light' fractions, indicating that organisms of these families were dominant users of glucose-derived carbon. The closest related cultivated species of labeled Clostridiaceae sequences were Clostridium beijerinckii, ‘C. chromoreductans', ‘C. corinoforum' and C. puniceum (Figure 4a). The closest related cultivated species of labeled Enterobacteriaceae-sequences were Buttiauxella spp., Kluyvera cryocrescens, Pantoea agglomerans, Raoultella spp. and Serratia fonticola (Figure 4b). Many closest related species of sequences affiliated with Firmicutes and Gammaproteobacteria have been recovered from soil, water, intestines or feces of animals (Figures 4a and b).
Figure 4.
Phylogenetic tree of 16S rRNA complementaryDNA sequences (bold) retrieved from the earthworm gut and reference sequences of the Firmicutes (a) and Gammaproteobacteria (b). Only representative sequences are shown. The values next to the branches represent the percentages of replicate trees (>50%) in which the associated taxa clustered together in the bootstrap test (10 000 replicates). Dots at nodes indicate confirmation of topology by AxML using the same data set. Labeled taxa are marked with stars. Methanosarcina barkeri (AF028692) was used as outgroup. Accession numbers are in parentheses. Quotation marks indicate nonvalidated taxa (Euzéby, 2010). Abbreviations: t0, at the start of incubation; t51, at 51 h of incubation; L, ‘light' fractions; H, ‘heavy' fractions. (a): The family Veillonellaceae was excluded because the sequence that affiliated with this family (t51_12H90) was shorter than 700 bp. The phylogenetic tree was calculated using the neighbor-joining method (50% minimum similarity filter; 651 valid positions between 104 and 815 of the 16S rRNA gene of E. coli). Bar indicates 0.01 estimated change per nucleotide. (b): The labeled sequence t51_13H112 (next cultivated species Serratia fonticola, AY236502, 99% 16S rRNA gene similarity) was excluded from tree calculation because it was shorter than 700 bp. The phylogenetic tree was calculated using the neighbor-joining method (50% minimum similarity filter; 698 valid positions between 101 and 816 of the 16S rRNA gene of E. coli). Bar indicates 0.05 estimated change per nucleotide.
Discussion
Glucose was conceived in this study to be a model trophic link in the mutualistic digestive system of earthworms (Brown et al., 2000). Glucose is one of the most efficient substrates for the conservation of energy and growth (Reid, 2005). Thus, the high availability of glucose and other saccharides in earthworm gut (Horn et al., 2003; Wüst et al., 2009a) provides ingested soil microbes with a nutritional oasis. The organic acid product profile in glucose-supplemented incubations of gut content (Table 1) was similar to that detected along the alimentary canal (Wüst et al., 2009a) and also to that of high-dilution most probable number of tubes of tryptic soy broth inoculated with earthworm gut contents (Ihssen et al., 2003).
Fermentative processes and general diversity in gut contents
The detection of lactate as a transient intermediate and propionate as an end product in unsupplemented incubations (Table 1) indicates that lactate might have been consumed by propionic acid bacteria (Gottschalk, 1988). However, equal amounts of propionate were formed in unsupplemented and glucose-supplemented incubations, suggesting that propionate fermentation was not stimulated by supplemental glucose and that propionate fermenters in the earthworm gut prefer carbon sources other than glucose or lactate. Indeed, sequences related to Propionibacteriaceae were detected before and after incubation of earthworm gut content with [13C]glucose, but were not labeled (Table 2), indicating that Propionibacteriaceae-related species did not significantly assimilate carbon from supplemental glucose.
Nearly half of the CO2 produced in glucose-supplemented treatments could be attributed to endogenous sources of carbon (Figure 2b), a result consistent with previous studies showing that gut contents are rich in readily available sources of organic carbon (Brown et al., 2000; Horn et al., 2003; Wüst et al., 2009a). Thus, microorganisms in gut contents consumed endogenous sources of carbon in addition to supplemental glucose. Although most of the organic carbon consumed during fermentation is dissimilated (that is, not assimilated; Gottschalk, 1988), the rapid dissimilation of supplemental glucose was concomitant to the labeling of rRNA, indicating that taxa capable of anaerobiosis in gut contents were poised to respond quickly to increased nutrient availability and assimilated organic carbon.
Active phyla identified by 16S rRNA analysis of earthworm gut contents included Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes, Gemmatimonadetes, Nitrospirae, Planctomycetes, (Alpha-, Beta-, Gamma- and Delta-) Proteobacteria, Tenericutes and Verrucomicrobia, taxa common to soils (Table 2). Alpha-, Beta- and Gamma-Proteobacteria, Bacteroidetes, Actinobacteria and Firmicutes are potentially dominant taxa in the earthworm gut, as determined on the basis of 16S rRNA gene analyses (Furlong et al., 2002; Singleton et al., 2003; Knapp et al., 2009). Furthermore, species related to Aeromonadaceae, Comamonadaceae, Enterobacteriaceae, Flavobacteriaceae, Moraxellaceae, ‘Paenibacillaceae', Pseudomonadaceae, Rhodocyclaceae, Sphingobacteriaceae and Actinobacteria have been isolated from the alimentary canals of earthworms (Ihssen et al., 2003; Horn et al., 2005; Byzov et al., 2009; Knapp et al., 2009). These findings show the diverse taxa that might be metabolically active in the earthworm gut, many of which might participate in polymer hydrolysis in situ (Martin-Carnahan and Joseph, 2005; Bernadet and Bowman, 2006; Priest, 2009).
Sequences obtained before incubation were assigned to 14 phyla, whereas sequences obtained after incubation with glucose were assigned to only 4 phyla, with Firmicutes and Proteobacteria being dominant (Table 2). Clostridiaceae and Enterobacteriaceae were the labeled families (Table 2). These glucose-assimilating taxa occur in soils (Hamberger et al., 2008; Degelmann et al., 2009; Schellenberger et al., 2010) and have robust metabolic capacities and doubling times that can range between 0.5 and 3 h (Francis et al., 2000; Wiegel et al., 2006), factors that likely contributed to their competitiveness.
Many fermentative or facultative microorganisms can reduce nitrate or nitrite (Cole, 1990; Umarov, 1990). Active genera included Aeromonas, Bacillus, Buttiauxella, Clostridium, Enterobacter, Flavobacterium, Paenibacillus and Pseudomonas (Table 2, Figures 4a and b), and earthworm gut isolates affiliated with these genera have the ability to reduce either nitrate or nitrite and produce N2O (Ihssen et al., 2003). Different regions of the alimentary canal yield different organic acid, H2 and N2O signatures (Wüst et al., 2009a), reflecting potential trophic links that might occur between fermentation and denitrification during gut passage (for example, when denitrifiers use fermentation products as a source of reductant).
Clostridiaceae
Most sequences derived from ‘heavy' fractions after [13C]glucose incubation were affiliated with the genus Clostridium sensu stricto (Collins et al., 1994; Figure 4a). Species of this genus are considered to be obligate anaerobes and can dissimilate glucose to acetate, butyrate, lactate, ethanol, H2 and CO2 (Rainey et al., 2009). The formation of these products was enhanced by glucose (Figure2 and Table 1). The closest related cultured species of labeled clostridial sequences, C. beijerinckii and C. puniceum, can produce substantial amounts of H2 in addition to butyrate, acetate, formate and butanol (Rainey et al., 2009). Although H2 is a trophic link to methanogenesis and acetogenesis (Drake et al., 2006, 2008; Hedderich and Whitman, 2006; Liu and Whitman, 2008; McInerney et al., 2008), methanogens and acetogens are not metabolically significant in the earthworm gut (Karsten and Drake, 1995).
Most electrons derived from supplemental glucose were recovered in lactate, but classic lactate-producing genera such as Lactococcus, Lactobacillus and Streptococcus were not detected, which is not surprising, as most species in these taxa are not considered to be common to soil (Teuber et al., 1992; Hammes and Hertel, 2009; Whiley and Hardie, 2009). Similarly, Lactovum, a lactate-forming genus isolated from acidic soil (Matthies et al., 2004; Drake, 2010), was not detected. Lactate can also be produced by saccharolytic clostridia and can be the main fermentation product of these organisms under certain conditions (Wiegel et al., 2006). For example, C. perfringens produces mainly lactate under carbon-rich conditions (Macfarlane and Macfarlane, 2003). Some sequences were closely affiliated with C. gasigenes and C. frigidicarnis (Figure 4a), species that can grow at low temperatures and secret hydrolytic exoenzymes (Broda et al., 1999, 2000; Akila and Chandra, 2003), indicating that psychrotolerant clostridia might be involved in the degradation of organic matter in the earthworm alimentary canal. ‘Peptostreptococcaceae' are closely related to Clostridiaceae (Ludwig et al., 2009 and Figure 4a) and were enriched during incubation (Table2 and Figure 4a). However, the high relative abundance of ‘Peptostreptococcaceae'-related sequences in the ‘light' fractions of the [13C]glucose treatment indicated that ‘Peptostreptococcaceae' used endogenous sources of organic carbon.
Enterobacteriaceae
Species of the family Enterobacteriaceae are facultative aerobes having both a respiratory and a fermentative metabolism (Brenner, 1992). Characteristic fermentation products of Enterobacteriaceae include formate, succinate, lactate, acetate, ethanol, H2 and CO2 (Brenner and Farmer, 2005), products consistent with those detected in glucose treatments (Figure2 and Table 1) and also with the apparent fermentation products detected along the earthworm alimentary canal (Wüst et al., 2009a). Most Enterobacteriaceae are able to reduce nitrate to nitrite (Brenner, 1992) and might therefore be at least partly responsible for the low nitrate concentrations in the alimentary canal compared with those of preingested soil (Horn et al., 2003; Wüst et al., 2009a). Several genera within the Enterobacteriaceae (Buttiauxella, Kluyvera, Pantoea, Raoultella and Serratia) were labeled with [13C]glucose-derived carbon (Figure 4b). Species of these genera are able to ferment glucose (Müller et al., 1996; Ihssen et al., 2003; Brenner and Farmer, 2005; Grimont and Grimont, 2005). Some labeled enterobacterial sequences were affiliated with Buttiauxella agrestis (Figure 4b), strains of which can reduce N2O to N2 (Kaldorf et al., 1993). Many labeled enterobacterial sequences were related to P. agglomerans (Figure 4b). P. agglomerans produces exoenzymes chitinase, pectinase and protease (Mahmoud et al., 2008), and might therefore contribute to the degradation of complex organic matter in the earthworm gut. The numerous anaerobic capabilities of P. agglomerans (Grimont and Grimont, 2005) might confer a selective advantage in O2-limited habitats similar to the earthworm gut. In addition to growing by fermentation, this organism is capable of dissimilating metals such as Fe(III) by using lactate, acetate and H2 as electron donors (Francis et al., 2000). The large amounts of Fe(II) in the earthworm alimentary canal suggest that Fe(III) reduction occurs in situ and could be driven by such fermentation products (Wüst et al., 2009a).
Limitations and conclusions
Microcosm experiments do not fully duplicate in situ conditions. Furthermore, a major constraint in interpreting stable isotope probing experiments is the potential labeling that can occur because of cross-feeding (Neufeld et al., 2007). Thus, the limitations of this study should be examined. The 1–10 dilution of gut contents necessitated for obtaining samples for chemical and molecular analyses diluted both taxa and endogenous nutrients, and cross-feeding could have resulted in labeling of taxa that did not directly assimilate glucose. For example, the apparent consumption of small amounts of formate and lactate in unsupplemented microcosms indicated that a limited amount of [13C]glucose-derived formate and lactate might have been consumed and thus assimilated. Also, only one of several saccharides potentially available in the gut (Wüst et al., 2009a) was evaluated. The above interpretations must therefore be qualified within the constraints of these limitations. Nevertheless, anoxia, 15 °C, 10 m glucose, neutral pH, and the relatively short incubation period approximated important parameters likely to be of importance to fermentation in the earthworm gut. In addition, if cross-feeding occurred in the microcosms, it could and likely does occur in situ. For example, acetate was a major fermentation product of earthworm gut content (Table 1) and might constitute an in situ trophic link between fermenters and denitrifiers, as it is a substrate for many denitrifiers (Tiedje, 1988). Thus, the labeling observed might theoretically reflect the type of interconnected events that occur during gut passage. However, midgut and hindgut contents contain only marginal amounts of nitrate and nitrite (Wüst et al., 2009a), and no N2O was produced in the stable isotope probing microcosms (data not shown), suggesting that cross-feeding of denitrifiers was very unlikely. Indeed, although denitrifying taxa were detected at the beginning of incubation, they were neither labeled nor detected at the end of 51 h of incubation (Table 2). Although we cannot fully exclude the possibility that cross-feeding occurred, it was likely minimal because of the relatively short incubation period and limited availability of alternative electron acceptors. Furthermore, the majority of 13C from [13C]glucose was recovered in fermentation products that accumulated with time, suggesting that subsequent assimilation of these products was minimal. Similarly, selective death and selective predation seem unlikely causes of the marked enrichment of Clostridiaceae and Enterobacteriaceae.
Within these constraints and considerations, the findings of this study suggest that Clostridiaceae and Enterobacteriaceae are subsets of diverse active taxa in the alimentary canal of the earthworm that are capable of consuming mucus- and plant-derived saccharides during gut passage, and illustrate the effect that gut passage might have on selectively stimulating fermentative taxa of ingested soil. Although a minimal cross-feeding might have occurred, the substantial labeling of Clostridiaceae and Enterobacteriaceae indicates that certain ingested obligate anaerobes and facultative aerobes from soil can concomitantly metabolize the same source of carbon. This possibility is consistent with the formation of products considered indicative of clostridial (for example, butyrate) and enterobacterial (for example, succinate) fermentations (Brenner and Farmer, 2005; Rainey et al., 2009) in the midgut and hindgut of the alimentary canal (Wüst et al., 2009a).
Acknowledgments
We thank Peter Depkat-Jakob for assistance with dissecting worms. Support for this study from the Deutsche Forschungsgemeinschaft (DR310/4-1) and the University of Bayreuth is gratefully acknowledged.
References
- Akila G, Chandra TS. A novel cold-tolerant Clostridium strain PXYL1 isolated from a psychrophilic cattle manure digester that secretes thermolabile xylanase and cellulase. FEMS Microbiol Lett. 2003;219:63–67. doi: 10.1016/S0378-1097(02)01196-5. [DOI] [PubMed] [Google Scholar]
- Bernadet JF, Bowman J.2006The genus FlavobacteriumIn: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H and Stackebrandt E (eds).The Prokaryotes3rd edn.Springer: New York; 481–531. [Google Scholar]
- Brenner DJ.1992Introduction to the family EnterobacteriaceaeIn: Balows A, Trüper HG, Dworkin M, Harder W and Schleifer KH (eds).The Prokaryotes2nd edn.Springer Verlag: New York; 2673–2695. [Google Scholar]
- Brenner DJ, Farmer JJ.2005Family I. EnterobacteriaceaeIn: Brenner DJ, Krieg NR and Staley JT (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York; 587–607. [Google Scholar]
- Broda DM, Lawson PA, Bell RG, Musgrave DR. Clostridium frigidicarnis sp nov., a psychrotolerant bacterium associated with ′blown pack′ spoilage of vacuum-packed meats. Int J Syst Bacteriol. 1999;49:1539–1550. doi: 10.1099/00207713-49-4-1539. [DOI] [PubMed] [Google Scholar]
- Broda DM, Saul DJ, Lawson PA, Bell RG, Musgrave DR. Clostridium gasigenes sp. nov., a psychrophile causing spoilage of vacuum-packed meat. Int J Syst Evol Microbiol. 2000;50:107–118. doi: 10.1099/00207713-50-1-107. [DOI] [PubMed] [Google Scholar]
- Brown GG, Barois I, Lavelle P. Regulation of soil organic matter dynamics and microbial activity in the drilosphere and the role of interactions with other edaphic functional domains. Eur J Soil Biol. 2000;36:177–198. [Google Scholar]
- Byzov BA, Nechitaylo TY, Bumazhkin BK, Kurakov AV, Golyshin PN, Zvyagintsev DG. Culturable microorganisms from the earthworm digestive tract. Microbiology. 2009;78:360–368. [PubMed] [Google Scholar]
- Cole JA.1990Physiology, biochemistry, and genetics of nitrate dissimilation to ammoniaIn: Revsbech NP and Sorensen J (eds).Denitrification in Soils and Sediment Plenum Press: New York; 57–76. [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- Degelmann DM, Kolb S, Dumont M, Murrell JC, Drake HL. Enterobacteriaceae facilitate the anaerobic degradation of glucose by a forest soil. FEMS Microbiol Ecol. 2009;68:312–319. doi: 10.1111/j.1574-6941.2009.00681.x. [DOI] [PubMed] [Google Scholar]
- Drake HL.2010LactovumIn: Holzapfel W and Wood BJB (eds).Lactic Acid Bacteria - Biodiversity and Taxonomy Blackwell Publishing Ltd.: Oxford; (in press). [Google Scholar]
- Drake HL, Gößner AS, Daniel SL.2008Old acetogens, new lightIn: Wiegel J, Maier RJ and Adams MWW (eds).Incredible Anaerobes: From Physiology to Genomics to Fuels New York Academy of Sciences: Boston; 100–128. [DOI] [PubMed] [Google Scholar]
- Drake HL, Horn MA. As the worm turns: the earthworm gut as a transient habitat for soil microbial biomes. Annu Rev Microbiol. 2007;61:169–189. doi: 10.1146/annurev.micro.61.080706.093139. [DOI] [PubMed] [Google Scholar]
- Drake HL, Küsel K, Matthies C.2006Acetogenic prokaryotesIn: Balows A, Trüper HG, Dworkin M, Harder W and Schleifer KH (eds).The Prokaryotes3rd edn.Springer: New York; 354–420. [Google Scholar]
- Edwards CA, Bohlen PJ.1996Biology and Ecology of Earthworms3rd edn.Chapman & Hall: London; 426pp. [Google Scholar]
- Euzéby JP.2010List of prokaryotic names with standing nomenclature http://www.bacterio.cict.fr/number.html .
- Felsenstein J. Confidence limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fischer K, Hahn D, Honerlage W, Zeyer J. Effect of passage through the gut of the earthworm Lumbricus terrestris L on Bacillus megaterium studied by whole cell hybridization. Soil Biol Biochem. 1997;29:1149–1152. [Google Scholar]
- Francis CA, Obraztsova AY, Tebo BM. Dissimilatory metal reduction by the facultative anaerobe Pantoea agglomerans SP1. Appl Environ Microbiol. 2000;66:543–548. doi: 10.1128/aem.66.2.543-548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong MA, Singleton DR, Coleman DC, Whitman WB. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl Environ Microbiol. 2002;68:1265–1279. doi: 10.1128/AEM.68.3.1265-1279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good IJ. The population frequency of species and the estimation of the population parameters. Biometrics. 1958;40:237–246. [Google Scholar]
- Gottschalk G.1988Bacterial metabolism2nd edn.Springer: New York [Google Scholar]
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol. 2000;66:5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont PAD, Grimont F.2005Genus XXIII. PantoeaIn: Brenner DJ, Krieg NR and Staley JT (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York; 713–720. [Google Scholar]
- Hamberger A, Horn MA, Dumont MG, Murrell JC, Drake HL. Anaerobic consumers of monosaccharides in a moderately acidic fen. Appl Environ Microbiol. 2008;74:3112–3120. doi: 10.1128/AEM.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes WP, Hertel C.2009Genus I. LactobacillusIn: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA et al. (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York; 465–511. [Google Scholar]
- Hartenstein F, Hartenstein E, Hartenstein R. Gut load and transit time in the earthworm Eisenia foetida. Pedobiologia. 1981;22:5–20. [Google Scholar]
- Hartenstein R, Amico L. Production and carrying-capacity for the earthworm Lumbricus terrestris in culture. Soil Biol Biochem. 1983;15:51–54. [Google Scholar]
- Hedderich R, Whitman WB.2006Physiology and biochemistry of the methane-producing ArchaeaIn: Dworkin MM, Falkow S, Rosenberg E, Schleifer KH and Stackebrandt E (eds).The Prokaryotes3rd edn.Springer: New York; 1050–1079. [Google Scholar]
- Horn MA, Ihssen J, Matthies C, Schramm A, Acker G, Drake HL. Dechloromonas denitrificans sp. nov., Flavobacterium denitrificans sp. nov., Paenibacillus anaericanus sp. nov. and Paenibacillus terrae strain MH72, N2O-producing bacteria isolated from the gut of the earthworm Aporrectodea caliginosa. Int J Syst Evol Microbiol. 2005;55:1255–1265. doi: 10.1099/ijs.0.63484-0. [DOI] [PubMed] [Google Scholar]
- Horn MA, Schramm A, Drake HL. The earthworm gut: an ideal habitat for ingested N2O-producing microorganisms. Appl Environ Microbiol. 2003;69:1662–1669. doi: 10.1128/AEM.69.3.1662-1669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert SH. Nonconcept of species diversity—critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- Ihssen J, Horn MA, Matthies C, Gößner A, Schramm A, Drake HL. N2O-producing microorganisms in the gut of the earthworm Aporrectodea caliginosa are indicative of ingested soil bacteria. Appl Environ Microbiol. 2003;69:1655–1661. doi: 10.1128/AEM.69.3.1655-1661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA, Janssen PH. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microbiol. 2003;69:7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldorf M, Linne von Berg KH, Meier U, Servos U, Bothe H. The reduction of nitrous oxide to dinitrogen by Escherichia coli. Arch Microbiol. 1993;160:432–439. doi: 10.1007/BF00245303. [DOI] [PubMed] [Google Scholar]
- Karsten GR, Drake HL. Comparative assessment of the aerobic and anaerobic microfloras of earthworm guts and forest soils. Appl Environ Microbiol. 1995;61:1039–1044. doi: 10.1128/aem.61.3.1039-1044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten GR, Drake HL. Denitrifying bacteria in the earthworm gastrointestinal tract and in vivo emission of nitrous oxide (N2O) by earthworms. Appl Environ Microbiol. 1997;63:1878–1882. doi: 10.1128/aem.63.5.1878-1882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp BA, Podmirseg SM, Seeber J, Meyer E, Insam H. Diet-related composition of the gut microbiota of Lumbricus rubellus as revealed by a molecular fingerprinting technique and cloning. Soil Biol Biochem. 2009;41:2299–2307. [Google Scholar]
- Küsel K, Drake HL. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl Environ Microbiol. 1995;61:3667–3675. doi: 10.1128/aem.61.10.3667-3675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E and Goodfellow M (eds).Nucleic acid techniques in bacterial systematics John Wiley & Sons Ltd.: Chichester, England; 205–248. [Google Scholar]
- Lavelle P, Bignell D, Lepage M, Wolters V, Roger P, Ineson P, et al. Soil function in a changing world: the role of invertebrate ecosystem engineers. Eur J Soil Biol. 1997;33:159–193. [Google Scholar]
- Lee KE.1985Earthworms. Their Ecology and Relationships with Soils and Land Use Academic Press: Sydney; 411pp. [Google Scholar]
- Liu Y, Whitman WB.2008Metabolic, phylogenetic, and ecological diversity of the methanogenic ArchaeaIn: Wiegel J, Maier RJ and Adams MWW (eds).Incredible Anaerobes: From Physiology to Genomics to Fuels 171–189. [DOI] [PubMed]
- Lüders T, Manefield M, Friedrich MW. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol. 2004;6:73–78. doi: 10.1046/j.1462-2920.2003.00536.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Schleifer KH, Whitman WB.2009Revised road map to the phylum FirmicutesIn: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA et al. (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York; 1–15. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- Mahmoud DAR, Mahmoud AA, Gomaa AM. Antagonistic activities of potato associated bacteria via their production of hydrolytic enzymes with special reference to pectinases. Res J Agric Biol Sci. 2008;4:575–584. [Google Scholar]
- Makeschin F.1997Earthworms (Lumbricidae: Oligochaeta): important promoters of soil development and soil fertilityIn: Benckiser G (ed).Fauna in soil ecosystems Marcel Dekker Inc.: New York; 173–223. [Google Scholar]
- Martin-Carnahan A, Joseph SW.2005Genus I. AeromonasIn: Brenner DJ, Krieg NR, Staley JT (eds). Bergey's Manual of Systematic Bacteriology, 2nd edn. Springer: New York; 557–578. [Google Scholar]
- Martin A, Cortez J, Barois I, Lavelle P. The production of intestinal mucus by earthworms—a key process in their interactions with the soil microflora. Rev Ecol Biol Sol. 1987;24:549–558. [Google Scholar]
- Matthies C, Gossner A, Acker G, Schramm A, Drake HL. Lactovum miscens gen. nov., sp. nov., an aerotolerant, psychrotolerant, mixed-fermentative anaerobe from acidic forest soil. Res Microbiol. 2004;155:847–854. doi: 10.1016/j.resmic.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Matthies C, Griesshammer A, Schmittroth M, Drake HL. Evidence for involvement of gut-associated denitrifying bacteria in emission of nitrous oxide (N2O) by earthworms obtained from garden and forest soils. Appl Environ Microbiol. 1999;65:3599–3604. doi: 10.1128/aem.65.8.3599-3604.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B, et al. 2008Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolismIn: Wiegel J, Maier RJ and Adams MWW (eds).Incredible Anaerobes: From Physiology to Genomics to Fuels New York Academy of Sciences: Boston; 58–72. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Meth Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Müller HE, Brenner DJ, Fanning GR, Grimont PA, Kämpfer P. Emended description of Buttiauxella agrestis with recognition of six new species of Buttiauxella and two new species of Kluyvera: Buttiauxella ferragutiae sp. nov., Buttiauxella gaviniae sp. nov., Buttiauxella brennerae sp. nov., Buttiauxella izardii sp. nov., Buttiauxella noackiae sp. nov., Buttiauxella warmboldiae sp. nov., Kluyvera cochleae sp. nov., and Kluyvera georgiana sp. nov. Int J Syst Bacteriol. 1996;46:50–63. doi: 10.1099/00207713-46-1-50. [DOI] [PubMed] [Google Scholar]
- Nanninga HJ, Gottschal JC.1985Amino acid fermentation and hydrogen transfer in mixed cultures FEMS Microbiol Ecol 31261–269.10719208 [Google Scholar]
- Neufeld JD, Dumont MG, Vohra J, Murrell JC. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb Ecol. 2007;53:435–442. doi: 10.1007/s00248-006-9125-x. [DOI] [PubMed] [Google Scholar]
- Priest FG.2009Genus I. PaenibacillusIn: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA et al. (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York; 269–295. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahemtulla F, Lovtrup S. Comparative biochemistry of invertebrate mucopolysaccharides .3. Oligochaeta and Hirudinea. Comp Biochem Physiol B, Biochem Mol Biol. 1975;50:627–629. doi: 10.1016/0305-0491(75)90101-7. [DOI] [PubMed] [Google Scholar]
- Rainey FA, Hollen BJ, Small A.2009Genus I. ClostridiumIn: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA et al (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York [Google Scholar]
- Reid SJ.2005Genetic organization and regulation of hexose and pentose utilization in the ClostridiaIn: Dürre P (ed).Handbook on Clostridia CRC Press: Boca Raton; 133–153. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schellenberger S, Kolb S, Drake HL. Metabolic responses of novel cellulolytic and saccharolytic agricultural soil Bacteria to oxygen. Environ Microbiol. 2010;12:845–861. doi: 10.1111/j.1462-2920.2009.02128.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Larget BR, Handelsman J. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol. 2004;70:5485–5492. doi: 10.1128/AEM.70.9.5485-5492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Hendrix PF, Coleman DC, Whitman WB. Identification of uncultured bacteria tightly associated with the intestine of the earthworm Lumbricus rubellus (Lumbricidae; Oligochaeta) Soil Biol Biochem. 2003;35:1547–1555. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Teuber M, Geis A, Neve H.1992The genus LactococcusIn: Balows A, Trüper HG, Dworkin MM, Harder W and Schleifer KH (eds).The Prokaryotes2nd edn.Springer: New York; 1482–1501. [Google Scholar]
- Tiedje JM.1988Ecology of denitrification and dissimilatory nitrate reduction to ammoniumIn: Zehnder AJB (ed).Biology of Anaerobic Microorganisms John Wiley and Sons: New York; 179–243. [Google Scholar]
- Trigo D, Barois I, Garvin MH, Huerta E, Irisson S, Lavelle P. Mutualism between earthworms and soil microflora. Pedobiologia. 1999;43:866–873. [Google Scholar]
- Umarov MM.1990Biotic sources of nitrous oxide in the context of the global budget of nitrous oxideIn: Bouwman AF (ed).Soils and the Greenhouse Effect John Wiley and Sons: Chichester; 263–268. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley RA, Hardie JM.2009Genus I. StreptococcusIn: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA et al (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York; 655–711. [Google Scholar]
- Wiegel J, Tanner R, Rainey FA.2006An introduction to the family ClostridiaceaeIn: Balows A, Trüper HG, Dworkin M, Harder W and Schleifer KH (eds).The Prokaryotes Springer: New York; 654–678. [Google Scholar]
- Wüst PK, Horn MA, Drake HL. In situ hydrogen and nitrous oxide as indicators of concomitant fermentation and denitrification in the alimentary canal of the earthworm Lumbricus terrestris. Appl Environ Microbiol. 2009a;75:1852–1859. doi: 10.1128/AEM.02745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst PK, Horn MA, Henderson G, Janssen PH, Rehm BHA, Drake HL. Gut-associated denitrification and in vivo emission of nitrous oxide by the earthworm families Megascolecidae and Lumbricidae in New Zealand. Appl Environ Microbiol. 2009b;75:3430–3436. doi: 10.1128/AEM.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Richter M, Peplies J, Euzéby J, Amann R, Schleifer KH, et al. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]