Abstract
Stoichiometry of microbial biomass is a key determinant of nutrient recycling in a wide variety of ecosystems. However, little is known about the underlying causes of variance in microbial biomass stoichiometry. This is primarily because of technological constraints limiting the analysis of macromolecular composition to large quantities of microbial biomass. Here, we use Raman microspectroscopy (MS), to analyze the macromolecular composition of single cells of two species of bacteria grown on minimal media over a wide range of resource stoichiometry. We show that macromolecular composition, determined from a subset of identified peaks within the Raman spectra, was consistent with macromolecular composition determined using traditional analytical methods. In addition, macromolecular composition determined by Raman MS correlated with total biomass stoichiometry, indicating that analysis with Raman MS included a large proportion of a cell's total macromolecular composition. Growth phase (logarithmic or stationary), resource stoichiometry and species identity each influenced each organism's macromolecular composition and thus biomass stoichiometry. Interestingly, the least variable peaks in the Raman spectra were those responsible for differentiation between species, suggesting a phylogenetically specific cellular architecture. As Raman MS has been previously shown to be applicable to cells sampled directly from complex environments, our results suggest Raman MS is an extremely useful application for evaluating the biomass stoichiometry of environmental microorganisms. This includes the ability to partition microbial biomass into its constituent macromolecules and increase our understanding of how microorganisms in the environment respond to resource heterogeneity.
Keywords: ecological stoichiometry, macromolecular composition, Raman microspectroscopy, resource allocation
Introduction
Microbial biomass stoichiometry (specifically carbon (C):nitrogen (N) and phosphorus (P) stoichiometry) is a primary determinant of whether mineral nutrients are sequestered in microbial biomass or released to the environment during decomposition (Manzoni et al., 2008). However, little is known about the how microbial physiology and environmental parameters interact to constrain the range and variance of microbial biomass stoichiometry. Microbial biomass is composed of a vast array of macromolecules, each containing a wide range of specific functions. These macromolecules can be assigned to relatively few classes (for example, carbohydrates, proteins and nucleic acids), each with a constrained elemental content that can be linked to its dominant element (Elser et al., 1996). Proteins are on average relatively rich in N (53% C, 17% N, 0% P by weight), nucleic acids are rich in P (32.7% C, 14.5% N and 8.7% P), while carbohydrates (37% C, 0% N, 0% P) are rich in C and contain no N or P (Sterner and Elser, 2002). Shifts in the relative concentration of these constituent macromolecule pools ultimately determine the stoichiometry of microbial biomass. From this perspective, carbohydrate content should be positively correlated with biomass C:P and C:N, protein content should be inversely correlated with biomass C:N, while nucleic acid content should be inversely correlated with both biomass C:P and N:P. While previous studies have shown a relationship between microbial biomass P and RNA content in culture (Makino et al., 2003; Makino and Cotner, 2004) and in the environment (Hall et al., 2009), few if any studies have evaluated the effect of changes in other macromolecular pools on microbial biomass stoichiometry in an ecological context. Determining how shifts in constituent macromolecules are related to changes in biomass stoichiometry will lead to a more mechanistic understanding of what controls or constrains microbial biomass stoichiometry in nature.
Microbial biomass stoichiometry has been shown to change in response to physical (for example, temperature), chemical (for example, resource stoichiometry) and physiological (for example, growth rate) factors (Makino et al., 2003; Makino and Cotner, 2004; Cotner et al., 2006). How microbial biomass changes in response to resource stoichiometry is of particular interest because the relationship between biomass stoichiometry and resource stoichiometry ultimately determines how microorganisms recycle limiting nutrients (Manzoni et al., 2008), which can markedly affect the growth and community composition of the surrounding organisms (Danger et al., 2007; Cherif and Loreau, 2009). The current dearth of information on the relationship between resource stoichiometry and microbial biomass stoichiometry is due to multiple logistical constraints. First, most environmental microorganisms cannot be cultured; therefore it is not possible to follow the response of their biomass stoichiometry to experimentally-manipulated resource treatments. Second, the resource pool of environmental microorganisms is notoriously hard to define, thus relating resource stoichiometry to biomass stoichiometry in situ is not feasible. Third, and perhaps most important, determining macromolecular biomass composition of microorganisms has traditionally required large amounts of biomass and therefore requires culturing or enrichment of the organisms of interest or analysis of undifferentiated microbial communities.
Various technological advances have helped to overcome these constraints and now allow for direct measurement of microorganisms and to some extent their in situ resource pool. For example, advances in quantitative chemical methods permit high-resolution analysis of the dissolved organic carbon pool in aquatic environments (Kim et al., 2006). Although not widely applied, it is also important to note that elemental analysis of single microbial cells is possible using x-ray microanalysis (Norland et al., 1995), thus avoiding the need to grow organisms in culture to determine biomass CNP. Also, the recent application of stable isotope mass spectroscopy to microbial ecology (see Wagner, 2009 for review) has allowed for analysis of microbial isotope at unprecedented levels of spatial resolution (30–50 nm). Complementary to these high resolution/single cell techniques for analysis of the microbial resource pool and microbial elemental composition, is Raman microspectroscopy (MS). Raman MS allows for analysis of macromolecular composition at the level of the single cell and therefore has the potential to directly evaluate how constituent macromolecules contribute to biomass stoichiometry, while avoiding the constraints imposed by isolation and culturing of environmental organisms. Although X-ray microanalysis and stable isotope mass spectroscopy can evaluate elemental and isotopic composition, respectively, Raman MS has the advantage of eliciting information on specific classes of macromolecules, and in some cases specific compounds (De Gelder et al., 2008) that compose microbial biomass.
Raman MS has been applied to the analyses of microorganisms for 30 years (Howard et al., 1980). However, it has only recently been rigorously applied to environmental microbial communities (Huang et al., 2007a, 2007b), and its potential to address key processes in environmental microbiology is only beginning to be recognized (Wagner, 2009). Here, we use Raman MS to evaluate the macromolecular composition of two bacteria, Verrucomicrobium spinosum and Pectobacterium carotovorum, grown on minimal media covering a broad range of C:N:P resource stoichiometry and harvested during log and stationary growth phase. Specifically we ask: Is macromolecular composition as determined by Raman MS comparable to macromolecular composition derived from traditional analytical methods? Does macromolecular composition as determined by Raman MS correlate with total biomass stoichiometry in the predicted manner? And finally, what is the influence of resource stoichiometry, growth phase and species identity on macromolecular composition of microbial biomass? Answering each of the above questions is an important first step in applying Raman MS to fundamental questions in microbial stoichiometry and to questions in microbial ecology in general.
Materials and methods
Experimental design and harvesting
V. spinosum Schlesner, 1987 (DSM 4136T) is a relatively slow growing (0.05–0.15 h−1, this study) member of the bacterial phylum Verrucomicrobia originally isolated from the epilimnion of a eutrophic lake (Schlesner, 1987). Members of the Verrucomicrobia comprise a major group of soil microorganisms representing up to 10% of the total bacteria in terrestrial environments (Sangwan et al., 2004). Pectobacterium carotovorum (subspecies carotovorum (American Type Culture Collection 100 no. 39048)) previously named E. carotovora is a relatively fast growing (0.10–0.43 h−1, this study) gammaproteobacterium from the family enterobacteriaceae. It is a plant pathogen (causative agent of ‘soft rot') capable of infecting a wide range of host tissues (Pérombelon, 1980). We selected these two environmental organisms as representatives of bacteria with distinct phylogenetic affiliations, habitats and growth strategies.
We grew each species in batch cultures (22 °C±1 °C) in a buffered minimal media containing varying amounts of C, N and P as described in Keiblinger et al. (2010). Briefly, C:N:P amendments resulted in four unique N:P resource ratios replicated at multiple high quantity and low quantity levels, for a total of twelve unique resource treatments (Table 1). This design allowed us to test both the effect of resource stoichiometry and resource quantity on each species' macromolecular composition. To test if the buffer could be used as a carbon or nitrogen source, we attempted to culture each species on the complete minimal media with the exception of glucose and separately in complete media with the exception of the –NO3−2 (V. spinosum) or –NH4+ (P. carotovorum). In each case, each species was unable to achieve any significant growth. During the main experiment, all cultures were harvested by centrifugation during logarithmic and stationary growth phase as determined for each resource treatment in a priori growth experiments. The growth experiments were identical to the main experiment only that each culture flask was sub-sampled 7–10 times over the entire growth phase and evaluated for optical density at 450 nm as described in Keiblinger et al. (2010).
Table 1. Concentration and molar ratios of each resource treatment.
| Treatment | C (m) | N (m) | P (m) | C:N | C:P | N:P |
|---|---|---|---|---|---|---|
| 1 | 150 | 12.5 | 0.25 | 12 | 600 | 50 |
| 2 | 150 | 12.5 | 0.025 | 12 | 6000 | 500 |
| 3 | 150 | 1.25 | 0.25 | 120 | 600 | 5 |
| 4 | 150 | 1.25 | 0.025 | 120 | 6000 | 50 |
| 5 | 150 | 0.125 | 0.25 | 1200 | 600 | 0.5 |
| 6 | 150 | 0.125 | 0.025 | 1200 | 6000 | 5 |
| 7 | 30 | 5 | 0.1 | 6 | 300 | 50 |
| 8 | 30 | 5 | 0.01 | 6 | 3000 | 500 |
| 9 | 30 | 0.5 | 0.1 | 60 | 300 | 5 |
| 10 | 30 | 0.5 | 0.01 | 60 | 3000 | 50 |
| 11 | 30 | 0.05 | 0.1 | 600 | 300 | 0.5 |
| 12 | 30 | 0.05 | 0.01 | 600 | 3000 | 5 |
Lists the molar concentration of each resource amendment for each treatment, as well as the intended resource ratios of each treatment at the beginning of the batch culture experiments. Treatments 1–6 mirror treatments 7–12 except they contain two- (N and P) to fivefold (C) more of each element than the higher numbered treatments. Therefore, treatments 1–6 are referred to as the low-quantity treatments and treatments 7–12 are referred to as the high-quantity treatments in the text.
Analytical methods
Raman analysis
For analysis with Raman MS the biomass pellet was preserved by washing with 50 ml of 130 m NaCl (vortexed, then centrifuged at 36 700 g for 15 min) in order to eliminate possible spectral interference of the medium and resuspended in 130 m NaCl. An aliquot of 200–500 μl of the suspension (depending on cell density) was taken and fixed with paraformaldehyde (final concentration 3%, 4 h, 4 °C; Amann, 1995). The cells were subsequently washed with phosphate-buffered saline, transferred to 300–800 μl (depending on cell density) of a 1:1 (volume:volume) mixture of 96% ethanol and phosphate-buffered saline and stored at−20 °C until analysis.
Raman MS analysis are described in detail in the Supplementary Material, briefly, 1–2 μl of the fixed cell suspension were dried on a calcium fluoride slide at 48 °C and dipped into particle-free water to remove salts and other solutes for 1–2 s before air drying. For each treatment we collected Raman spectra from 40 individual cells (20 from each of two replicate flasks) using a LabRAM HR800 high-resolution confocal Raman microscope (HORIBA Jobin-Yvon, Irvine, CA, USA) with an attached Olympus BX-41 microscope (Mountainview, CA, USA). From each spectrum we extracted peak heights at locations, which were previously reported to represent specific macromolecules belonging to one of the three classes of interest, that is, proteins, carbohydrates and nucleic acids (Table 2).
Table 2. Subset of Raman MS peaks used for each analysis.
| Wave number (cm−1) | Code | Macromolecular assignment | Reference | PC 1 loading | PC 2 loading |
|---|---|---|---|---|---|
| 440 | ca 1 | carbohydrates | De Gelder et al., 2007 | 0.761 | 0.323 |
| 481 | ca 2 | carbohydrates | Goral and Zichy, 1990 | 0.912 | 0.354 |
| 665 | na 1 | nucleic acids (guanine) | (De Gelder et al., 2007; Maquelin et al., 2002) | −0.652 | 0.351 |
| 723 | na 2 | nucleic acids (adenine) | (De Gelder et al., 2007; Maquelin et al., 2002) | −0.769 | 0.348 |
| 785 | na 3 | nucleic acids (cytosine, uracil) | (De Gelder et al., 2007; Maquelin et al., 2002) | −0.736 | 0.498 |
| 854 | ca 3 | carbohydrates | De Gelder et al., 2007 | 0.884 | 0.133 |
| 936 | ca 4 | carbohydrates | De Gelder et al., 2007 | 0.949 | 0.102 |
| 1004 | pr 1 | phenylalanine in proteins | De Gelder et al., 2007 | −0.572 | −0.754 |
| 1030–1130 | a 5 | carbohydrates, mainly -C-C- (skeletal) | Schuster et al., 2000 | 0.710 | −0.287 |
| ca 6 | carbohydrates, mainly | Schuster et al., 2000 | 0.931 | 0.372 | |
| ca 7 | carbohydrates, mainly | Schuster et al., 2000 | 0.915 | 0.402 | |
| ca 8 | carbohydrates, mainly | Schuster et al., 2000 | 0.944 | 0.144 | |
| 1254 | pr 2 | proteins (amide III random) | Maquelin et al., 2002 | −0.823 | −0.046 |
| 1575 | na 4 | nucleic acids (guanine, adenine) | (De Gelder et al., 2007; Maquelin et al., 2002) | −0.881 | 0.058 |
| 1660–1670 | pr 3 | proteins (amide I) | Maquelin et al., 2002 | −0.821 | −0.518 |
Lists each of the peaks from the Raman microspectrascopy spectra that were included in the analyses, the position of each peak, their literature source and the factor loading of each peak for the principal component analysis on each principal component.
Analytical chemistry of macromolecule composition
Nucleic acids
RNA and DNA were measured by fluorometry using the fluorescent stain RiboGreen (Molecular Probes Invitrogen, Carlsbad, CA, USA) Makino and Cotner, 2004). Centrifuged biomass samples stored in RNAlater (Ambion, AM7020, Austin, TX, USA) at−80 °C were first thawed to room temperature and separated from RNAlater by centrifugation (32 200 g for 15 min). Following removal of the supernatant, ∼1 mg of biomass (wet weight) was transferred into a 2 ml eppendorf tube, resuspended in standard buffer, and sonicated (at 35 kHz, 8 min).
Samples were diluted to an appropriate concentration, aliquoted into a microplate and incubated with RNase and DNase for digestion, or without enzyme for determination of total nucleic acid content (1 h, ∼20 °C). Previous experiments determined that both the length of the experiment and the enzyme activity were sufficient to maximize the degradation of each nucleic acid (data not shown). After digestion, we added 100 μl of 200 × diluted RiboGreen to each well of a 96-well black polystyrene microplate and incubated plates in the dark for 5 min before measuring fluorescence (excitation 480 nm, emission 520 nm, TECAN infinite M200, Maennedorf, Switzerland). To calculate the amount of RNA and DNA in each sample, we compared the difference in fluorescence between undigested and digested samples to concurrently determined standard curves prepared and measured simultaneously in each plate (Invitrogen Quant iT RiboGreen Reagent Kit for RNA Lambda DNA Standard, Invitrogen cat no. R-11490). We quantified RNA at several time points during the sample processing and saw no significant degradation during the time it took us to analyze the samples or for several hours thereafter.
Protein and carbohydrate analysis
Frozen biomass pellets stored at −80 °C were freeze-dried (−50 °C, 20 μbar) and weighed before being resuspended in Milli-Q water, and pulse-sonicated in an ice-bath for 100 s with a ‘microtip' sonicator (Branson Sonifier W-250D, Danbury, CT, USA). Sonicated samples were diluted with 0.1 M Tris-HCl buffer (pH 7.5) to a concentration of ∼50 μg biomass ml−1, and analyzed using the Bradford assay (Bradford, 1976) taking bovine gamma globuline as a standard.
For carbohydrate analysis, biomass pellets were freeze-dried, weighed, sonicated (as described above) and diluted with distilled water to reach a concentration of ∼1 mg cells ml−1. Diluted samples (0.5 ml) were mixed with 0.5 ml of phenol solution (5 g per 100 ml) and 2.5 ml of concentrated H2SO4 (95%) and incubated (50–60 °C, 1 h). Samples, glucose standards and blanks were cooled in an ice bath before photometric measurement at an absorbance of 488 nm with final carbohydrate content expressed in glucose equivalents (Daniels et al., 2007).
Biomass CNP analysis
Total P content of bacterial biomass was determined through acid extraction with HNO3 (65%) and HClO4 (70%) and subsequent photometric analysis (Schinner et al., 1993). Total biomass N and C content were measured using an elemental analyzer (EA1110, CE Instruments, Milan, Italy).
Statistical analysis
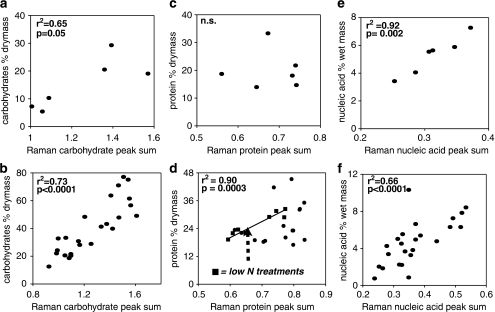
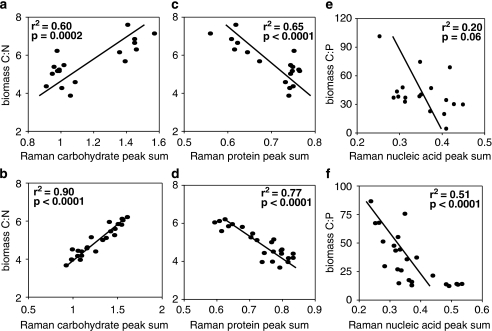
We reduced each Raman spectrum to a series of 15 peaks, which had been previously identified in the literature (Table 2). On the basis of their identification we grouped these peaks into one of three macromolecular classes (three proteins, eight carbohydrates and four nucleic acids). For each class of macromolecules we computed the sum of the respective peak heights for each cell. Mean sums of peak heights (total of 40 cells per treatment) were computed and tested for correlation (Pearson's R) with macromolecular content determined by analytical chemistry. The relationship between macromolecular composition and biomass stoichiometry was tested using simple linear regression, using JMP software (SAS Institute, Cary, NC, USA). For each analysis we only compared microbial biomass sampled from the same flask during the same experiment. Experiments for V. spinosum were repeated on multiple dates but analysis of wet chemistry was only conducted for a single date. Therefore, we had fewer observations for V. spinosum than for P. carotovorum. This resulted in fewer data points to compare macromolecular composition determined by Raman MS with that determined using traditional analytical methods for V. spinosum than P. carotovorum (Figure 1). This was also true, although to a lesser extent, for comparison of biomass stoichiometry and macromolecular composition as determined by Raman MS (Figure 2).
Figure 1.
Compares macromolecular composition measured using analytical chemistry with that generated from Raman microspectroscopy (sum of mean peak height for all peaks in each macromolecular category). V. spinosum are presented in the top row (a, c and e) and P. carotovorum are presented in the bottom row (b, d and f).
Figure 2.
Compares biomass stoichiometry (atomic ratios) with relative macromolecular composition generated from Raman microspectroscopy (sum of mean peak height for all peaks in each macromolecular category). Lines represent least square regressions with associated r2 and P-values. V. spinosum data are presented in the top row (a, c and e) and P. carotovorum data are presented in the bottom row (b, d and f).
To assess the effect of each treatment on macromolecular composition the data mean sum of peak heights from 15 peaks (Table 2) was used as input for multivariate analyses. We used Principal component analysis (PCA) and Canonical discriminant analysis (CDA) as unconstrained and constrained ordination techniques, respectively. The CDA was carried out on a full-factorial design defined by two species × two growth phases × two resource quantities × two resource qualities (as N:P levels). This was achieved by combining the two lower (0.5 and 5) and the two higher (50 and 500) resource treatments. Combination of nutrient treatments in order to simplify analyses was supported by the results of the PCA suggesting the formation of two resource N:P groups (upper two resource N:P levels versus lower two levels) rather than a continuous response along a resource N:P gradient. The CDA was complemented by non-parametric permutational MANOVA (Anderson, 2001) on the basis of Euclidean distance. Variation of macromolecular composition among cells from a given treatment was determined by computing Euclidean distances (Legendre and Legendre, 2003) to a multivariate mean (the centroid) on the basis of the 15 standardized Raman peaks (see Supplementary Material for details). All multivariate statistics were carried out using the statistical language environment R (R-Development-Core-Team, 2005).
Results
Variance in resource stoichiometry resulted in significant phenotypic plasticity of each organism's growth rate. Specifically, V. spinosum showed a threefold difference in growth rate (ranging from 0.05–0.15 h−1) and nearly 300-fold difference in biomass yield (0.003 and 1.03 g dry mass l−1). P. carotovorum had a more constrained but still variable physiology growing between 0.1–0.43 h−1 and yielding 0.18–0.35 g dry mass l−1.
We compared macromolecular composition determined by Raman MS with traditional methods by plotting values from each method against each other (Figure 1). For each species carbohydrate content, as determined by Raman MS, was significantly and positively correlated with carbohydrate content determined by chemical analysis (Figures 1a and b). Similarly, nucleic acid content determined by each method was also significantly and positively correlated (Figures 1e and f). Protein content determined by Raman MS, however, did not correlate with protein content as determined by the Bradford assay for either species (Figures 1c and d). For P. carotovorum, when only the low resource N treatments (treatments 5, 6, 11 and 12) were considered there was a significant correlation between protein content determined by each method (Figure 1f). It was not possible to do the same analysis for V. spinosum because of missing data in the low resource N treatment groups.
We next evaluated relationships between each of the three macromolecular classes and biomass stoichiometry to see if analysis with Raman MS could be used to partition total biomass stoichiometry as hypothesized. For each species, carbohydrate content was positively and significantly correlated with biomass C:N (Figures 2a and b). Similarly, proteins were significantly and negatively correlated with C:N, as hypothesized (Figures 2c and d). The nucleic acid—biomass C:P relationship were in the hypothesized direction for both species, however in both cases the relationship was noticeably weaker. Specifically, for V. spinosum there was a weak, negative but not significant correlation between nucleic acid content and biomass C:P (Figure 2e). Although for P. carotovorum the relationship was negative and significant (Figure 2f).
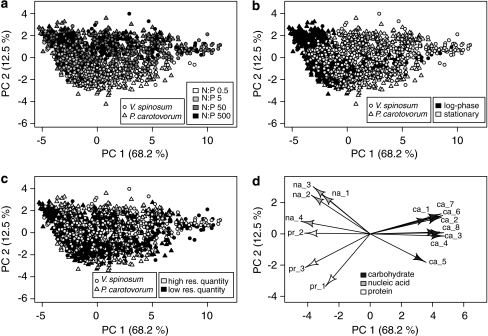
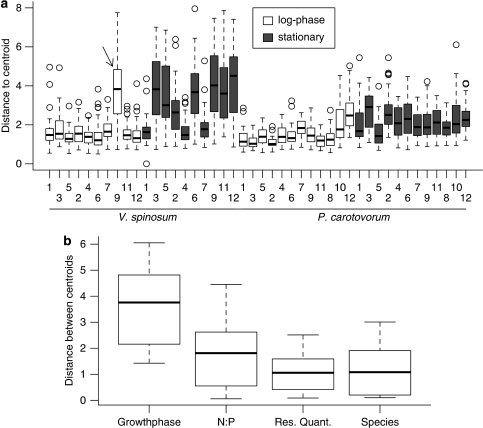
By using PCA we investigated how macromolecular composition differed between growth phase, resource treatment and species. The first two principal components (PC 1 and 2, Table 2) explained 80.7% of variation in Raman MS-derived macromolecular composition with PC 1 alone explaining over two-thirds of the variation (Figure 3). Raman peaks within a macromolecular class (carbohydrates, proteins and nucleic acids) loaded in similar directions in PC space, but loaded in different directions from each other (Figure 3d). Specifically, carbohydrates were positively associated with PC 1, whereas nucleic acids and proteins loaded negatively on PC 1. In addition, differences in nucleic acids and proteins were the cause of the majority of the separation along PC 2 (Figure 3d). Coding data points (that is, single cells) by the various treatments showed a shift in growth phases along PC 1 (Figure 3b), and a separation of resource N:P levels along PC 2 (Figure 3a). Thus, a shift from the log to the stationary phase was associated with an increase in carbohydrates, while cells growing logarithmically were richer in nucleic acids and proteins (Figure 3b). Also, variation in macromolecular composition was higher for cells in stationary growth phase than for the logarithmically growing cells, which were more constrained in macromolecular composition (Figure 4). This result was further corroborated by the higher distances to centroids in Euclidean space (a metric that compares multivariate variation within a treatment) for stationary cells relative to logarithmic cells (Figure 5). A two-way ANOVA on these distances confirmed a significant difference in variation of macromolecular composition between growth phases (F1,1996=470.3, P<0.001) and also indicated a significant difference in variation between species (F1,1696=99.4, P<0.001) with V. spinosum cells having greater variability in macromolecular composition than P. carotovorum cells (Figures 4 and 5). A significant interaction term (F1,1696=80.5, P<0.001) between species and growth phase was supported by Tukey (Honestly Significant Difference) post-hoc tests, indicating significant differences between each species and growth phase combination (all P<0.001), except for log-phase in which the composition of each species was not significantly different from each other (Table 3).
Figure 3.
Principal component analysis (PCA) was computed on 15 Raman peaks belonging to specific macromolecule classes. Scores on first and second principal component, commonly explaining 80.7% of data variation, color coded by (a) resource N:P, (b) growth phase and (c) resource quantity. (d) PCA loadings (correlations of macromolecules with the principal components) with absolute values >0.25 on either of two principal components indicate contributions of the various macromolecules to the ordination. Although carbohydrates, and protein 2 and 3 load primarily on PC 1, nucleic acids and the remaining proteins load about equivalently on both PC 1 and 2. Loadings are scaled by a factor of five.
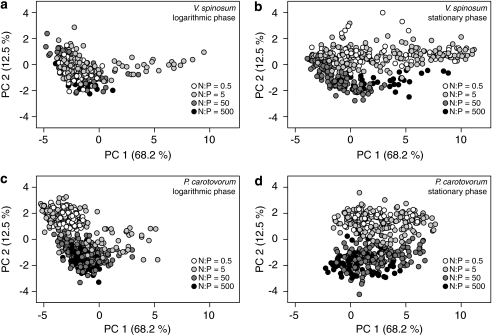
Figure 4.
Shows the effect of resource stoichiometry on macromolecular composition as determined by Raman microspectrascopy. Data is sorted in each panel by log and stationary phase and by species. Logarithmic phase for (a) V. spinosum and (b) P. carotovorum, stationary phase for (c) V. spinosum and (d) P. carotovorum.
Figure 5.
Analyses of distances indicating differences in macromolecular composition. (a) For each unique treatment (Table 1) multivariate distances of individual cells to the treatment centroid in Euclidean space were computed based on the 16 Raman peaks. Histograms of distances to the centroid express the variation of macromolecular composition among individual cells of the various treatments. Data of one treatment (marked by an arrow) was excluded to allow computation of the two-way ANOVA testing effects of growth phase and species. (b) For each factor, distances between centroids were calculated while holding all other factors constant. All boxplots show median (black line), 1st and 3rd quartiles (box), and 5 and 95% quantiles (whiskers).
Table 3. Permutational non-parametric ANOVA comparing individual treatments.
| Effect | V. spinosum |
P. carotovorum |
||
|---|---|---|---|---|
| log-phase | stationary phase | log-phase | stationary phase | |
| Resource quantity | F1,376=5.9 | F1,396=3.6 | F1,476=56.7 | F1,476=5.6 |
| P <0.01 | P < 0.05 | P < 0.001 | P < 0.001 | |
| N:P resource quality | F1,376=8.4 | F1,396=92.2 | F1,476=216.0 | F1,476=160.4 |
| P < 0.01 | P < 0.001 | P < 0.001 | P < 0.001 | |
| Interaction | F1,376=36.6 | F1,396=4.7 | F1,476=38.3 | F1,476=10.9 |
| P < 0.001 | P < 0.05 | P < 0.001 | P < 0.001 | |
| Post-hocs | all tests significant | one test nonsignificant*,a | one test nonsignificant*,b | all tests significant |
| Fmin=8.5 | Fmin=11.1 | Fmin=54.8 | Fmin=8.2 | |
| Pmax=0.006 | Pmax=0.006 | Pmax=0.006 | Pmax=0.006 | |
*No effect of resource quantity at low resource N:P (F1,238=1.4 P=1.0).
*No effect of resource quantity at high resource N:P (F1,238=3.6 P=0.06).
Separate analyses were run for each species and each growth phase because of differences in dispersion between species and growth phases. Given are F-values (subscripts are degrees of freedom) and significances (from 1000 permutations) for effects of resource quantity, resource N:P (as condensed two-level factor) and the interaction effect. Because of significant interaction effects, all analyses were followed by post-hoc multiple pair-wise comparisons using a Bonferroni adjustment of significance values. All post-hoc tests were significant except two (see footnote in table). The last line of the table gives minimum F and maximum P-values for all significant post-hoc tests.
The PCA plots also suggested that cells grouped distinctly between the two lower and the two higher resource N:P treatments. To evaluate this we condensed resource N:P levels to a two-level factor (high N:P=50 and 500 and low N:P=0.5 and 5). A permutational ANOVA (Table 3) found macromolecular composition to be significantly different between high and low resource N:P levels, with a stronger effect of resource N:P on biomass composition of P. carotovorum and in logarithmic phase. These differences in macromolecular composition driven by resource N:P separated primarily along PC 2 with high resource N:P associated with higher abundance of proteins and lower abundance of nucleic acids, as would be expected on the basis of stoichiometric principles (Figure 4). This pattern was clear for both species in stationary phase (Figures 4c and d). In logarithmic phase, a similar pattern was found for P. carotovorum with clear shifts between resource N:P levels along PC 2, but not for V. spinosum, which showed very stable macromolecular composition in logarithmic phase (Figures 4a and b).
We further analyzed the selected Raman spectra with CDA to evaluate if we could use the Raman MS data to discriminate between species and levels of resource quantity, which we were unable to do with PCA (Table 4). We related the dimensions hosting variability (that is, the PCA axes) to those hosting treatment differentiation (that is, the CDA axes) by projecting PC axes onto the CDA ordination plots (Figures 6c and d).
Table 4. Results of the CDA analysis.
| Canonical axis | Eigenvalue | Canonical R | Percentage variance among groups | Cumulative percentage | χ2 value | Df | Significance |
|---|---|---|---|---|---|---|---|
| CD_1 | 4.02 | 0.9 | 40.5 | 40.5 | 8536.4 | 225 | <0.001 |
| CD_2 | 3.65 | 0.89 | 36.7 | 77.2 | 5755.1 | 196 | <0.001 |
| CD_3 | 1.05 | 0.72 | 10.6 | 87.8 | 3105.8 | 169 | <0.001 |
| CD_4 | 0.38 | 0.52 | 3.8 | 91.6 | 1864.5 | 144 | <0.001 |
| CD_5 | 0.33 | 0.42 | 3.3 | 94.9 | 1311.0 | 121 | <0.001 |
| CD_6 | 0.21 | 0.4 | 2.1 | 97.0 | 825.5 | 100 | <0.001 |
| CD_7 | 0.13 | 0.3 | 1.3 | 98.3 | 494.9 | 81 | <0.001 |
Results of canonical discriminant analysis (CDA) using the factors: species, growth phase, resource quantity and resource N:P in a combined factor as grouping criterion. For the first seven canonical axes the table gives Eigenvalues, canonical correlation coefficients, percentage of among group variation explained by each canonical axis, cumulative percentages of variation explained and the results of tests of significance for each axis. In all, 14 of a maximum possible of 15 canonical axes were significant. The first four axes explained more than 91% of among group variation.
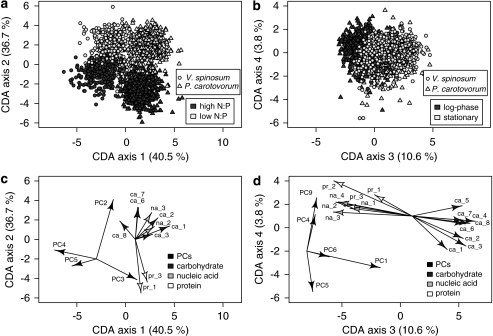
Figure 6.
Canonical discriminant analysis (CDA) was computed on 16 Raman peaks belonging to specific macromolecule classes. Scores on CDA axes 1 and 2 (a), and 3 and 4 (b), respectively. Symbols are shaded according to major factor discriminated by the respective CDA axes. Although CDA axis 1 discriminates strongly between species, CDA axis 2 and 3 separate resource N:P quality and growth phase, respectively. Structure coefficients (correlations of macromolecules with the CDA axes) indicate which macromolecules discriminate best between the various groupings on CDA axes 1 and 2 (c), and 3 and 4 (d), respectively. Also shown are correlations of principal components (PCs, black arrows) with CDA axes. All coefficients scaled by a factor of seven.
The first CDA axis explained over 40% of among group variation and was able to successfully classify species with a success rate of 96.7% (Figure 6a). Species primarily differed in the abundance of a subset of the carbohydrate peaks (ca 1, ca 2, ca 3 and ca 8, as defined in Table 2) and P. carotovorum was generally enriched in nucleic acids and proteins relative to V. spinosum. Interestingly, CDA axis 1 was found to correlate with PC 3, 4 and 5 (Figure 6c) indicating that the differences between species were not because of the most variable Raman peaks but rather those that changed little across resource treatments.
CDA axis 2 explained a similar amount of variation (36.7%) as CDA axis 1 and successfully discriminated between high and low resource N:P with a success rate of 89.7% (Figure 6a). Differences between resource treatments were again associated with differences in proteins and nucleic acid content and to a lesser extent some carbohydrate peaks (Figure 6c). Also, differences between resource N:P levels were greater for P. carotovorum than for V. spinosum and cells in the high resource N:P treatments had higher protein content and lower nucleic acid content than cells in the low resource N:P treatments, corroborating the results of the PCA (Figure 6a).
Additional CDA axes had limited but not negligible predictive power. CDA axis 3 explained a comparably small amount of between-treatment variation (∼10.6%) but it was able to correctly assign growth phases (Figure 6b) with a success rate of ∼76%. CDA axis 3 was positively associated with carbohydrates and negatively with proteins and nucleic acids (Figure 6d) and correlated with PC 1, supporting the conclusion of the PCA that the majority of variation in macromolecular composition was because of differences in growth phases.
Discrimination in resource quantity was not visible along any CDA axis (Supplementary Figure 1), consistent with the PCA results (Figure 3c), even though permutational ANOVA did identify significant effects of resource quantity for all species and growth phases (Table 3). However, these were comparably weak (low F-values) and classification success for resource quantity (∼66% success rate) was close to, but still significantly different from, the random success rate of 50% (Table 5). When using all CDA axes with significant discriminatory power for a leave-one-out-cross-validation scheme, overall classification success was maximal for species and resource N:P levels (both >90%), followed closely by growth phase with approximately 82% (Supplementary Figure 1, Table 5).
Table 5. Classification success of the canonical discriminant analysis (CDA).
| CDA classification success (%) | Expected classification success upon random group allocation (%) | |
|---|---|---|
| Combined factor (16 factorial combinations) | 53.8 | 6.6 |
| Species | 98.6 | 50.5 |
| Growth phase | 82.9 | 50.0 |
| Resource quantity | 66.4 | 50.3 |
| Resource N:P level | 93.3 | 50.5 |
Classification success of the canonical discriminant analysis (CDA) following a leave-one-out-cross-validation scheme. Percentage of correctly classified cells are given for the combined factors used as grouping criterion in the CDA, and the four factors (species, growth phase, resource quantity and resource N:P), separately. The first column gives percentages of cells correctly classified to each level of the respective factor using all axes of the CDA identified as significant (see Table 4). The second column gives expected classification success on random group allocation analytically computed from the respective group sample sizes. Permutational tests indicated that achieved classification success was significantly different from expected success for all factors (all P <0.001).
Discussion
Our results show that analysis of a subset of identified peaks from the Raman spectra was sufficient to link variance in constituent macromolecular pools to variance in biomass stoichiometry. With minor exceptions, macromolecular composition as determined by Raman MS correlated well with macromolecular composition determined by traditional methods and showed predictable relationships with microbial biomass stoichiometry. In addition, Raman MS gave information indicative of an organism's resource environment and physiological status (that is, growth phase) and provided insight into how microbes respond to differences in resource stoichiometry. This suggests that analysis with Raman includes the majority of the macromolecule pools responsible for variance in biomass stoichiometry and is potentially a useful tool for evaluating the ecology of environmental microorganisms.
The relationships between macromolecular compositions as analyzed by Raman MS and traditional analytical techniques showed that Raman MS gave a reliable signal of macromolecular composition for carbohydrates and nucleic acids but not for proteins (Figure 1). We propose three possible explanations for the differences between protein content as determined by Raman MS versus that determined by the Bradford assay. First, we used peak identities that had been assigned in the literature (Table 2). These identifications came from a variety of organisms, which did not include either species considered here. Therefore, it is possible that the peaks we identified as proteins may actually represent macromolecules other than proteins in our organisms. Similarly, it is possible that non-protein macromolecules occurred in the range of the Raman spectra that also included one or more protein peaks used in the analysis and had a masking effect on the identified peak(s). Third, protein peaks we used from the Raman spectra may have only represented a portion of the total protein content of the entire cell, that is, variance in protein content was driven by proteins not identified in the subset of peaks we used in this analysis. This third possibility is at least partly corroborated by the close correlation between the sum of Raman protein peak heights and protein concentration determined by the Bradford assay for low N treatments but not for high N treatments (Figure 1d). This suggests that when excess N was present, cells may have synthesized protein not identified by the peaks in Table 2, for example, storage proteins. However, the sum of Raman protein peak heights was negatively and significantly correlated to biomass C:N (Figures 2c and d) and responded to N:P resource treatments in a manner consistent with stoichiometric predictions (Figure 3a and 4). These final two results suggest that even in the absence of agreement with the Bradford assay Raman MS captured the portion of the protein pool responsible for variance in biomass N and those that were most likely to respond to N additions.
Although protein and carbohydrate content derived from analysis with Raman MS showed significant relationships with C:N as predicted (Figures 2a–d), the negative relationship between nucleic acid content and biomass C:P was relatively weaker for both species and only significant for P. carotovorum (Figures 2e and f). The weaker relationship between biomass P and nucleic acid content has two probable sources. First, RNA and DNA were analyzed per unit wet mass. Therefore, some of the variation in the nucleic acid—biomass P relationship may have been because of differences in water content of the biomass used for nucleic acid analyses. In addition, it is possible that some of the RNA in the samples analyzed for RNA quantity by chemical analyses had disproportionate RNAse activity compared with those samples analyzed by Raman MS. However, we did conduct a time-series analysis where we measured RNA at several intervals during sampling handling and preparation and saw no decrease in RNA quantity during the time it took to prepare the samples and for several hours beyond that period (data not shown). Therefore, it is unlikely that RNA in the samples processed for chemical analysis experienced disproportionate RNA degradation compared with those samples processed for Raman MS. More likely, bacteria are known to accumulate significant quantities of P as polyphosphates when P is available in excess of immediate biomass requirements (Kornberg et al., 1999). The relationship between biomass P and nucleic acid content has been shown to decouple when cells are grown on P-rich media (Makino and Cotner, 2004). However separating the data by growth phase or by P-treatment did not reveal an effect of either on the nucleic acid—biomass C:P relationship (data not shown). Regardless, it is still likely that for some treatments nucleic acids were not the main pool of P in the cell, and therefore a weaker relationship between biomass C:P and nucleic acid content would be expected.
In addition to a clearer understanding of the contribution of macromolecular composition to biomass stoichiometry, analysis with Raman MS gave insight into how variance in resource stoichiometry affected microbial physiology. Previous studies have shown that cells in logarithmic and stationary phase differed significantly in their Raman spectra but the differences were not greater than the differences between species (Huang et al., 2004). In our study, PCA and CDA analyses revealed greater variation of macromolecular composition between growth phases and between resource treatments than between species (Figure 3). Moreover, cells in stationary phase had highly variable macromolecular composition, showing significant population heterogeneity (Figure 5). These results suggest that microorganisms sampled from the natural environment, where many cells are quiescent (for example, in stationary phase) and resources are heterogeneous, are unlikely to be taxonomically distinguishable on the basis of Raman MS spectra alone. Although an unconstrained analysis (PCA) was unable to differentiate between species, the CDA using just the two species as a grouping criterion was able to accurately assign species identity to a cell in 97.9% of the cases. This constrained CDA allowed to ask in which manner the macromolecular composition of species differed from each other.
Successful species differentiation by the CDA was because of differences in the least variable Raman peaks. This result has interesting and somewhat intuitive implications. If differences between phylogenetically distinct organisms are present in macromolecules that show low phenotypic plasticity (that is, are conserved across a variable resource environment), this suggests that there may be a basic cellular architecture that defines a species, or at least is conserved within a given phylum. Although this is somewhat intuitive, that is, species are ‘different', it suggests that analysis with Raman MS using a limited subset of identified peaks from the complete Raman spectra, can identify in which manner the macromolecular composition of species differ from one another. This provides independent empirical and mechanistic support for the hypothesis that individual species should differ in their biomass stoichiometry (Makino et al., 2003; Danger et al., 2008). Such linkages between phylogeny and biomass composition are aided by the ability of Raman MS to be coupled with fluorescence in situ hybridization and to detect shifts in isotopic composition of different macromolecules, thus lending itself to isotope-labeled substrate studies (Huang et al., 2007b). The combination of fluorescence in situ hybridization and Raman MS allows information on macromolecular composition to be collected from a well-defined group of target organisms and to identify compositional variability within and between microbial populations. In addition, studies with isotope-labeled substrates can identify how specific resources alter the macromolecular composition of microbial populations organisms, thus linking group specific resource use to shifts in community biomass stoichiometry.
In addition to differences in their least variable peaks, species also differed in their range of macromolecular composition, that is, phenotypic plasticity, in a manner that was consistent with their physiology. Over the full range of resource treatments V. spinosum had a more variable macromolecular composition during stationary phase and a more constrained macromolecular composition during logarithmic phase (Figure 4). These differences in biomass composition are consistent with the significantly (F=18.04, P<0.0001) lower mean growth rate of V. spinosum (0.08 h−1) compared with P. carotovorum (0.20 h−1). Variation in composition of logarithmically growing cells was spread primarily along the second PC indicating differences in protein and nucleic acid content. Compared with the heterogeneous P. carotovorum population, logarithmically growing V. spinosum cells had a relatively high protein and low nucleic acid content independent of resource N:P level (Figure 4), indicating that it may be incapable of realizing a fast growth response and invests into protein-based cellular machinery. These results are consistent with our general understanding of each organism's growth strategy. On an average, fast-growing organisms show a high investment in nucleic acids to maximize growth (Elser et al., 2003). This general pattern among microorganisms provides a physiological basis that links life history to biomass stoichiometry through macromolecular composition. Similar relationships between life history, macromolecular composition and biomass stoichiometry have been identified in metazoans (Gorokhova et al., 2002), however these links have rarely been considered in microorganisms, in part because of the difficulty of evaluating differences in macromolecular composition between microbial species.
The CDA and the PCA both suggest that although species were indistinguishable in an unconstrained analysis, actively growing cells had Raman spectra that were statistically distinguishable from quiescent cells, even when multiple species from a heterogeneous resource environment were considered simultaneously. Although there was significant overlap between the two groups, in the unconstrained PCA log and stationary cells could be visually differentiated (Figure 3b). Using only growth phase as a grouping criterion, the CDA was able to successfully classify 81.2% of all cells. From a biogeochemical perspective, understanding the proportion of the microbial community that is actively growing is a valuable parameter and provides a complementary technique to staining methods that evaluate cellular activity (for example, Lopez-Amores et al., 1998). The ability to visualize cells directly before Raman MS analysis allows for complementary staining (to indicate cellular activity) and Raman MS analysis of undefined microbial communities.
Finally, the position of the loading vectors in principal component space suggested additional information about microbial physiology and ecology. The orthogonal positioning of the proteins and nucleic acids along the second PC indicated that cells with higher protein content had relatively lower nucleic acid content. To some extent both species were able to allocate resources to either proteins or nucleic acids but less so to both macromolecular classes simultaneously. We interpret this as a strategic response to the resource environment and possibly evidence of a physiological trade-off. While low resource N:P treatments led to an investment in nucleic acids, presumably to maximize growth, high resource N:P resulted in investment into proteins, perhaps to increase resource P acquisition. In both species, an increase in protein content was accompanied by a decrease in nucleic acid content, and vice-versa. As proteins and nucleic acids also loaded negatively along the first PC, some cells in our study were rich in both nucleic acids and proteins. However, the cells that were richest in proteins were not also the richest in nucleic acids, consistent with a physiological trade-off. Such trade-offs are well-studied ecological phenomena that have been shown to structure communities in time and space (Tilman, 1982). A study of marine algae revealed that optimization of macromolecules for either light-harvesting, growth or resource acquisition provided a mechanistic explanation of mean algal biomass stoichiometry in marine ecosystems (Klausmeier et al., 2004). Although the study presented here provides only limited evidence of a trade-off between an investment in nucleic acids versus an investment in proteins in these species, it does suggest Raman MS may be a useful tool to evaluate the presence of such trade-offs in a wide variety of microorganisms.
In conclusion, analysis of a subset of defined peaks within the Raman spectra gave insight into how resource stoichiometry affected microbial biomass composition, metabolism and ecology. Differences in macromolecular composition as determined by Raman MS correlated with species level traits and physiology. The link between macromolecular composition as determined by Raman MS and elemental biomass composition suggests that Raman is a useful tool for evaluating underlying variance in microbial biomass stoichiometry. This ability is further complemented by the ability of Raman MS to be coupled with phylogenetic identification and labeled substrate studies (Huang et al., 2007b) thus, providing the opportunity to link stoichiometric analyses with phylogenetic identification at the level of the single cell. The application of Raman MS to environmental microorganisms allows ecological stoichiometry, an increasingly utilized body of ecological theory, to be applied to microbial ecology in the absence of culture enrichment, making it complementary to molecular techniques currently driving the microbial revolution.
Acknowledgments
This study is a contribution from the research network MICDIF supported by the Austrian Research Fund (FWF) through Grants S10007-B17 to TB and S10002-B17 to HD The current version of this manuscript has been greatly improved by comments from G Kowalchuk and four anonymous reviewers.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amann RI.1995In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes Molecular Microbial Ecology ManualAkkeman ADC, , van Elsas JD, de Bruigin FJKluwer Academic Publishers: Dortrecht; 1–15. [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cherif M, Loreau M. When microbes and consumers determine the limiting nutrient of autotrophs: a theoretical analysis. Proc R Soc Lond B. 2009;276:487–497. doi: 10.1098/rspb.2008.0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotner JB, Makino W, Biddanda BA. Temperature affects stoichiometry and biochemical composition of Escherichia coli. Microb Ecol. 2006;52:26–33. doi: 10.1007/s00248-006-9040-1. [DOI] [PubMed] [Google Scholar]
- Danger M, Oumarou C, Benest D, Lacroix G. Bacteria can control stoichiometry and nutrient limitation of phytoplankton. Funct Ecol. 2007;21:202–210. [Google Scholar]
- Danger M, Daufresne T, Lucas F, Pissard S, Lacroix G. Does Liebig's law of the minimum scale up from species to communities. Oikos. 2008;117:1741–1751. [Google Scholar]
- Daniels L, Hanson RS, Phillips J.2007Total carbohydrates by phenol reaction. Methods for General and Molecular MicrobiologyReddy CA, Beveridge TJ, Breznak JA et al.Chemical Analysis ASM Press: Washington, D.C [Google Scholar]
- De Gelder J, Willemse-Rix D, Scholtes MJ, Sanchez JI, Maquelln K, Vandenabeele P, et al. Monitoring Poly(3-hydroxydutyrate) production in Cupriavidus necator DSM 428 (H16) with Raman Spectroscopy. Anal Chem. 2008;80:2155–2160. doi: 10.1021/ac702185d. [DOI] [PubMed] [Google Scholar]
- De Gelder J, De Gussem K, Vandenabelle P, De Vos P, Moens L. Methods for extracting biochemical information from bacterial Raman spectra: an explorative study on Cupriavidus metallidurans. Analytica Chimica Acta. 2007;585:234–240. doi: 10.1016/j.aca.2006.12.050. [DOI] [PubMed] [Google Scholar]
- Elser JJ, Dobberfuhl DR, Mackay NA, Schampel JH. Organism size, life history, and N:P stoichiometry. BioScience. 1996;46:674–684. [Google Scholar]
- Elser JJ, Acharya K, Kyle M, Cotner JB, Makino W, Markow TA, et al. Growth rate-stoichiometry couplings in diverse biota. Ecol Lett. 2003;6:936–943. [Google Scholar]
- Goral J, Zichy V. Fourier transform Raman studies of materials and compounds of biological importance. Spectrochim Acta. 1990;46A:253–275. [Google Scholar]
- Gorokhova E, Dowling TE, Weider LJ, Crease TJ, Elser JJ. Functional and ecological significance of rDNA intergenic spacer variation in a clonal organism under divergent selection for production rate. Proc R Soc Lond B. 2002;269:2373–2379. doi: 10.1098/rspb.2002.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall EK, Dzialowski AR, Stoxen SM, Cotner JB. The effect of temperature on the coupling between phosphorus and growth in lacustrine bacterioplankton communities. Limnol Oceanogr. 2009;54:880–889. [Google Scholar]
- Howard WFJ, Nelson WH, Sperry JF. A resonance Raman method for the rapid detection and identification of bacteria in water. App Spectrosc. 1980;34:72–75. [Google Scholar]
- Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol. 2007a;9:1878–1889. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- Huang WE, Bailey MJ, Thompson IP, Whiteley AS, Spiers AJ. Single-cell Raman spectral profiles of Pseudomonas fluorescence SBW25 reflects in vitro and in planta metabolic history. Microb Ecol. 2007b;53:414–425. doi: 10.1007/s00248-006-9138-5. [DOI] [PubMed] [Google Scholar]
- Huang WE, Griffiths R, Thompson IP, Bailey MJ, Whiteley AS. Raman microscopic analysis of single microbial cells. Anal Chem. 2004;76:4452–4458. doi: 10.1021/ac049753k. [DOI] [PubMed] [Google Scholar]
- Keiblinger KM, Hall EK, Szukics U, Haemmerle I, Ellersdorfer G, Sterflinger K, et al. 2010The effect of resource quantity and resource stoichiometry on microbial carbon use efficiency FEMS Microbiol Ecoldoi: 10.1111/j.1574-6941.2010.00913.x [DOI] [PubMed]
- Kim S, Kaplan LA, Hatcher PG. Biodegradable dissolved organic matter in a temperate and a tropical stream determined from ultra-high resolution mass spectrometry. Limnol Oceanogr. 2006;51:1054–1063. [Google Scholar]
- Klausmeier CA, Litchman E, Daufresne T, Levin SA. Optimal nitrogen-to-phosphorus stoichiometry of phytoplankton. Nature. 2004;429:171–175. doi: 10.1038/nature02454. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Legendre L, Legendre P. Numerical Ecology. Elsevier Science: Amsterdam; 2003. [Google Scholar]
- Lopez-Amores R, Comas J, Garcia MT, Vives-Rego J. Use of the 5-cyano-2,3-ditolyl tetrazolium chloride reduction test to asses respiring marine bacteria and grazing effects by flow cytometry during linear alkylbenzene sulfonate degradation. FEMS Microbiol Ecol. 1998;27:33–42. [Google Scholar]
- Makino W, Cotner JB. Elemental stoichiometry of a heterotrophic bacterial community in a freshwater lake: implications for growth and resource dependent variations. Aquat Microb Ecol. 2004;34:33–41. [Google Scholar]
- Makino W, Cotner JB, Sterner RW, Elser JJ. Are bacteria more like plants or animals? Growth rate and resource dependence of bacterial C:N:P stoichiometry. Funct Ecol. 2003;17:121–130. [Google Scholar]
- Manzoni S, Jackson RB, Trofymow JA, Porporato A. The global stoichiometry of nitrogen mineralization. Science. 2008;321:684–686. doi: 10.1126/science.1159792. [DOI] [PubMed] [Google Scholar]
- Maquelin K, Kirschner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, et al. Identification of medically relevant microorganisms by virbrational spectroscopy. J Microbiol Methods. 2002;51:255–271. doi: 10.1016/s0167-7012(02)00127-6. [DOI] [PubMed] [Google Scholar]
- Norland S, Fagerbakke K, Heldel M. Light element analysis of individual bacteria by X-ray microanalysis. Appl Environ Microbiol. 1995;61:1357–1362. doi: 10.1128/aem.61.4.1357-1362.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérombelon MCM. Ecology of the soft rot Erwinias. Ann Rev Phytopathol. 1980;18:361–387. [Google Scholar]
- R-Development-Core-Team . R: A language and environment for statistical computing, R Foundation for statistical computing. R-Development-Core-Team: Vienna, Austria; 2005. [Google Scholar]
- Sangwan P, Chen X, Hugenholtz P, Janssen PH. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl Environ Microbiol. 2004;70:5875–5881. doi: 10.1128/AEM.70.10.5875-5881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinner F, Öhlinger R, Kandeler E, Margesin R. Bodenbiologische Arbeitsmethoden. Springer Labor: Berlin, Germany; 1993. [Google Scholar]
- Schlesner H. Verrucomicrobium spinosum gen. nov., sp. nov,: a fimbriated prosthecate bacterium. System Appl Microbiol. 1987;10:54–56. [Google Scholar]
- Schuster KC, Reese I, Urlaub E, Gapes JR, Lendl B. Multidimensional information on the chemical composition of single bacterial cells by confocal Raman microspectroscopy. Anal Chem. 2000;72:5529–5534. doi: 10.1021/ac000718x. [DOI] [PubMed] [Google Scholar]
- Sterner RW, Elser JJ. Ecological stoichiometry: The biology of the elements from the molecules to the biosphere. Princeton University Press: Princeton, New Jersey; 2002. [Google Scholar]
- Tilman DG. Resource competition and community structure. Princeton University Press: Princeton; 1982. [PubMed] [Google Scholar]
- Wagner M. Single cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411–429. doi: 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.