Abstract
Salmonella enterica Typhimurium remains undigested in the food vacuoles of the common protist, Tetrahymena. Contrary to its interaction with Acanthamoeba spp., S. Typhimurium is not cytotoxic to Tetrahymena and is egested as viable cells in its fecal pellets. Through microarray gene expression profiling we investigated the factors in S. Typhimurium that are involved in its resistance to digestion by Tetrahymena. The transcriptome of S. Typhimurium in Tetrahymena phagosomes showed that 989 and 1282 genes were altered in expression compared with that in water and in LB culture medium, respectively. A great proportion of the upregulated genes have a role in anaerobic metabolism and the use of alternate electron acceptors. Many genes required for survival and replication within macrophages and human epithelial cells also had increased expression in Tetrahymena, including mgtC, one of the most highly induced genes in all three cells types. A ΔmgtC mutant of S. Typhimurium did not show decreased viability in Tetrahymena, but paradoxically, was egested at a higher cell density than the wild type. The expression of adiA and adiY, which are involved in arginine-dependent acid resistance, also was increased in the protozoan phagosome. A ΔadiAY mutant had lower viability after passage through Tetrahymena, and a higher proportion of S. Typhimurium wild-type cells within pellets remained viable after exposure to pH 3.4 as compared with uningested cells. Our results provide evidence that acid resistance has a role in the resistance of Salmonella to digestion by Tetrahymena and that passage through the protist confers physiological advantages relevant to its contamination cycle.
Keywords: enteric pathogen, grazing, human pathogen, protozoa, resistance, vacuole
Introduction
Grazing by phagocytic protozoa is a major factor in shaping bacterial populations in aquatic, soil, and anthropogenic ecosystems (Pace, 1988; Barker and Brown, 1994). Bacteria that resist grazing by protozoa may show increased environmental fitness (Hahn and Hofle, 2001). Grazing resistance may occur through pre-ingestional adaptations involving development of oversized cells, surface masking or microcolony formation, and through post-ingestional adaptations that include development of toxin release, digestional resistance and/or intracellular growth (Matz and Kjelleberg, 2005). Indeed, the intracellular pathogens Mycobacterium avium, Chlamydia pneumoniae, Listeria monocytogenes and Legionella pneumophila replicate in the digestive vacuoles (phagosomes) of Acanthamoeba castellanii (Ly and Muller, 1990; Cirillo et al., 1997; Essig et al., 1997; Abu Kwaik et al., 1998) whereas Salmonella enterica serovars Dublin and Typhimurium multiply in Acanthamoeba rhysodes and Acanthamoeba polyphaga (Gaze et al., 2003; Tezcan-Merdol et al., 2004).
The resistance of certain intracellular pathogens to digestion by protozoa may coincidentally facilitate their ability to cause disease in their eukaryotic hosts. Phagocytosis in free-living protozoa shares basic mechanisms with that in human phagocytic cells, and conditions within the protozoan food vacuoles overlap with those in the macrophage phagosome (Lock et al., 1987; Jacobs et al., 2006; Cosson and Soldati, 2008). The interaction between pathogens and predatory amoebae has been implicated in the maintenance of bacterial virulence in various pathogens (Molmeret et al., 2005). For example, L. pneumophila and M. avium both show increased infectivity of human macrophages after passage through A. castellanii (Cirillo et al., 1994, 1997). On the basis of similarities in L. pneumophila mechanisms of infection of amoebae and mammalian cells, Molmeret et al. (2005) suggested that the intracellular lifestyle of human pathogens may have resulted from their adaptation to replication within free-living amoebae.
We have reported previously that S. Typhimurium can survive digestion by Tetrahymena at high rates and is released as viable cells in its fecal pellets in which it has enhanced survival compared with cells remaining undigested and free in suspension (Brandl et al., 2005). This is in contrast with L. monocytogenes, which is digested by the protist and detected infrequently in its fecal pellets (Brandl et al., 2005; Gourabathini et al., 2008). In addition, S. Typhimurium does not decrease the viability of Tetrahymena during its intravacuolar passage (Gourabathini et al., 2008). Therefore, its interaction appears to be different than with Acanthamoeba spp., which causes death of the protist (Gaze et al., 2003; Tezcan-Merdol et al., 2004; Feng et al., 2009). S. Typhimurium is an intracellular pathogen that has evolved specific mechanisms for its persistence and replication in eukaryotic cells. It is currently unclear whether the ability of the pathogen to resist digestion by Tetrahymena involves the same adaptations used to survive the phagocytic process of other eukaryotic cells.
In this study, we investigated the interaction of S. Typhimurium with Tetrahymena by microarray analysis of gene expression in S. Typhimurium cells residing in the Tetrahymena digestive vacuole. The global transcriptional response of this human pathogen to the Tetrahymena vacuolar environment indicates that it experiences conditions in the protozoan phagosome that overlap with those in macrophages and epithelial cells. In addition, cell viability assays showed that S. Typhimurium requires acid stress tolerance for survival to phagocytosis by Tetrahymena.
Materials and methods
Strains, plasmids and culture conditions
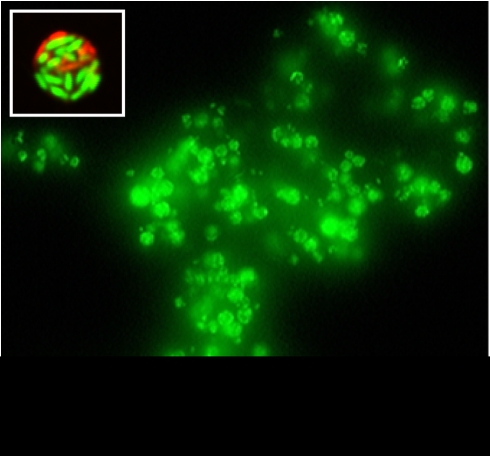
All strains and plasmids used in this study are listed in Table 1. Tetrahymena sp. MB125 was isolated from soil in California, belongs to an unknown species of this genus and has been described previously (Brandl et al., 2005). It was used in this study because of its release of numerous large fecal pellets containing viable S. enterica cells upon grazing on this pathogen (Figure 1).
Table 1. Strains and plasmids used in this study.
| Strain name | Description | Reference |
|---|---|---|
| MB125 | Tetrahymena spp. isolated from wet soil in California | Brandl et al., 2005 |
| MB282 | S. enterica serovar Typhimurium SL1344 | Hoiseth and Stocker, 1981 |
| MB676 | SL1344-derived; mgtC ORF replaced with CAT cassette | This study |
| MB681 | SL1344-derived; adiA and adiY ORFs replaced with CAT cassette | This study |
| MB692 | MB676-derived; complemented with pMTB687 | This study |
| MB694 | MB681-derived; complemented with pMTB688 | This study |
| Plasmid | ||
| pKD3 | Used in lambda Red-mediated recombination; contains CAT cassette | Datsenko and Wanner, 2000 |
| pKD46 | Lambda-Red recombinase expression plasmid | Datsenko and Wanner, 2000 |
| pBBR1MCS-5 | Broad-range cloning vector; gentR | Kovach et al., 1995 |
| pMTB687 | pBBR1MCS-5 ligated with mgtC ORF | This study |
| pMTB688 | pBBR1MCS-5 ligated with adiA–adiY ORFs | This study |
Figure 1.
Epifluorescence micrograph of a large aggregate of fecal pellets released by Tetrahymena sp. upon feeding on S. Typhimurium and stained with SYTO 9 (Invitrogen). The inset shows a single optical scan through a Live/Dead BacLight-stained fecal pellet containing live (green) and dead (red) S. Typhimurium cells. The micrograph was captured with a Leica SP5 AOTF confocal microscope (Leica Microsystems, Wetzlar, Germany).
S. enterica serovar Typhimurium SL1344 strains MB676 and MB681 are derivatives of SL1344 with a deletion in mgtC and adiAY, respectively. Mutants MB676 and MB681 were created using the lambda Red recombinase system (Datsenko and Wanner, 2000) to replace mgtC and adiAY, respectively, with the chloramphenicol acetyltransferase (CAT) cassette. All primers used for this procedure and others in this study are listed in Supplementary Table S1. The mgtC and adiAY deletions were confirmed by PCR. Complemented mutant strains MB692 and MB694 were constructed by transforming MB676 and MB681 with the stably maintained plasmid pBBR1MCS-5 (Kovach et al., 1995) containing mgtC or adiAY, respectively. Briefly, mgtC or adiAY were PCR-cloned from strain SL1344 into pBBR1MCS-5 using primers KpnI+mgtC_F/PstI+mgtC_R or KpnI+adiA_F/PstI+adiY_R. Plasmid pBBR1MCS-5 containing either mgtC (pMTB687) or adiAY (pMTB688) was electroporated into mutant MB676 or MB681, respectively. Transformants were isolated on LB agar containing gentamycin (15 μg ml−1) and X-gal. S. enterica SL1344 and derivatives were cultured in LB broth containing streptomycin (30 μg ml−1), and chloramphenicol (20 μg ml−1) and gentamycin (15 μg ml−1), as appropriate.
Co-culture conditions
Tetrahymena MB125 was grown in 2/3 strength Plate Count Broth (2/3 PCB) (Becton Dickinson, Franklin Lakes, NJ, USA) for 2 days at 28 °C, with agitation at 50 r.p.m. It was then centrifuged at 200 × g for 2 min and repeatedly washed with half volumes of sterile deionized H2O (dH2O), incubating the cells for 15 min during each wash to minimize lysis by osmotic shock followed by centrifugation. The cell concentration was measured with a hemacytometer and was adjusted to 1 × 105 cells ml−1. SL1344 cells were cultured to the mid-log phase of growth in LB broth at 28 °C and washed twice in sterile dH2O. The OD600 of the suspension was adjusted to 0.2 and the suspension was combined with that of washed Tetrahymena cells at a bacteria:ciliate ratio of 1000:1 in sterile dH2O. The mixed suspensions were incubated at 28 °C and 50 r.p.m. for 3 h until most bacteria were ingested by Tetrahymena. This was determined by staining the suspension with SYTO 9 (Invitrogen, Carlsbad, CA, USA) (10 μ final) for 30 min at 23 °C in the dark and visualized by epifluorescence microscopy with a Leica DMR microscope.
RNA extraction
Eight co-cultures of 50 ml were centrifuged at 800 × g for 2 min. The resulting pellet was enriched for Tetrahymena cells containing ingested SL1344 cells as most uningested free bacteria were not pelleted and remained in suspension. The supernatant was quickly removed and the pellet was resuspended in ice-cold lysis buffer (0.5% SDS, 19% ethanol and 1% phenol in H2O), on the basis of the protocol by Eriksson et al. (2003). The suspension containing lysed Tetrahymena cells was centrifuged at 3200 × g for 5 min at 4 °C. The bacterial pellet was stored at −80 °C. RNA extraction was performed with the Promega SV Total RNA Isolation kit per the manufacturer's specifications, except that bacterial pellets were first treated with 50 μg ml−1 of lysozyme (Fisher Scientific, Pittsburgh, PA, USA). RNA from two different sources was used as control. The first control RNA was isolated from 50 ml of SL1344 cells incubated in sterile dH2O (OD600=0.2) for 3 h, similar to the SL1344 cells mixed with Tetrahymena. The second control RNA was isolated from 20 ml of SL1344 cells grown to the mid-log phase of growth in LB broth. Both suspensions were centrifuged at 3200 × g for 5 min at 4 °C, followed by addition of lysis buffer, centrifugation and RNA extraction, as described above. Three biological replicates were used for each type of suspension. RNA integrity was assessed with the Agilent 2100 Bioanalyzer and only RNA of high quality was used for microarray hybridizations.
Microarray analysis
Custom, whole-genome S. enterica LT2 arrays were prepared at the Western Regional Research Center. Each array contained 4360 PCR-generated open-reading frames (including 104 from virulence plasmid pSLT) from S. enterica serovar Typhimurium strain LT2 printed onto Ultra-GAPS glass slides (Corning, Corning, NY, USA) with a Gene Machine Omnigrid Accent Arrayer (Genomic Solutions, Ann Arbor, MI, USA). Labeled nucleotides were prepared and hybridized to arrays as described previously (Kyle et al., 2010) on the basis of the method developed by Eriksson et al. (2003), with a few modifications. Briefly, 20 μg of RNA from co-cultures and controls was reverse-transcribed and Cy3-dCTP (GE Healthcare, Waukesha, WI, USA) incorporated into cDNA using the Fairplay III Microarray Labeling Kit (Stratagene, La Jolla, CA, USA). A 2-μg volume of SL1344 genomic DNA was labeled with Cy5-dCTP (GE Healthcare) using Klenow (New England Biolabs, Ipswich, MA, USA) and served as the reference signal on the arrays. Labeling efficiency was determined with a Nanodrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA). All cDNA solutions were adjusted to the same label concentration in the Pronto! cDNA hybridization solution (Corning) and were hybridized to the arrays overnight at 42 °C according to the manufacturer's protocol. For each experimental condition, cDNA was prepared from three replicate suspensions (biological replicates) and each cDNA was hybridized to three replicate arrays (technical replicates) at random among several slides. The slides were washed and then scanned with an Axon Genepix 4000b scanner and the Axon Genepix Pro 4.1 software (Molecular Devices, Sunnyvale, CA, USA).
To address spatial dye effects or any disparity in the amounts of spotted cDNA, data were normalized by setting the median log (Cy3/Cy5 signals) of all of the spots on the same array to zero. Biological replicates were then analyzed by analysis of variance (P<0.05) with the Genespring version 7 software (Agilent Technologies, Santa Clara, CA, USA). From these data sets, only genes that were differentially regulated at least two-fold and for which the mean change in expression passed the Benjamini–Hochberg False Discovery Rate test (P<0.05) were included in gene lists. Gene lists were generated for comparison of expression in SL1344 cells in Tetrahymena phagosomes with that in (1) H2O and (2) LB broth, and for comparison of expression in H2O versus that in LB broth. All final gene lists are presented as Supplementary Table S2. Genes of particular interest were those that were differentially regulated in S. Typhimurium in Tetrahymena vacuoles versus both in H2O and in LB broth.
Determination of cell viability and population density in fecal pellets
After co-incubation of SL1344 and Tetrahymena MB125 as described above, the suspension was filtered through Millipore black Isopore membranes (0.2 μm pore size, 25 mm diameter) and cell viability assessed as described previously (Brandl et al., 2005). Briefly, filters were gently submerged in H2O to allow Tetrahymena cells to swim away from the filter leaving behind free bacteria and pellets. The filters were removed from the water and drained. For assessment of bacterial acid resistance, the filters were submerged again in H2O at pH 3.4 for 1 h. The filters were rinsed by submersion in H2O five times and drained.
Viability staining of cells on the filters was performed with Live/Dead BacLight (Invitrogen) by placing the filter into the stain solution (1.5 μl of each dye per ml) for 25 min at 24 °C in the dark. The filters were rinsed by submersion in H2O and cells were viewed under an epifluorescence microscope with a fluorescein filter. The ratio of dead cells (red fluorescence) to viable cells (green fluorescence) was estimated in at least 50 fecal pellets in each of three replicate co-cultures. This ratio was estimated also for 10–30 cells external to each fecal pellet to assess the proportion of viable cells among cells remaining uningested and free in the mixed suspension. Figure 1 shows pellets produced by Tetrahymena and the presence of live and dead S. Typhimurium cells in the pellets, as shown by Live/Dead BacLight stain.
The viability of the ΔadiAY and ΔmgtC mutants, of the complemented mutants and of the wild-type SL1344 in fecal pellets was assessed as described above. Cell density in pellets was estimated for the ΔmgtC mutant, the complemented mutant and the wild-type strain by counting the total cell number per pellet for at least 50 pellets. For strain comparisons, an aliquot of the bacterial inoculum was stained with Live/Dead BacLight to ensure that viability did not differ among strains before preparation of the co-cultures with Tetrahymena. Cell viability and density data were analyzed statistically with a two-tailed t-test or a one-way analysis of variance followed by Tukey's multiple comparison test, with P<0.05. All tests were performed with the Prism software version 5.02 (GraphPad Software, La Jolla, CA, USA). Experiments were performed at least twice.
Results
Transcriptional profile of S. Typhimurium in Tetrahymena phagosomes
The gene expression profile of S. Typhimurium SL1344 residing in Tetrahymena food vacuoles was compared with that of cells incubated in H2O or in LB broth. Because S. Typhimurium cells were harvested from the food vacuoles 3 h after the start of co-incubation of the two microorganisms in water, it is likely that a considerable percentage of pathogen cells spent a significant amount of time in water before falling prey to the protist. Therefore, we considered incubation of S. Typhimurium in water without the protist as the most appropriate control to identify genes differentially regulated in Tetrahymena phagosomes. However, because microarray studies of the S. Typhimurium transcriptome in other eukaryotic cells used LB broth as a control (Eriksson et al., 2003; Hautefort et al, 2008), we included LB broth as an additional control to compare our results with previously published data. Our microarray analysis showed that expression of 989 and 1282 genes changed at least two-fold in the Tetrahymena phagosome compared with in water and in LB medium, respectively (Supplementary Table S2). Overall, more genes were downregulated (520 and 811) than upregulated (469 and 471) in the Tetrahymena phagosome versus in H2O and in LB broth, respectively (Figure 2 and Supplementary Table S2). Notable exceptions, as illustrated by Categories of Orthologous Genes (COG) categories with H2O as the control environment, include upregulated genes involved in energy production and conversion, nucleotide transport and metabolism, translation, cell motility and intracellular trafficking and secretion (Figure 2). Also of interest is the higher number of upregulated than downregulated genes that have a role in replication, recombination and repair, and in cell wall/membrane biogenesis. A close examination of the detailed gene lists showed that phagosome conditions induced numerous genes involved in anerobiosis. These included the hydrogenase operons hyc, hyd and hyp, and the reductase genes dmsAB and frdBA (Table 2).
Figure 2.
Differential expression of genes within Categories of Orthologous Genes (COG) in S. Typhimurium cells residing in Tetrahymena vacuoles compared with that in H2O, as shown by microarray analysis. The bars represent the percentage of genes with a change in transcription of at least two-fold within a given category.
Table 2. Select categories of S. Typhimurium genes upregulated at least two-fold in the Tetrahymena phagosome compared with that in water and in LB broth, as determined by microarray analysis.
| Locus | Name |
Fold change |
Function | |
|---|---|---|---|---|
| vs H2O | vs LB | |||
| Anaerobic energy generation/alternate electron acceptors | ||||
| STM0964 | dmsA | 17.87 | 14.40 | Anaerobic DMSO reductase subunit-A |
| STM0965 | dmsB | 11.79 | 18.94 | Anaerobic DMSO reductase subunit-B |
| STM1538 | hydA | 2.36 | 4.31 | Putative Ni-Fe hydrogenase-1 large subunit |
| STM1539 | hydB | 3.16 | 6.22 | Putative Ni-Fe hydrogenase-1 small subunit |
| STM2063 | phsC | 2.33 | 3.50 | H2S production from thiosulfate |
| STM2065 | phsA | 4.78 | 5.46 | H2S production from thiosulfate |
| STM2529 | 7.27 | 7.14 | Putative anaerobic DMSO (dimethylsulfoxide) reductase | |
| STM2530 | 8.69 | 8.51 | Putative anaerobic DMSO reductase | |
| STM2843 | hydN | 5.30 | 6.85 | Formate dehydrogenase-H, [4Fe-4S] ferredoxin subunit |
| STM2845 | hycI | 5.07 | 5.80 | Protease involved in processing the C-terminal end of HycE |
| STM2846 | hycH | 6.62 | 5.59 | Processing of HycE (part of the formate–hydrogen–lyase (FHL) complex) |
| STM2848 | hycF | 5.66 | 7.87 | Hydrogenase-3, putative quinone oxidoreductase |
| STM2850 | hycD | 3.42 | 5.64 | Hydrogenase-3, membrane subunit (part of FHL complex) |
| STM2852 | hycB | 4.86 | 7.62 | Hydrogenase-3, Fe-S subunit (part of FHL complex) |
| STM2853 | hycA | 3.78 | 6.13 | Transcriptional repressor of hyc and hyp operons |
| STM2854 | hypA | 5.57 | Functions as nickel donor for HycE of hydrogenlyase-3 in FHL complex | |
| STM2855 | hypB | 8.84 | 2.79 | Hydrogenase-3 accessory protein, assembly of metallocenter |
| STM2856 | hypC | 3.93 | 2.80 | Putative hydrogenase expression/formation protein |
| STM2857 | hypD | 8.62 | 3.09 | Putative hydrogenase expression/formation protein |
| STM2858 | hypE | 3.77 | 2.98 | Putative hydrogenase expression/formation protein |
| STM2859 | fhlA | 2.69 | 2.55 | Formate hydrogenlyase transcriptional activator for fdhF, hyc and hyp operons |
| STM3143 | hybG | 2.20 | 2.25 | Hydrogenase-2 operon protein |
| STM3144 | hybF | 3.81 | 2.84 | Putative hydrogenase-2 expression/formation protein |
| STM3145 | hybE | 3.28 | 2.64 | Hydrogenase-2 operon protein |
| STM3146 | hybD | 4.70 | 3.52 | Putative processing element for hydrogenase-2 |
| STM3147 | hybC | 4.64 | 2.95 | Hydrogenase-2, large subunit |
| STM3148 | hybB | 8.96 | 5.20 | Putative cytochrome Ni/Fe component of hydrogenase-2 |
| STM3149 | hybA | 9.29 | 5.92 | Putative hydrogenase operon protein |
| STM3150 | hypO | 11.29 | 4.61 | Hydrogenase-2, small subunit |
| STM4285 | fdhF | 17.52 | 14.14 | Formate dehydrogenase |
| STM4342 | frdB | 2.54 | 3.18 | Fumarate reductase, Fe-S subunit |
| STM4343 | frdA | 3.20 | Fumarate reductase, flavoprotein subunit | |
| Virulence and antimicrobial resistance genes | ||||
| Virulence plasmid | ||||
| pSLT012 | orf7 | 4.07 | 3.43 | Putative bacterial regulatory protein, luxR family |
| pSLT013 | pefI | 5.25 | 6.94 | Transcriptional regulator of pef operon |
| pSTL096 | trbE | 2.29 | 2.16 | Conjugative transfer |
| PhoP–PhoQ-activated genes | ||||
| STM0628 | pagP | 3.56 | Lipid-A palmitoyl transferase required for resistance to antimicrobial peptides | |
| STM1242 | envE | 2.41 | Putative envelope protein | |
| STM1246 | pagC | 2.73 | 2.16 | Putative envelope protein required for survival in macrophages |
| STM1254 | 4.37 | 5.51 | Putative outer membrane lipoprotein | |
| STM1867 | pagK | 2.23 | 2.47 | Putative virulence protein |
| STM2781 | virK | 6.67 | Putative virulence protein | |
| STM2782 | mig-14 | 6.94 | Required for virulence and resistance to antimicrobial peptides | |
| STM3763 | mgtB | 3.32 | 4.46 | Mg2+-transporting ATPase |
| STM3764 | mgtC | 81.98 | 73.67 | Putative ion homeostasis protein required for persistence in macrophages |
| Other virulence genes | ||||
| STM1091 | sopB | 2.02 | Inositol polyphosphatase required for entry into intestinal epithelial cells | |
| STM1414 | ssaV | 3.87 | 5.66 | Type-III secretion system apparatus protein |
| STM1602 | sifB | 2.84 | 3.08 | Secreted effector |
| Antimicrobial resistance | ||||
| STM1518 | marB | 2.33 | Multiple antibiotic resistance protein | |
| STM1519 | marA | 5.76 | Transcriptional activator of defense systems, multiple antibiotic resistance protein | |
| STM1520 | marR | 8.58 | 3.89 | Transcriptional repressor of mar operon, multiple antibiotic resistance protein |
| STM1571 | yddG | 4.17 | 3.73 | Required for efflux of methyl viologen, a quaternary ammonium compound |
| STM2814 | emrA | 5.06 | Multidrug resistance protein | |
| Osmotic stress | ||||
| STM0705 | kdpB | 5.15 | 9.31 | P-type ATPase, high-affinity potassium transport system, B-chain |
| STM0706 | kdpA | 9.01 | 11.67 | P-type ATPase, high-affinity potassium transport system, A-chain |
| STM1732 | ompW | 9.99 | Porin involved in osmoregulation | |
| Acid stress | ||||
| STM4295 | adiY | 4.12 | 6.81 | Transcriptional activator of adiA (AraC/XylS family) |
| STM4296 | adiA | 6.20 | 5.71 | Arginine decarboxylase, catabolic; inducible by acid |
We also observed increased expression of various virulence genes, several of which are associated with the Salmonella-containing vacuole of the macrophage (Eriksson et al., 2003; Faucher et al., 2006). These included the PhoP/PhoQ-regulated genes pagC, pagK, envE, virK, mgtB and mgtC (Table 2). Additional Salmonella-containing vacuole-associated virulence genes upregulated in the Tetrahymena phagosome were those encoding the Salmonella Pathogenicity Island-2 (SPI-2) type-III secretion system apparatus protein SsaV and the secreted effector proteins SifB and SopB (Faucher et al., 2006) (Table 2).
Other genes with increased expression in Tetrahymena have a role in acid stress-response (adiAY) and in antibiotic or antimicrobial resistance (marRAB, emrA and yddG) (Table 2). yddG codes for a porin involved in the efflux of methyl viologen, which generates oxygen radicals (Santiviago et al., 2002). In addition, a small number of upregulated genes are involved in osmotic stress, namely kdpA and kpdB, and ompW. The kdp operon is induced upon osmotic upshift through loss of turgor (Balaji et al., 2005), whereas ompW encodes a porin involved in osmoregulation. However, there is also evidence that similar to YddG, OmpW exports methyl viologen and may work in conjunction with YddG (Gil et al., 2007).
When examining expression changes in S. Typhimurium in Tetrahymena phagosomes with LB broth as a control environment, we identified additional genes that are upregulated also in macrophages and epithelial cells. Supplementary Table S3 lists a subset of 146 and 92 genes that are upregulated in Tetrahymena versus in LB and which were reported previously to also increase in expression after 8 h in J774-A.1 macrophages and after 2 h in HeLa cells, respectively (Supplementary Table S1 in reference Hautefort et al., 2008). Commonalities in differentially regulated genes in the latter cells and in Tetrahymena were observed at other incubation times as well, but the number of overlapping upregulated genes was smaller. Of the genes that showed increased expression in both Tetrahymena and macrophages, 53% were upregulated also in HeLa cells (Supplementary Table S3). Common Tetrahymena- and macrophage-upregulated genes in comparison to LB included, but were not limited to, the following categories: SPI-2 (ssrA, ssaB/G/H/I/L/V/R and sscA); oxidative stress (ycfR, trxC and ibpB—in addition to dps, yfiA and katG, which are involved in oxidative stress response also but were not upregulated in macrophages); osmotic stress (osmB); SOS response (uvrB and umuC); multidrug resistance (emrD); phosphate starvation (psiF); Mg2+ transport (mgtB); anerobic metabolism (hydN, hycA and fhlA) and 49 hypothetical proteins (Supplementary Table S3). It is noteworthy that as observed in macrophages, the iron acquisition genes entABCE were highly downregulated in Tetrahymena. Along with downregulation of sitABCD, this suggests the presence of iron and manganese in the protozoan phagosome.
Passage through Tetrahymena induces an acid stress response
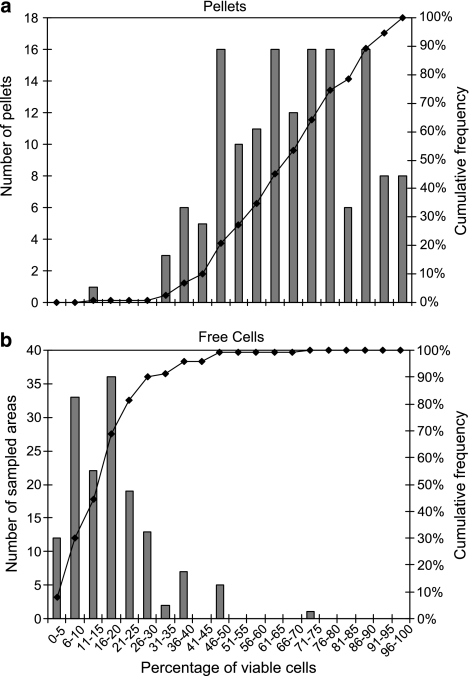
To determine whether the acidic conditions in Tetrahymena digestive vacuoles induce an adaptive tolerance to low pH in S. Typhimurium, we measured the viability of strain SL1344 cells in fecal pellets released by the protist and subsequently exposed to acidic conditions. The mean proportion of viable cells in pellets after acid treatment at pH 3.4 was 67.7% with a standard deviation (s.d.) of 1.47, and was significantly greater than that among cells remaining uningested and free in suspension (7.7% s.d., 0.95) (t-test, P<0.0001). The frequency distribution of percentage viability among the two cell types clearly illustrates a shift toward greater viability of SL1344 in the pellets as compared with that of free cells upon exposure of both to acid stress (Figure 3). Although the percentage of viable cells in individual pellets was variable after exposure to pH 3.4, with the exception of one outlier pellet, it did not reduce below 31% (Figure 3a). By contrast, the percentage viability among free cells after acid treatment was lower than 31% in 90% of the areas sampled on the filter (Figure 3b).
Figure 3.
Frequency distribution of percent viable S. Typhimurium cells in individual Tetrahymena fecal pellets and in sampled areas of free uningested cells after exposure to pH 3.4. Cell viability was assessed with the Live/Dead BacLight stain. The bars represent the number of pellets (a) and the number of sampled areas (for free cells) (b), with a proportion of viable S. Typhimurium cells in a given range. The data on the solid line, plotted against the right y-axis, represent the cumulative frequency of observations in each distribution.
Arginine-dependent acid resistance in S. Typhimurium within Tetrahymena phagosomes
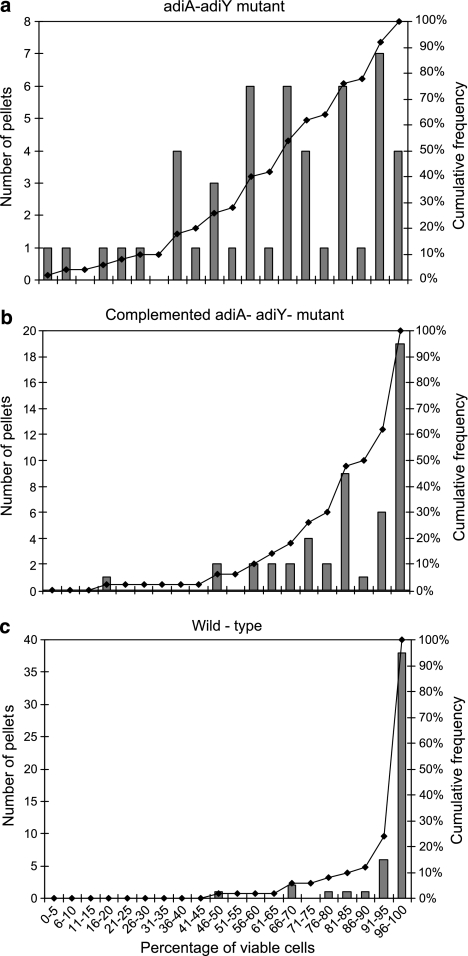
The viability of the SL1344 ΔadiAY mutant (MB681) and of its complemented strain (MB694) was compared to that of the wild-type strain in Tetrahymena fecal pellets. After a 3-h incubation of these strains with Tetrahymena, MB681 showed a mean percent viability in pellets of 66.03% (s.d., 3.53), versus 84.82% (s.d., 2.49) and 95.50% (s.d., 1.49) for MB694 and the wild-type strain, respectively. Although complete mortality of MB681 was not observed, MB681 had lower survival in the Tetrahymena phagosome than MB694 and the wild-type strain (Tukey's multiple comparison test, P<0.05). The percentage viable cells among free cells uningested by Tetrahymena did not differ significantly between the three strains (analysis of variance, P=0.59). The distribution of percentage viable MB681 cells in pellets was broad, ranging from 100% mortality in one pellet to over 96% viable cells in others (Figure 4a). By contrast, the proportion of viable cells of the complemented mutant (MB694) in the pellets did not reduce below 46% (with the exception of one outlier), and in half of the pellets the percentage of viable cells was greater than 85% (Figure 4b). The wild-type strain had the highest viability of all three strains, with 76% of the pellets containing at least 96% of viable cells (Figure 4c).
Figure 4.
Frequency distribution of percent viable S. Typhimurium cells in individual fecal pellets released by Tetrahymena during co-culture of the two microorganisms, as assessed with the Live/Dead BacLight stain. The number of individual pellets with a proportion of viable S. Typhimurium cells in a given range (bars) and the cumulative frequency of pellets across the range of percent viable cells (solid line, right y-axis) are illustrated for the S. Typhimurium ΔadiA ΔadiY mutant (MB681) (a), the complemented mutant (MB694) (b) and the wild-type strain (c).
Role of MgtC in S. Typhimurium cell density in Tetrahymena fecal pellets
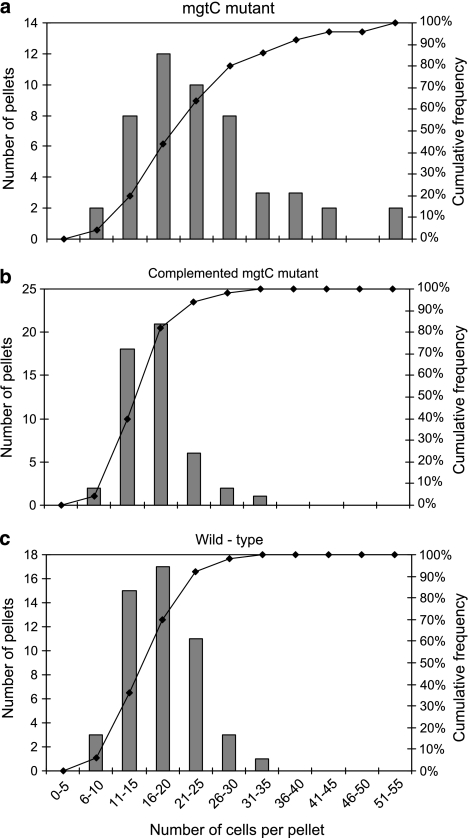
The macrophage virulence factor MgtC was investigated for its potential role in the digestion resistance of the pathogen. Whereas mgtB encodes an Mg2+ transporter, mgtC is involved in regulating membrane potential (Gunzel et al. 2006). With an increase in expression of 82- and 74-fold compared with that in water and LB broth, respectively, mgtC was one of the most highly induced genes in Tetrahymena (Table 2). The cell density and viability of the ΔmgtC mutant (MB676), its complemented strain (MB692) and the wild-type strain of SL1344 were compared in the fecal pellets produced by co-culture of each strain with Tetrahymena. Live/Dead staining of the cells in the fecal pellets showed that their viability did not differ between each strain (data not shown). However, the mean cell density per pellet for the mutant MB676 (24.3; s.d., 1.5) was significantly greater than that of the wild-type (17.8; s.d., 0.7) and the complemented mutant MB 692 (17.3; s.d., 0.7) (Tukey's multiple comparison test, P<0.05). Compared with the wild-type and the complemented mutant, the frequency distribution of the number of mutant MB676 cells in the pellets was shifted toward a higher cell density (Figure 5).
Figure 5.
Frequency distribution of the number of S. Typhimurium cells in individual fecal pellets released by Tetrahymena during co-culture of the two microorganisms, as assessed with the Live/Dead BacLight stain. The number of individual pellets with an S. Typhimurium cell density in a given range (bars) and the cumulative frequency of pellets across the range of cell density (solid line, right y-axis) are illustrated for the S. Typhimurium ΔmgtC mutant (MB676) (a), the complemented mutant (MB692) (b) and the wild-type strain (c).
Discussion
As intracellular pathogens have evolved to resist or escape phagosomal conditions, grazing protozoa may represent an environmental reservoir for these pathogens. Understanding the interaction between pathogenic bacteria and protozoan grazers may further our understanding of the factors that allow persistence of pathogens in the environment. We have reported previously that the intracellular pathogen, S. enterica, can remain undigested in Tetrahymena and that most cells remain viable upon their release in its fecal pellets (Brandl et al., 2005). The viability of cells within nascent fecal pellets is thus a direct result of survival within the Tetrahymena phagosome.
Bacteria that resist degradation in Tetrahymena phagosomes must counter harsh conditions such as acidification from proton-translocating ATPases, oxidative stress caused by reactive oxygen species, the presence of hydrolytic enzymes and reduced oxygen tension (Fok and Allen, 1975; Jacobs et al., 2006). Our global transcriptional analysis of S. Typhimurium SL1344 cells in Tetrahymena phagosomes showed extensive upregulation of the hyc, hyp, hyd and hyb hydrogenase operons, which function in anerobiosis (Vignais and Colbeau, 2004), thus, indicating a metabolic shift to an anaerobic lifestyle. Hyc and Hyp are involved in fermentative H2 evolution as part of the formate H2 lyase (FHL) complex, whereas Hyd and Hyb are linked to respiratory fumarate reduction (Richard et al., 1999; Zbell et al., 2007; Zbell and Maier, 2009). Deletion of hyd and hyb in S. Typhimurium results in colonization deficiency in a mouse model (Maier et al., 2004). Upregulation of the terminal reductases for fumurate and dimethylsulfoxide in the phagosome suggests that oxygen is highly limiting and alternate terminal electron acceptors may be preferred by S. Typhimurium. Overall, this metabolic adaptation, along with increased translation and expression of genes that function in cell replication and wall/membrane biogenesis, may be indicative of the ability of S. Typhimurium to grow in the Tetrahymena phagosome. Further evidence of such a phenomenon still needs to be obtained.
Several S. Typhimurium genes that are upregulated in Tetrahymena phagosomes as compared with that in H2O are also induced in macrophages. These included the antimicrobial resistance genes marRAB and emrA, of which increased expression was observed in the macrophage Salmonella-containing vacuole (Eriksson et al., 2003). Also noteworthy is the higher transcription of several virulence genes in the Tetrahymena phagosomes, including those of the PhoP–PhoQ regulon (pagP/C/K, envE, virK, mig-14 and mgtBC) and others belonging to SPI-1 (sopB) and SPI-2 (ssaV, sifB). The PhoP–PhoQ system is required for survival of S. Typhimurium and expression of SPI-2 genes within macrophages (Miller et al., 1989; Fass and Groisman, 2009). PagP is involved in remodeling of the lipid-A domain of lipopolysaccharide (Bishop, 2005), whereas Mig-14 and VirK promote resistance to antimicrobial peptides produced in macrophages (Brodsky et al., 2005). Low amounts of Ca2+ and Mg2+ (Groisman, 2001), acidic pH (Prost et al., 2007) and antimicrobial peptides (Bader et al., 2005) are the environmental cues for PhoPQ-mediated regulation. Thus, upregulation of mgtB, adiAY and a variety of genes involved in antimicrobial resistance correlates well with the activation of the PhoPQ regulon in the Tetrahymena phagosome.
Using LB culture medium as a common control environment, we compared the microarray data obtained in this study with that reported by Hautefort et al. (2008) regarding S. Typhimurium gene expression in macrophages and epithelial cells. This comparative analysis provided evidence that the pathogen experiences physicochemical conditions in the Tetrahymena phagosome that overlap with those encountered in macrophages and epithelial cells. Commonalities between the protist vacuoles and vacuoles of at least one of the two other cell types on the basis of transcriptional profiles include conditions of acid, oxidative and osmotic stress, low magnesium and phosphate concentrations, presence of antimicrobials and conditions inducing the SOS response and SPI-2. This overall response of S. Typhimurium to a variety of stresses in intravacuolar environments may underlie its ability to resist protozoan digestion. Of particular interest are the 49 hypothetical proteins that are part of the transcriptional signature of the pathogen in both the protist and macrophages, and which may represent proteins with unknown function that are crucial to the survival of S. Typhimurium in phagocytic cells.
Despite this overlap in transcriptional profile, the considerably larger sets of S. Typhimurium genes upregulated in macrophages and HeLa cells (Hautefort et al., 2008) indicate that significant differences also exist. S. enterica does not have any detectable cytotoxic effect in Tetrahymena (Brandl et al., 2005; Gourabathini et al., 2008), in contrast to Acanthamoeba spp., which are killed by S. enterica and other pathogenic species (Abu Kwaik et al., 1998; Gaze et al., 2003; Tezcan-Merdol et al., 2004; Matz et al., 2008; Feng et al., 2009). Hence, the role of SPI-2 and other virulence determinants in its resistance to digestion by Tetrahymena is less clear than their potential pathogenic function during interaction with Acanthamoeba rhysodes, in which SPI genes are also induced (Feng et al., 2009). The possibility remains that increased expression of virulence genes in S. Typhimurium in Tetrahymena is simply a response to environmental signals present also in host phagocytic cells, particularly low pH (Yu et al., 2010). Further investigation of the response of Tetrahymena to this enteric pathogen may provide more insight into their interaction.
mgtC is one of the most highly upregulated S. Typhimurium genes in macrophages (Eriksson et al., 2003) and in HeLa cells (Hautefort et al., 2008), and is required for its long-term phagosomal survival (Alix and Blanc-Potard, 2007). Similarly, mgtC had the highest differential expression in our study, yet the ΔmgtC mutant was as viable as the wild-type strain in Tetrahymena pellets, suggesting that it was not impaired for survival in its phagosome. Possibly, the passage time of the pathogen before release at the cytoproct, which we estimated to be approximately 1 h (Brandl et al., 2005), is not sufficiently long for this mutation to affect cell survival in Tetrahymena. Paradoxically, the ΔmgtC mutant had a greater cell density than the wild type in the fecal pellets. Complementation of the mutant caused lower cell density in the pellets, thus supporting a role for MgtC in this phenotype. Because growth in low-Mg2+ medium causes cell elongation and aggregation of MgtC-minus mutants (Rang et al., 2007), Tetrahymena may have ingested cell aggregates during feeding on this mutant in H2O, leading to a greater cell density in its food vacuoles. However, it is unclear if Tetrahymena would be able to feed on such aggregates because of their size.
In light of the increased expression in Tetrahymena phagosomes of two genes belonging to the arginine-dependent acid tolerance pathway, we investigated the effect of S. Typhimurium's passage through Tetrahymena on its subsequent acid resistance while in fecal pellets. S. enterica gains resistance to acid stress at pH 3–4 after adaptation to mild acidic conditions of pH 4.5–5.8 (Foster, 1995; Audia et al., 2001; Audia and Foster, 2003). The digestive process in Tetrahymena pyriformis involves a decrease in vacuolar pH to 5.5–6.0 after 5 min, eventually reaching 3.5–4.0 after 1 h (Nilsson, 1977). The viability of S. Typhimurium cells exposed to pH 3.4 was enhanced in Tetrahymena fecal pellets compared with that of non-ingested cells in the same suspension. This suggested that the pathogen adapted to acidic conditions during its passage through Tetrahymena and thereby gained long-term protection from acidic stress in the external environment. The deletion of adiAY, which are part of the arginine decarboxylase system for extreme acid resistance in S. enterica and function under acidic pH in anaerobic environments (Kieboom and Abee, 2006; Alvarez-Ordonez et al., 2010), significantly decreased the viability of the pathogen in Tetrahymena. It is noteworthy that adiY is upregulated also under the acidic pH of macrophages (Eriksson et al., 2003). Viability of the ΔadiAY mutant in the pellets was partially restored by complementation, supporting a role for this acid stress response in the pathogen's resistance to digestion by Tetrahymena and providing further evidence for the presence of anaerobic conditions in Tetrahymena food vacuoles.
We have previously reported that ciliated protozoa isolated from bagged leafy vegetables sold at the marketplace can release viable S. enterica cells in pellets in vitro and that Tetrahymena has the ability to produce such pellets while grazing on S. enterica inoculated onto plants in the laboratory (Gourabathini et al., 2008). As the postprandial pH in the stomach ranges from 2.5 to 4.9 (Simonian et al., 2005), a gain in acid resistance of S. enterica by means of passage through Tetrahymena and egestion in its fecal pellets may enhance its survival in the human host. Hence, adaptation to low pH and to a range of other stresses, as shown by transcriptional profiling, may mediate the passage of this food-borne pathogen through Tetrahymena and thereby contribute to its contamination cycle.
Acknowledgments
We thank Steven Huynh for technical assistance. This work was supported by CSREES-NRI Grant 2007-03116 and by funds from USDA ARS CRIS projects 5325-42000-044-00D and 5325-42000-045-00D.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abu Kwaik Y, Gao LY, Stone BJ, Venkataraman C, Harb OS. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix E, Blanc-Potard AB. MgtC: a key player in intramacrophage survival. Trends Microbiol. 2007;15:252–256. doi: 10.1016/j.tim.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ordonez A, Fernandez A, Bernardo A, Lopez M. Arginine and lysine decarboxylases and the acid tolerance response of Salmonella Typhimurium. Int J Food Microbiol. 2010;136:278–282. doi: 10.1016/j.ijfoodmicro.2009.09.024. [DOI] [PubMed] [Google Scholar]
- Audia JP, Foster JW. Acid shock accumulation of sigma S in Salmonella enterica involves increased translation, not regulated degradation. J Mol Microbiol Biotechnol. 2003;5:17–28. doi: 10.1159/000068717. [DOI] [PubMed] [Google Scholar]
- Audia JP, Webb CC, Foster JW. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int J Med Microbiol. 2001;291:97–106. doi: 10.1078/1438-4221-00106. [DOI] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Balaji B, O'Connor K, Lucas JR, Anderson JM, Csonka LN. Timing of induction of osmotically controlled genes in Salmonella enterica serovar Typhimurium, determined with quantitative real-time reverse transcription-PCR. Appl Environ Microbiol. 2005;71:8273–8283. doi: 10.1128/AEM.71.12.8273-8283.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J, Brown MRW. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;240:1253–1259. doi: 10.1099/00221287-140-6-1253. [DOI] [PubMed] [Google Scholar]
- Bishop RE. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol Microbiol. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- Brandl MT, Rosenthal BM, Haxo AF, Berk SG. Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl Environ Microbiol. 2005;71:1562–1569. doi: 10.1128/AEM.71.3.1562-1569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky IE, Ghori N, Falkow S, Monack D. Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol Microbiol. 2005;55:954–972. doi: 10.1111/j.1365-2958.2004.04444.x. [DOI] [PubMed] [Google Scholar]
- Cirillo JD, Falkow S, Tompkins LS. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Essig A, Heinemann M, Simnacher U, Marre R. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl Environ Microbiol. 1997;63:1396–1399. doi: 10.1128/aem.63.4.1396-1399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E, Groisman EA. Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol. 2009;12:199–204. doi: 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci USA. 2006;103:1906–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Hsiao YH, Chen HL, Chu C, Tang P, Chiu CH. Apoptosis-like cell death induced by Salmonella in Acanthamoeba rhysodes. Genomics. 2009;94:132–137. doi: 10.1016/j.ygeno.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Fok AK, Allen RD. Cytochemical localization of peroxisomes in Tetrahymena pyriformis. J Histochem Cytochem. 1975;23:599–606. doi: 10.1177/23.8.51038. [DOI] [PubMed] [Google Scholar]
- Foster JW. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit Rev Microbiol. 1995;21:215–237. doi: 10.3109/10408419509113541. [DOI] [PubMed] [Google Scholar]
- Gaze WH, Burroughs N, Gallagher MP, Wellington EM. Interactions between Salmonella typhimurium and Acanthamoeba polyphaga, and observation of a new mode of intracellular growth within contractile vacuoles. Microb Ecol. 2003;46:358–369. doi: 10.1007/s00248-003-1001-3. [DOI] [PubMed] [Google Scholar]
- Gil F, Ipinza F, Fuentes J, Fumeron R, Villarreal JM, Aspee A, et al. The ompW (porin) gene mediates methyl viologen (paraquat) efflux in Salmonella enterica serovar Typhimurium. Res Microbiol. 2007;158:529–536. doi: 10.1016/j.resmic.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Gourabathini P, Brandl MT, Redding KS, Gunderson JH, Berk SG. Interactions between food-borne pathogens and protozoa isolated from lettuce and spinach. Appl Environ Microbiol. 2008;74:2518–2525. doi: 10.1128/AEM.02709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA. The pleiotropic two-component regulatory system PhoP–PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzel D, Kucharski LM, Kehres DG, Romero MF, Maguire ME. The MgtC virulence factor of Salmonella enterica serovar Typhimurium activates Na+,K+ATPase. J Bacteriol. 2006;188:5586–5594. doi: 10.1128/JB.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Hofle MG. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Ecol. 2001;35:113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x. [DOI] [PubMed] [Google Scholar]
- Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, et al. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008;10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Jacobs ME, DeSouza LV, Samaranayake H, Pearlman RE, Siu KW, Klobutcher LA. The Tetrahymena thermophila phagosome proteome. Eukaryot Cell. 2006;5:1990–2000. doi: 10.1128/EC.00195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieboom J, Abee T. Arginine-dependent acid resistance in Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:5650–5653. doi: 10.1128/JB.00323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Kyle JL, Parker CT, Goudeau D, Brandl MT. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl Environ Microbiol. 2010;76:1375–1387. doi: 10.1128/AEM.02461-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Öhman L, Dahlgren C. Phagocytic recognition mechanisms in human granulocytes and Acanthamoeba castellanii using type 1 fimbriated Escherichia coli as phagocytic prey. FEMS Microbiol Lett. 1987;44:135–140. [Google Scholar]
- Ly TM, Muller HE. Ingested Listeria monocytogenes survive and multiply in protozoa. J Med Microbiol. 1990;33:51–54. doi: 10.1099/00222615-33-1-51. [DOI] [PubMed] [Google Scholar]
- Maier RJ, Olczak A, Maier S, Soni S, Gunn J. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect Immun. 2004;72:6294–6299. doi: 10.1128/IAI.72.11.6294-6299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Kjelleberg S. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 2005;13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Matz C, Moreno AM, Alhede M, Manefield M, Hauser AR, Givskov M, et al. Pseudomonas aeruginosa uses type III secretion system to kill biofilm associated amoebae. ISME J. 2008;2:843–852. doi: 10.1038/ismej.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JR. On food vacuoles in Tetrahymena pyriformis GL. J Euk Microbiol. 1977;24:502–507. [Google Scholar]
- Pace ML. Bacterial mortality and the fate of bacterial production. Hydrobiologia. 1988;159:41–49. [Google Scholar]
- Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, et al. Activation of the bacterial sensor kinase PhoP by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Rang C, Alix E, Felix C, Heitz A, Tasse L, Blanc-Potard AB. Dual role of the MgtC virulence factor in host and non-host environments. Mol Microbiol. 2007;63:605–622. doi: 10.1111/j.1365-2958.2006.05542.x. [DOI] [PubMed] [Google Scholar]
- Richard DJ, Sawers G, Sargent F, McWalter L, Boxer DH. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the (NiFe) hydrogenases 1 and 2 of Escherichia coli. Microbiology. 1999;145 (Part 10:2903–2912. doi: 10.1099/00221287-145-10-2903. [DOI] [PubMed] [Google Scholar]
- Santiviago CA, Fuentes JA, Bueno SM, Trombert AN, Hildago AA, Socias LT, et al. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol Microbiol. 2002;46:687–698. doi: 10.1046/j.1365-2958.2002.03204.x. [DOI] [PubMed] [Google Scholar]
- Simonian HP, Vo L, Doma S, Fisher RS, Parkman HP. Regional postprandial differences in pH within the stomach and gastroesophageal junction. Dig Dis Sci. 2005;50:2276–2285. doi: 10.1007/s10620-005-3048-0. [DOI] [PubMed] [Google Scholar]
- Tezcan-Merdol D, Ljungstrom M, Winiecka-Krusnell J, Linder E, Engstrand L, Rhen M. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl Environ Microbiol. 2004;70:3706–3714. doi: 10.1128/AEM.70.6.3706-3714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais PM, Colbeau A. Molecular biology of microbial hydrogenases. Curr Issues Mol Biol. 2004;6:159–188. [PubMed] [Google Scholar]
- Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. pH sensing by intracellular Salmonella induces effector translocation. Science. 2010;328:1040–1043. doi: 10.1126/science.1189000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbell AL, Benoit SL, Maier RJ. Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology. 2007;153:3508–3516. doi: 10.1099/mic.0.2007/009027-0. [DOI] [PubMed] [Google Scholar]
- Zbell AL, Maier RJ. Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl Environ Microbiol. 2009;75:1456–1459. doi: 10.1128/AEM.02064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.