Abstract
Enhanced biological phosphorus removal (EBPR) activated sludge communities enriched in ‘Candidatus Accumulibacter' relatives are widely used in wastewater treatment, but much remains to be learned about molecular-level controls on the EBPR process. The expression of genes found in the carbon and polyphosphate metabolic pathways in Accumulibacter was investigated using reverse transcription quantitative PCR. During a normal anaerobic/aerobic EBPR cycle, gene expression exhibited a dynamic change in response to external acetate, oxygen, phosphate concentrations and probably internal chemical pools. Anaerobic acetate addition induced expression of genes associated with the methylmalonyl-CoA pathway enabling the split mode of the tricarboxylic acid (TCA) cycle. Components of the full TCA cycle were induced after the switch to aerobic conditions. The induction of a key gene in the glyoxylate shunt pathway was observed under both anaerobic and aerobic conditions, with a higher induction by aeration. Polyphosphate kinase 1 from Accumulibacter was expressed, but did not appear to be regulated by phosphate limitation. To understand how Accumulibacter responds to disturbed electron donor and acceptor conditions, we perturbed the process by adding acetate aerobically. When high concentrations of oxygen were present simultaneously with acetate, phosphate-release was almost completely inhibited, and polyphosphate kinase 1 transcript abundance decreased. Genes associated with the methylmalonyl-CoA pathway were repressed and genes associated with the aerobic TCA cycle exhibited higher expression under this perturbation, suggesting that more acetyl-CoA was metabolized through the TCA cycle. These findings suggest that several genes involved in EBPR are tightly regulated at the transcriptional level.
Keywords: enhanced biological phosphorus removal, ‘candidatus accumulibacter', gene expression, RT-qPCR
Introduction
Enhanced biological phosphorus removal (EBPR) is widely used to remove phosphorus (P) from wastewater in order to prevent the receiving water body eutrophication. By cycling activated sludge under sequential anaerobic and aerobic conditions in conventional EBPR processes, some microorganisms can accumulate large amounts of polyphosphate (polyP). These microorganisms are thus referred to as ‘polyP accumulating organisms' (PAOs; Oehmen et al., 2007). When fed with volatile fatty acids under anaerobic conditions, PAOs take them up and convert them to polyhydroxyalkanoates (PHAs), presumably using the energy provided by polyP degradation, resulting in inorganic phosphate (Pi) release. When switched to aerobic conditions, PAOs oxidize PHAs, providing energy for growth and polyP formation, leading to P removal from the wastewater.
Using the 16S rRNA gene as a genetic marker, a bacterial group phylogenetically close to Rhodocyclus tenuis in the Betaproteobacteria, named ‘Candidatus Accumulibacter phosphatis' (henceforth referred to as Accumulibacter; Hesselmann et al., 1999), has been identified as a primary PAO in acetate or propionate-fed lab-scale reactors (Hesselmann et al., 1999; Crocetti et al., 2000; McMahon et al., 2002; Pijuan et al., 2004) and several full-scale wastewater treatment plants (Zilles et al., 2002; Kong et al., 2004; He et al., 2008). The polyP kinase 1 (ppk1) gene was used to reveal a fine-scale population structure within the Accumulibacter lineage, which is comprised of two major types, each consisting of several clades (He et al., 2007; Peterson et al., 2008). Clades IA and IIA were found as the dominant Accumulibacter in acetate-fed lab-scale reactors (He et al., 2007). The complete Accumulibacter clade IIA genome sequence and partial sequence of clade IA was obtained through metagenomic sequencing of lab-scale EBPR sludges (Garcia Martin et al., 2006).
PolyP formation is the key step in EBPR. In some bacteria, environmental Pi limitation or carbon starvation are triggers for polyP accumulation (Wanner, 1996). In activated sludge, polyP accumulation appears to be promoted by physical/temporal separation of electron donor (usually volatile fatty acids) and acceptor (oxygen or nitrate; Mulkerrins et al., 2004; Barnard and Abraham, 2006). In practice, deteriorated EBPR performance was sometimes caused by simultaneous presence of electron donor and acceptor. For example, the alleviated oxygen concentrations in the anaerobic zone because of perturbations, such as heavy rainfalls could lead to poor P removal (Shehab et al., 1996; Brdjanovic et al., 1998). Furthermore, omitting the anaerobic phase (referred to as ‘continuous aeration' by some wastewater engineers) eventually resulted in loss of EBPR activity, presumably due to Accumulibacter metabolic shift from PAO to non-PAO mode (Pijuan et al., 2006). Therefore, we are interested in how PAOs, particularly Accumulibacter, regulate their gene expression in response to relative electron donor and acceptor availabilities during a normal EBPR cycle, as well as under perturbed conditions. Upon being taken up, acetate is converted to acetyl-CoA, which can be fluxed into multiple pathways (Figure 1). Therefore, we chose the key gene(s) associated with these routes (Figure 1) and polyP metabolism, attempting to identify which pathways were up- or down-regulated under conditions tested. Reverse transcription quantitative PCR (RT-qPCR) was employed to monitor gene expression during a normal EBPR cycle at a sub-hourly resolved timescale. We also compared gene expression under normal conditions to the perturbed conditions with simultaneous presence of electron donor and acceptor, namely ‘oxygen-acetate contact' in this study specifically.
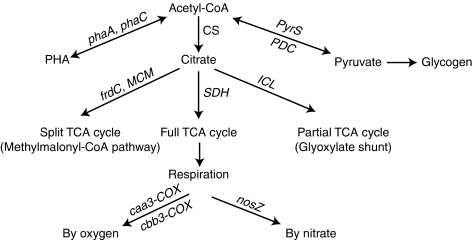
Figure 1.
Multiple routes for acetyl-CoA entering the carbon metabolic pathways based on the Accumulibacter clade IIA genome-based metabolic reconstruction (Garcia Martin et al., 2006). The key genes in these pathways are shown as their abbreviations used in this study, and were studied using RT-qPCR. See Table 1 for the full names of the enzymes and gene symbols.
Materials and methods
Reactor profiling and chemical analysis
A lab-scale acetate-fed sequencing batch reactor was operated under cyclic anaerobic/aerobic (normal EBPR) conditions, as described in He et al. (2006). Under steady-state operating conditions (represented by relatively constant total suspended solid, volatile suspended solid, soluble Pi concentrations at the end of the anaerobic and aerobic phases), we monitored the chemical profiles in the bulk liquid during an EBPR cycle. Soluble Pi was measured by an ascorbic acid method (Standard Method 4500-P E; APHA, 1995). Acetate, NO3-N and NO2-N were measured by a Shimadzu high performance liquid chromatography (Shimadzu Co., Columbia, MD, USA) equipped with an Alltech Previal Organic Acid Column (Alltech Associates, Deerfield, IL, USA) with the UV detector setting at 210 nm for acetate and 214 nm for NO3-N and NO2-N. Total suspended solid and volatile suspended solid were measured according to Standard Methods 2540B and 2540E, respectively (APHA, 1995).
Oxygen–acetate contact batch test
At the end of the aerobic phase, 500 ml of sludge was taken from the reactor to establish two 250 ml batches: a control batch where normal operation was resumed and a treatment batch for oxygen-acetate contact (‘aerated batch'). Nitrogen gas was sparged to establish an oxygen-free environment in the control batch. For the treatment batch, air was delivered to maintain the aerobic conditions. Once anaerobic or aerobic conditions were established, indicated by the dissolved oxygen (DO) concentration, nutrient and acetate feeds were added to both batches to reach initial Pi and acetate concentrations of 7 mg per -P and 100 mg per -acetate, respectively, comparable to their concentrations in the reactor. The pH was controlled at 7.5±0.1 by manually adding HCl or Na2CO3. The control batch was incubated anaerobically for 100 min following nutrient and acetate feed addition. After that, nitrogen sparging was switched to aeration. The profiles of soluble Pi and acetate in both batches, and DO in the treatment batch were measured.
Accumulibacter population characterization
The Accumulibacter percentage in the bacterial community was estimated by fluorescence in situ hybridization with PAOMIX probes (Crocetti et al., 2000), with the total bacterial community enumerated by 4′,6-diamidino-2-phenylindole staining (He et al., 2008). To estimate the relative abundances of Accumulibacter clades within the lineage, genomic DNA from sludge was extracted using the PowerSoil DNA Isolation Kit with bead beating (Mo-Bio Laboratories Inc., Carlsbad, CA, USA). Quantitative real-time PCR (qPCR) with Accumulibacter clade-specific primers targeting ppk1 was performed (He et al., 2007).
Primer design
In addition to the Accumulibacter 16S rRNA and clade-specific ppk1 primers (ppk1-IA and ppk1-IIA for clades IA and IIA, respectively; He et al., 2007), new primers were designed for genes hypothesized to be involved in carbon transformations and the electron transfer chain (Figure 1), to simultaneously target both clades IA and IIA (Table 1). Genes from clade IIA (binned as ‘Candidatus Accumulibacter phosphatis' on IMG/M (http://img.jgi.doe.gov/cgi-bin/m/main.cgi)) and genes likely from clade IA (see Supplementary Text 1 and Supplementary Table S1) were aligned with homologs from bacteria that share relatively high DNA sequence identity with Accumulibacter. DNA fragments unique to Accumulibacter were identified and further compared with sequences in Genbank to verify the specificity for Accumulibacter. PCR condition optimization procedures are described in Supplementary Text 2.
Table 1. Summary of primers used in this study.
| Gene | Gene symbol | Abbreviations in this study | Reference gene OID in MGM/M | Gene length (bp) | Primer target site | Primer sequence (5′-3′) | Amplicon length (bp) | Ta (oC)a | PCR efficiency |
|---|---|---|---|---|---|---|---|---|---|
| Pyruvate dehydrogenase complex | pdh | PDC | 2001099980 | 2184 | 383–405 | GGATTCACAACTACATTCGCTGG | 137 | 63 | 0.763 |
| 500–519 | GTGCCAGAACCACGAGAAGC | ||||||||
| Pyruvate synthase (pyruvate flavodoxin/ferredoxin oxidoreductase) | porC | PyrS | 2001062970 | 642 | 112–131 | ATTGCTGCSTTTGAGGAAGG | 227 | 61 | 0.746 |
| 319–338 | CTGGTGTTGATCAGGATGTA | ||||||||
| Citrate synthase | gltA | CS | 2001061560 | 1293 | 650–672 | TGGTGCATGCCCTGGACACCATC | 184 | 63 | 0.87 |
| 811–833 | TCGAGCATCTGCAGGCAGGCCTC | ||||||||
| Isocitrate lyase | aceA | ICL | 2001006860 | 1299 | 282–302 | TGCCGACAACAACTCGTACGC | 284 | 61 | 0.751 |
| 543–565 | AAGCCAGTTGATCTTCCCAGTGC | ||||||||
| Succinate dehydrogenase/ fumarate reductase, Fe-S protein subunit | sdhB | SDH | 2001061540 | 714 | 236–258 | TCACCGAGATCAAGGACCTCAAG | 206 | 63 | 0.655 |
| 421–441 | CTCGTAGAGGCCGTTGAGTTC | ||||||||
| Fumarate reductase subunit C | frdC | frdC | 2001024420 | 399 | 51–72 | GAATCCGTTCTTCGTCGAGTAC | 209 | 61 | 0.741 |
| 240–259 | TGAACCAGGTGTAGGTGTGG | ||||||||
| Methylmalonyl-CoA mutase, N-terminal domain/subunit | mutA | MCM | 2001062840 | 2019 | 638–660 | TCGAGCGCATCTCGTTCTTTGTC | 207 | 61 | 0.745 |
| 820–844 | CGATTCTCTGSACATTGTTTTCCGG | ||||||||
| Acetyl-CoA acetyltransferase | phaA | phaA | 2001070320 | 1182 | 158–178 | TGATGGGCTGCTGCCTGATG | 154 | 61 | 0.683 |
| 289–311 | AGCAGTTCGTCRTGCGCGATCAT | ||||||||
| Poly(3-hydroxyalanoate) synthase | phaC | phaC | 2001070280 | 1113 | 499–520 | CTCAATCCAGACAAGATCAGCC | 178 | 63 | 0.786 |
| 657–676 | ACGACTCGTTGAGCACGGTG | ||||||||
| Heme/copper-type cytochrome/ quinol oxidases, subunit 1 | ctaD | caa3-COX | 2001010230 | 1608 | 800–819 | ACATTCTGTTGCTGCCGATC | 209 | 63 | 0.808 |
| 989–1008 | CAGGGAGATGAACATCGTGC | ||||||||
| Cbb3-type cytochrome oxidase, subunit 1 | ccoN | cbb3-COX | 2001063240 | 1422 | 843–862 | GGGCGGCATGATCAACGGYG | 250 | 65 | 0.869 |
| 1069–1092 | CGGCAACAGGTAGTAGATCGAACC | ||||||||
| Nitrous oxide reductase | nosZ | nosZ | 2001002170 | 2298 | 1119–1139 | CGAATCGAAGGATCCCTACGG | 194 | 61 | 0.746 |
| 1293–1312 | CGCTGATCTTGTCGAGCACC | ||||||||
| Polyphosphate kinase from Accumulibacter clade IA | ppk1 | ppk1-IA | 2000024060 | 1437b | 79–96 | GACGAAGAAGCGGTCAAG | 408 | 61 | 0.83 |
| 469–486 | AACGGTCATCTTGATGGC | ||||||||
| Polyphosphate kinase from Accumulibacter clade IIA | ppk1 | ppk1-IIA | 2001025850 | 2121 | 893–912 | AGTTCAATCTCACCGAGAGC | 105 | 61 | 0.85 |
| 980–997 | GGAACTTCAGGTCGTTGC | ||||||||
| 16S rRNA from total Accumulibacter | 16S | 2001008560 | 1262b | 287–303 | CCAGCAGCCGCGGTAAT | 350 | 65 | 0.81 | |
| 616–636 | GTTAGCTACGGCACTAAAAGG |
Ta, Annealing temperature.
The predicted gene length is not available because of its partial sequence in IMG/M. The number indicated is the length from the currently available partial gene sequence fragment.
RNA extraction and purification
Cell pellets were collected at intervals indicated in Figures 2 and 3 for RT-qPCR analysis, by centrifugation at 8 000 × g for 3 min at room temperature, followed by immediate flash-freezing in an ethanol/dry ice bath, and storage at −80 °C. RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA, USA), including a 2 min bead-beating step with a spherical ceramic bead at the speed setting of 3 on a Mini-beadbeater (BioSpec Products, Bartlesville, OK, USA). An on-column DNase I digestion was applied to remove DNA contamination (Qiagen). A second round of DNase digestion was performed using TURBO DNA-free DNase kit (Ambion, Austin, TX, USA) to further remove trace DNA contamination. RNA concentration was estimated using microspectrophotometry (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, DE, USA).
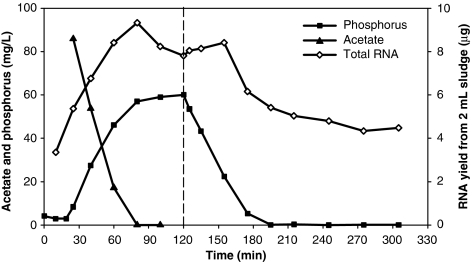
Figure 2.
Phosphorus and acetate concentrations during a normal EBPR cycle, along with total RNA yield extracted from 2 ml of biomass. A total of 16 biomass samples were collected for RT-qPCR analyses at time points indicating corresponding total RNA yield (open diamonds). The vertical dashed line indicates the switch from the anaerobic to the aerobic phase.
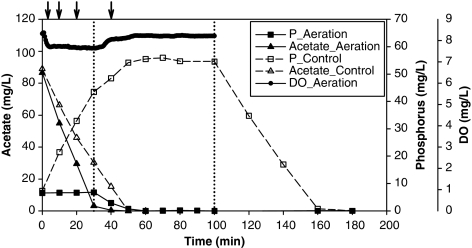
Figure 3.
Acetate (y axis on the left) and phosphorus (the first y axis on the right) concentrations from the aerated and control batches, and dissolved oxygen (DO) concentrations (the second y axis on the right) from the aerated batch. The first vertical dashed line indicates the transition from the ‘feast' to the ‘famine' phase in the aerated batch, which is defined by the acetate concentrations, and the second vertical dashed line indicates the switch from the anaerobic to the aerobic phase in the control batch. Arrows indicate the time points when samples were collected for RT-qPCR analyses.
RT-qPCR
Complementary DNA (cDNA) was generated from 1–5 μg of total RNA, primed by random hexamers (SuperScript III first-strand synthesis system, Invitrogen, Carlsbad, CA, USA). The reaction was terminated by incubation at 85 °C for 5 min and RNase H treatment was performed to degrade RNA in RNA:DNA hybrids. To quantify 16S rRNA, cDNA was diluted by 800-fold with TE buffer (10 m Tris, 1 m EDTA, pH 8) to fit in the dynamic range of PCR. For other genes, cDNA was diluted by threefold with TE buffer. Subsequently, 1 μl of diluted cDNA was applied as the template in qPCR. All quantifications were performed in triplicate. RNA samples without reverse transcription were used as no RT (reverse transcription) controls to evaluate DNA contamination for all primers tested.
qPCR was conducted on an iCycler using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) with a total reaction volume of 15 μl. Except for ppk1 and 16S rRNA genes (reaction conditions described in He et al. (2007)), primers and betaine were added to 400 n and 0.5 respectively. The standard curve generation is described in Supplementary Text 2. All qPCR programs consisted of an initial 3-min denaturation at 95 °C, followed by 45 cycles of denaturing at 94 °C for 30 s, annealing for 45 s and extension at 72 °C for 30 s.
For all RNA samples tested with all primer sets, the average of the minimal difference in Ct (threshold cycle) values between the cDNA and no RT control was 15 cycles, indicating the DNA contamination was negligible after two rounds of DNase digestion. Therefore, the RT-qPCR assays had very low detection limits (<10 copies per reaction for all primer sets, except for citrate synthase (CS) and ppk1-IIA, for which the limits were ∼50 copies per reaction).
Data analysis
The estimated cDNA copy number per qPCR reaction was first normalized by the sampled sludge volume (see Supplementary Text 3). This effectively assumes equivalent RNA extraction efficiencies across samples. The transcription profile during an EBPR cycle was obtained by normalizing the transcript level of each gene at any time point to the first time point. Differential expression between the aerated and control batches was obtained by pairwise comparison of the transcript abundance at each individual sampling point.
Results
Accumulibacter population characterization
During the study period, total Accumulibacter abundance was estimated to be higher than 70% of total cells, and IIA was the dominant clade relative to total Accumulibacter (Table 2).
Table 2. Summary of population structure, reaction kinetics and stoichiometry.
| Experiment | Total Acc (%)a | IA (%)b | IIA (%)b | Prel rate (mmol gVSS−1 h−1) | Pupt rate (mmol gVSS−1 h−1) | Acupt rate (mmol gVSS−1 h−1) | Prel/Acupt (P-mol /C-mol) |
|---|---|---|---|---|---|---|---|
| Reactor transcriptional profiling | 76±11 | 15±2 | 85±2 | 3.9 | 3.6 | 3.7 | 0.52 |
| Aerated batch | 74±13 | 28±4 | 72±4 | 0.03 | 1.36 | 5.2 | 0.003 |
| Control batch | 74±13 | 28±4 | 72±4 | 4.2 | 3.3 | 3.6 | 0.59 |
Abbreviation: VSS, volatile suspended solid.
The percentage of total Accumulibacter in the bacterial community, determined using Accumulibacter-targeted fluorescence in situ hybridization and 4′,6-Diamidino-2-phenylindole staining of total cells.
The percentage of clade IA or IIA relative to total Accumulibacter (the sum of clades IA and IIA), determined using quantitative PCR.
Reactor chemical profiling during a normal EBPR cycle
Soluble P and acetate concentrations during an EBPR cycle from a steady-state bioreactor were monitored (Figure 2). Acetate was fully consumed within the first 60 min after its addition, with a P-release to acetate-uptake ratio (Prel/Acupt) of 0.52 P-mol/C-mol. P was completely taken up 75 min after switching to aeration. Nitrate was <0.1 mg l−1 as NO3-N and nitrite was below detection (0.001 mg l−1 as NO2-N). Standard EBPR performance measures in the form of uptake and release kinetics and stoichiometry were calculated (Table 2).
Oxygen–acetate contact batch test
Batch tests were conducted to investigate the effects of simultaneous presence of oxygen and acetate (Figure 3). The EBPR activity in the control batch was comparable to that from the reactor profiling experiment (Table 2). In the aerated batch, P-release was almost negligible (<0.5 mg per -P in total) and the acetate-uptake rate was markedly higher than in the control. P-uptake activity was observed after the cells entered the famine phase, although at a lower rate than the control (Table 2).
Transcriptional profiling during a normal EBPR cycle
To study gene transcription during a cycle, we collected biomass samples 5 min after acetate addition, 5 min after switching to aeration and every 15–30 min during the cycle, resulting in a total of 16 time points, with total RNA yield profile shown in Figure 2. The first sample was collected 10 min before acetate addition. Transcript abundances in subsequent samples were normalized to this time point. Supplementary Figure S1 lists the individual transcription profiles of all genes studied. Based on the DO, soluble acetate and P concentrations, we divided an EBPR cycle into four phases: anaerobic feast, anaerobic famine, P-rich aerobic famine and P-depleted aerobic famine (as indicated by vertical dashed lines in Figure 4 and Supplementary Figure S1).
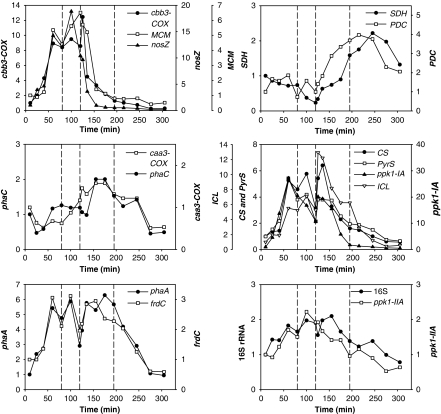
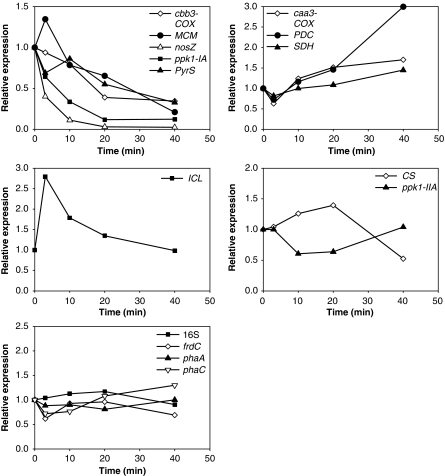
Figure 4.
Transcription profiles categorized based on trends of change in transcript abundance during a normal EBPR cycle. The y axes show transcript levels normalized to the first sampling time point. Profiles were categorized based on clusters identified in Supplementary Figure S2. The three vertical dashed lines divide the entire EBPR cycle into four phases: anaerobic feast, anaerobic famine, P-rich aerobic famine and P-depleted aerobic famine based on the DO, soluble acetate and P concentrations.
Based on the trend in transcript abundance (without considering the scale of the fold change), some genes exhibited similar transcriptional patterns. These were grouped into categories in Figure 4, based on the clustering pattern generated from the heatmap function in the R software package (R Development Core Team, 2008; Supplementary Figure S2). For example, genes nitrous oxide reductase (nosZ), cbb3-type cytochrome oxidase (cbb3-COX) and methylmalonyl-CoA mutase (MCM) formed an anaerobically upregulated gene category. Transcripts from genes pyruvate dehydrogenase complex (PDC) and succinate dehydrogenase (SDH) increased in abundance during the early aerobic phase. The abundance of ppk1-IA, CS, pyruvate synthase (PyrS) and isocitrate lyase (ICL) transcripts increased with acetate addition, but decreased during the anaerobic famine phase. When DO was provided, their transcript abundances increased again within the first 5–10 min, and started to drop with low levels maintained for the rest of the cycle. Genes Acetyl-CoA acetyltransferase (phaA) and fumarate reductase (frdC) had similar profiles, but the major decrease in transcript abundance in the aerobic phase occurred after P was completely taken up.
Supplementary Figure S1 also indicates the maximal fold change (MFC) in transcript abundance for each gene, which is the ratio of the maximal to minimal transcript abundance across a cycle. Gene nosZ experienced the largest change (110-fold), followed by cbb3-COX (51-fold) and ppk1-IA (50-fold), which was much higher than the fourfold MFC of ppk1-IIA. The Accumulibacter-16S rRNA level varied with a MFC of 2.7 during a cycle. Although this MFC was the smallest among the genes studied, it was statistically significant (t-test P-value P<0.05). The profile was similar to the total RNA yield profile (Figure 2; correlation coefficient=0.86). Further discussion of rRNA abundance profile can be found in Supplementary Text 4.
Transcriptional change responding to oxygen–acetate contact
RNA samples were collected from the aerated and control batches at the same time points to conduct pairwise comparisons (Figure 3). The relative expression over time under perturbed conditions was compared with the control (Figure 5). When DO was provided with acetate, genes cbb3-COX, nosZ, MCM, ppk1-IA and PyrS had statistically lower transcript levels than in the control at maximal difference (P<0.05). Notably, a 41-fold and 8-fold maximal difference was observed for nosZ and ppk1-IA, respectively. Genes caa3-COX (Heme/copper-type cytochrome oxidase), PDC and SDH had higher transcript levels in the treatment, relative to the control at maximal difference (P<0.05). ICL transcripts exhibited the largest difference between the treatment and control at 3 min after acetate addition (P=0.01), and transcript levels remained higher in the treatment during the first 10 min (P=0.02), coincident with its sharp increase immediately after switching to aeration during a normal EBPR cycle (Supplementary Figure S1). Under both treatment and control conditions, CS transcription appeared to be induced after acetate addition, and a slightly higher (but statistically insignificant) increase was observed in the aerated batch during the feast phase. On the contrary, ppk1-IIA transcript levels were lower during the feast phase (P=0.01), but the difference was not significant after the aerated batch entered the famine phase where cells started to uptake Pi. The transcript levels of the remaining genes remained relatively unchanged (maximal difference <1.5-fold and P>0.05), except for frdC, which had a lower transcript level initially in the aerated batch (P<0.05).
Figure 5.
Relative expression levels in the aerated batch comparative to the control by pairwise comparison of transcript abundances at each sampling time point. Relative expression higher than 1 indicates induction and lower than 1 indicates repression in the aerated batch compared with the control. Among these genes, 16S, frdC, phaA and phaC expression was not statistically different between the aerated and control batches (maximal difference <1.5-fold and P>0.05).
Discussion
Gene transcription patterns
Electron donor and acceptor decoupling, along with the cyclic alteration of feast/famine and anaerobic/aerobic conditions, generate a unique niche for bacteria capable of taking up organic carbon anaerobically for subsequent aerobic energy generation, which may differentiate their regulation in response to oxygen availabilities from fermentation or regular aerobic respiration. As transcription is the first level and major point of regulation in prokaryotes, we studied the transcription of some key genes in EBPR metabolism, to understand (1) under normal EBPR conditions how Accumulibacter regulates its gene expression, as compared with non-EBPR model bacteria, and (2) under perturbed conditions with simultaneous electron donor and acceptor presence, how Accumulibacter changes its gene expression, as compared with the normal EBPR mode.
We first point out that we generally assume that an increase in transcript abundance equates to upregulation, and a decrease equates to downregulation. We acknowledge that alternate mechanisms may explain changes in transcript abundance, although we are measuring the transcript concentration at any time point and not transcript flux. Similarly, we sometimes interpret an increase in transcript abundance to imply the operation of an associated biochemical pathway at that time point, acknowledging that genes could be up- or down-regulated in response to particular environmental cues, making an enzyme or pathway available for a subsequent phase. We caution readers to remember, for example, that anaerobic upregulation does not necessarily mean that the enzyme is essential for anaerobic EBPR activity. However, we follow the most parsimonious interpretation (expression begets activity in that phase).
In aerobic and facultative anaerobic bacteria, Fnr and ArcA/ArcB are two major global regulators controlling gene expression under different oxygen availabilities (Lynch and Lin, 1996). Many studies have been conducted on model bacteria to compare aerobic and anaerobic transcript profiling. For example, when Escherichia coli was switched from anaerobic to aerobic conditions, transcripts involved in anaerobic metabolism decreased in abundance and transcripts associated with aerobic metabolisms such as the tricarboxylic acid (TCA) cycle and respiratory chain increased (Partridge et al., 2006). Our results suggest that nosZ, cbb3-COX and MCM were upregulated in the anaerobic phase during normal EBPR conditions, and repressed by the perturbation with oxygen–acetate contact. In a separate experiment, we found that nosZ transcription was induced by nitrate presence under anaerobic conditions (unpublished data). These suggest the availabilities of oxygen and nitrogen oxides form two layers of regulation on nosZ, as observed in Pseudomonas stutzeri (Korner and Zumft, 1989). Gene cbb3-COX encodes a high-affinity cytochrome C oxidase in model organisms (Richardson, 2000) and its expression was negatively controlled by high DO concentrations in our study, suggesting that cbb3-COX is also a high-affinity cytochrome C oxidase in Accumulibacter, only expressed under microaerobic conditions. MCM catalyzes the conversion of succinyl-CoA to methylmalonyl-CoA, and together with frdC, they form the methylmalonyl-CoA pathway, allowing the left arm of the TCA cycle to operate in reverse. This pathway is important in EBPR metabolic models that involve anaerobic TCA cycle operation, because it alleviates FADH2 (reduced form of flavin adenine dinucleotide) accumulation due to lack of external electron acceptor (Oehmen et al., 2007). In many bacteria, frdC is under positive control by Fnr (Gunsalus and Park 1994; Lynch and Lin 1996). Similarly, we observed an initial rapid increase in frdC transcript levels during the anaerobic phase, but it did not decrease significantly until P concentrations were very low in the late aerobic phase. Taken together, the increased MCM and frdC transcript abundances in the anaerobic phase suggest the methylmalonyl-CoA pathway is anaerobically upregulated in EBPR.
Genes PDC and SDH were upregulated in the early aerobic phase, but their transcript abundances decreased in the late aerobic phase, probably associated with regulation by internal carbon pools. Gene caa3-COX also exhibited higher transcript levels in the early aerobic phase, but its change was relatively small. Correspondingly, these three genes were induced under the oxygen–acetate contact perturbation. Among them, caa3-COX encodes a low-affinity cytochrome C oxidase (Richardson, 2000) and was expected to be turned on in the aerobic phase when the DO level was high. SDH converts succinate to fumarate with FADH2 generation. This gene is negatively controlled by Fnr and ArcA in E. coli (Gunsalus and Park, 1994; Lynch and Lin, 1996), thus inhibiting the forward direction of the TCA cycle under anaerobic conditions. Its aerobic induction in Accumulibacter may indicate that the forward full TCA cycle was upregulated when oxygen was present. PDC catalyzes oxidative decarboxylation of pyruvate to acetyl-CoA with NADH generation mostly in aerobic bacteria, and the reaction is generally regarded to be irreversible (Patel and Roche, 1990). In E. coli, PDC is also negatively regulated by Fnr or ArcA (Lynch and Lin, 1996). Accumulibacter PDC might be under a similar regulation based on our observed aerobically increased expression. Different from PDC, PyrS catalyzes the reversible conversion between pyruvate and acetyl-CoA using ferredoxin or flavodoxin, instead of NADH as electron carriers, and its forward reaction to generate acetyl-CoA from pyruvate is more common in anaerobes (Menon and Ragsdale, 1997). PyrS exhibited the highest expression in the anaerobic phase, which seems consistent with its general role in anaerobic pyruvate metabolism in model anaerobic bacteria.
Several genes, including PyrS, had an initial increase in transcript abundance followed by a decrease in both phases. Among them, CS facilitates the entrance of acetyl-CoA into the TCA cycle and ICL is the key gene in the glyoxylate shunt pathway, which is necessary in many bacteria when growing on acetate as the sole carbon source to replenish C4 sources for cell biosynthesis (Kornberg, 1966). Both CS and ICL are negatively regulated by ArcA in E. coli (Lynch and Lin, 1996), which is seemingly different from Accumulibacter's ‘biphasic' induction. Similar ‘biphasic' induction patterns were also observed in phaA and ppk1-IA, which can catalyze reversible reactions of PHA and polyP synthesis and degradation, respectively. Their expression patterns suggest they are involved in both the formation and degradation processes in Accumulibacter, although the exact factor(s) triggering their induction in the two phases are not clear. It was suggested that ppk1 was under Pho regulon control in Klebsiella aerogenes (Kato et al., 1993) and Acinetobacter baumannii (Gavigan et al., 1999), with transcription being induced by Pi limitation. If control mechanisms were similar in Accumulibacter, ppk1 transcript abundance would be expected to increase in the Pi-depleted aerobic famine phase. However, both ppk1-IA and ppk1-IIA did not exhibit such a pattern. Instead, both had higher expression when Pi in the bulk liquid was increasing. Therefore, ppk1 expression in Accumulibacter must be subjected to a different control, which may be a critical regulatory component in EBPR physiology. Furthermore, the difference in ppk1 expression patterns between the clade IA and IIA may reflect some physiological differentiation between these two clades, based on different regulatory mechanisms.
Gene phaC encodes a PHA synthase and was found in the proximity of phaA in the genome, but encoded in separate operons. The difference in their expression patterns suggests different regulatory mechanisms for these two genes, although their products are both involved in PHA metabolism. Further discussion on the observed difference in their regulation can be found in Supplementary Text 5.
We note that in our previous metatranscriptomic analysis using microarrays, we only compared the early anaerobic and aerobic phases, defined as 15 min after acetate addition and switching to aeration (He et al., 2010). We found that the majority of the genes tested did not change expression levels by more than twofold. Indeed, when comparing only these two time points, RT-qPCR analysis of the selected genes largely agreed with the microarray results. However, when inspecting the temporally resolved profiles, distinct transcriptional patterns emerged. This suggests that gene expression during an EBPR cycle was dynamic, presumably because of the short half-life of mRNA and the fluctuating environmental conditions in EBPR. It also highlights the need to use more finely resolved temporal analyses (for example, minutes versus hours) to reveal transcription-level changes in gene expression.
Relating transcription patterns to existing EBPR metabolic models
EBPR metabolic models suggest that the reducing equivalents to form PHA anaerobically from acetyl-CoA are provided by glycogen degradation (the Mino model (Mino et al., 1987)) and/or the TCA cycle (the Comeau/Wentzel model (Comeau et al., 1986; Wentzel et al., 1986)). Three modes were proposed for anaerobic TCA cycle operation, namely the full (Comeau et al., 1986), partial (bypass through the glyoxylate shunt; Louie et al., 2000) and split modes (right arm of the TCA cycle operating forward and left arm backward through the methylmalonyl-CoA pathway; Hesselmann et al., 2000). Only the split mode involving the methylmalonyl-CoA pathway can explain how reduced quinone is re-oxidized without external electron acceptor through the reaction catalyzed by frdC (Oehmen et al., 2007). Recently, a number of studies have addressed the contribution of the TCA cycle in Accumulibacter (Burow et al., 2008; Wexler et al., 2009; Zhou et al., 2009). For example, Burow et al. (2008) showed increased ICL expression after acetate addition and switching to aeration. They also observed decreased acetate-uptake and PHA-synthesis rates when the glyoxylate pathway was inhibited, thus suggesting a role for this pathway in anaerobic PHA formation. Wexler et al. (2009) used radiolabeled proteomics to show that the glyoxylate pathway was important in the aerobic phase for both type I and II Accumulibacter, whereas the other TCA cycle modes were more important in the anaerobic phase for type II, probably contributing to reducing power generation for PHA synthesis.
From the transcriptional profiling during a normal EBPR cycle, we identified that the key gene (CS) in the TCA cycle was induced both anaerobically by acetate and aerobically when first switching to aeration. Concurrent with the former induction is methylmalonyl-CoA pathway upregulation associated with the split mode of the TCA cycle. When provided with oxygen, the forward full operational mode of the TCA cycle was induced, indicated by SDH transcript increase. The ICL was induced by both acetate addition and aeration, agreeing with the study by Burow et al. (2008). Its expression pattern suggests that the partial mode through the glyoxylate shunt may be active in both phases, likely more active in the early aerobic phase, indicated by its higher induction by oxygen (Supplementary Figure S1), supporting the conclusion by Wexler et al. (2009) that the glyoxylate pathway is more important during the aerobic phase.
Besides the TCA cycle, the glycolytic pathway along with the conversion of pyruvate to acetyl-CoA also contributes to NADH generation. This process is reversed when PHA is oxidized to acetyl-CoA aerobically to replenish glycogen storage. It was hypothesized that PDC was responsible for anaerobic conversion of pyruvate to acetyl-CoA, and PyrS was responsible for the opposite process in the aerobic phase (Garcia Martin et al., 2006). PyrS exhibited some biphasic induction with the highest expression in the anaerobic phase, and thus might also be involved in the anaerobic process, although this would require explaining how the reduced ferredoxin from the PyrS activity is re-oxidized. On the other hand, PDC was only induced by oxygen, as in well-studied heterotrophs such as E. coli, but contrary to its hypothesized role in anaerobic EBPR metabolism. The expression patterns of PyrS and PDC may suggest the regulation of enzyme activities by intracellular pools of acetyl-CoA, NADH and ferredoxin, and/or regulation at the translational level may be the major control. Alternately, as described above, upregulation in one phase may provide active enzyme primarily for use in a subsequent phase.
Sludge response to oxygen–acetate contact
Several previous studies showed that EBPR activities were still maintained when acetate was fed under aerobic conditions (Guisasola et al., 2004; Pijuan et al., 2005, 2006; Ahn et al., 2007), where significant aerobic P-release was observed, coupled with acetate-uptake, similar to the anaerobic mode. By contrast, we observed negligible aerobic P-release. The discrepancy may have resulted from the relative availability of DO. In our study, the DO level during the entire testing period was higher than 7.6 mg l−1 (Figure 3). As comparison, in the studies by Guisasola et al. (2004) and Pijuan et al. (2005, 2006), the DO was only higher than 2 mg l−1; and especially the study by Ahn et al. (2007) clearly showed that DO quickly dropped to ∼10% of saturation (<1 mg l−1) during the feast phase, where P-release occurred. It was suggested that when the influent chemical oxygen demand was relatively high (for example, >60–120 mg l−1) and DO in the bulk liquid was not sufficiently high (for example, <4 mg l−1), the aerobic region of sludge flocs only occurred at the surface layer because of the rapid microbial oxygen utilization (Li and Bishop, 2004). Therefore, it is important to examine whether inside the sludge flocs was truly ‘aerobic'.
Comparing transcription patterns under aerobic acetate addition to the normal anaerobic counterpart, the full TCA cycle (indicated by SDH) and the partial TCA cycle through the glyoxylate shunt (by ICL) were expressed at higher levels when acetate was aerobically available, as was the aerobic electron transfer chain (by caa3-COX). In contrast, the split TCA cycle via the methylmalonyl-CoA pathway (indicated by MCM) was expressed at a lower level compared with the anaerobic counterpart. Therefore, a large fraction of acetyl-CoA probably flowed to the full and partial TCA cycles with oxygen as the terminal electron acceptor. During the feast phase, ppk1 transcription in both clades IA and IIA was repressed, suggesting that the decreased P-release was not likely because of simultaneous P-release and uptake. This is supported by the in situ ecophysiology characterization of Accumulibacter using FISH-MAR (microautoradiography) with 33Pi, which suggested that when acetate was present together with DO or nitrate, no P-uptake by Accumulibacter could occur (Kong et al., 2004). Thus, a large portion of energy for acetate-uptake was derived from aerobic operation of the TCA cycle, as this is a more efficient way to capture energy than through the transformation of PHA and polyP (Maurer et al., 1997). The transcription of phaA and phaC did not seem to be altered under this perturbation, suggesting that their transcription was not directly regulated by DO concentrations.
It is generally accepted that good EBPR activity requires physical/temporal separation of electron donor and acceptor (Mulkerrins et al., 2004; Barnard and Abraham, 2006), and EBPR performance deterioration in full-scale facilities had been associated with alleviated DO in the anaerobic zone (Shehab et al., 1996). However, lab-scale sludges enriched in Accumulibacter still exhibited EBPR activities under continuous aeration (Guisasola et al., 2004; Pijuan et al., 2005, 2006; Ahn et al., 2007). This raised a question on the role of the anaerobic conditions in EBPR: necessary to flux acetyl-CoA to PHA formation coupled with polyP metabolisms, and/or just as a selective force to eliminate organisms that cannot uptake acetate anaerobically? Under the continuous aeration conditions, glycogen transformation during a cycle decreased (Pijuan et al., 2006; Ahn et al., 2007), suggesting that the reducing power for PHA synthesis relied more on the TCA cycle. An immediate decrease in DO levels was observed after acetate addition, indicating that electron transfer to oxygen occurred. This process oxidizes FADH2, thus making the methylmalonyl-CoA pathway unnecessary, consistent with the decrease in MCM expression. Taking the chemical and transcriptional evidence together, a fraction of the TCA cycle was coupled to electron transfer to oxygen, providing energy required for acetate-uptake, in addition to that from polyP degradation. Compared with previous studies, an extremely low P-release was observed in our study, coincident with a much higher DO level. Therefore, we hypothesize that the extent to which acetyl-CoA utilization is coupled to PHA formation/polyP degradation versus to aerobic electron transfer might partly depend on the acetate-uptake rate relative to the rate of electron transfer to external electron acceptor.
In summary, we found gene expression during an EBPR cycle exhibited dynamic change. The transcriptional patterns suggested that the TCA cycle was induced by the acetate addition in the anaerobic phase, as well as by switching to aeration, with the former induction associated with the split mode and the latter with the full mode of the TCA cycle. The partial mode through the glyoxylate pathway was expressed in both phases, with a higher induction upon first exposure to oxygen. The regulation of gene expression in central carbon metabolism was largely similar to model bacteria, with some differences, such as the biphasic induction of CS and ICL. Also different from model bacteria, ppk1 from Accumulibacter did not seem to be regulated by Pi limitation, and ppk1 transcriptional patterns suggested ppk1 may be functional in both polyP degradation and synthesis. The transcriptional patterns of PDC and PyrS were not consistent with the EBPR model prediction, and thus the biochemical transformation between acetyl-CoA and pyruvate in Accumulibacter may not be regulated at the transcriptional level. When high concentrations of oxygen were present simultaneously with acetate, P release was almost completely inhibited, together with the repression of ppk1 expression. The split TCA cycle was repressed and the full and partial TCA cycle was increased, suggesting a larger portion of acetate was probably utilized through the TCA cycle as compared with the anaerobic counterpart.
Acknowledgments
The project was funded by the National Science Foundation (BES 0332136) and the University of Wisconsin Graduate School. We thank Forrest I Bishop for operating the bioreactors and other technical support when this study was conducted. We also thank Jason J Flowers and Natalia Ivanova for helpful discussion.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ahn J, Schroeder S, Beer M, McIlroy S, Bayly RC, May JW, et al. Ecology of the microbial community removing phosphate from wastewater under continuously aerobic conditions in a sequencing batch reactor. Appl Environ Microbiol. 2007;73:2257–2270. doi: 10.1128/AEM.02080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA . Standard Methods for the Examination of Water and Waste Water. Washington, DC: American Public Health Association, American Water Works Association and Water Environment Federation: Washington, DC; 1995. [Google Scholar]
- Barnard JL, Abraham K. Key features of successful BNR operation. Water Sci Technol. 2006;53:1–9. doi: 10.2166/wst.2006.400. [DOI] [PubMed] [Google Scholar]
- Brdjanovic D, Slamet A, van Loosdrecht MCM, Hooijmans CM, Alaerts GJ, Heijnen JJ. Impact of excessive aeration on biological phosphorus removal from wastewater. Water Res. 1998;32:200–208. [Google Scholar]
- Burow LC, Mabbett AN, Blackall LL. Anaerobic glyoxylate cycle activity during simultaneous utilization of glycogen and acetate in uncultured Accumulibacter enriched in enhanced biological phosphorus removal communities. ISME J. 2008;2:1040–1051. doi: 10.1038/ismej.2008.45. [DOI] [PubMed] [Google Scholar]
- Comeau Y, Hall KJ, Hancock REW, Oldham WK. Biochemical model for enhanced biological phosphorus removal. Water Res. 1986;20:1511–1521. [Google Scholar]
- Crocetti GR, Hugenholtz P, Bond PL, Schuler A, Keller J, Jenkins D, et al. Identification of polyphosphate accumulating organisms and the design of 16S rRNA-directed probes for their detection and quantitation. Appl Environ Microbiol. 2000;66:1175–1182. doi: 10.1128/aem.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Martin H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- Gavigan JA, Marshall LM, Dobson ADW. Regulation of polyphosphate kinase gene expression in Acinetobacter baumannii 252. Microbiology-Uk. 1999;145:2931–2937. doi: 10.1099/00221287-145-10-2931. [DOI] [PubMed] [Google Scholar]
- Guisasola A, Pijuan M, Baeza JA, Carrera J, Casas C, Lafuente J. Aerobic phosphorus release linked to acetate uptake in bio-P sludge: process modeling using oxygen uptake rate. Biotechnol Bioeng. 2004;85:722–733. doi: 10.1002/bit.10868. [DOI] [PubMed] [Google Scholar]
- Gunsalus RP, Park SJ. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- He S, Gu AZ, McMahon KD. Fine-scale differences between Accumulibacter-like bacteria in enhanced biological phosphorus removal activated sludge. Water Sci Technol. 2006;54:111–117. doi: 10.2166/wst.2006.378. [DOI] [PubMed] [Google Scholar]
- He S, Gall DL, McMahon KD. ‘Candidatus Accumulibacter' population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl Environ Microbiol. 2007;73:5865–5874. doi: 10.1128/AEM.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Gu AZ, McMahon KD. Progress toward understanding the distribution of Accumulibacter among full-scale enhanced biological phosphorus removal systems. Microb Ecol. 2008;55:229–236. doi: 10.1007/s00248-007-9270-x. [DOI] [PubMed] [Google Scholar]
- He S, Kunin V, Haynes M, Martin HG, Ivanova N, Rohwer F, et al. 2010Metatranscriptomic array analysis of ‘Candidatus Accumulibacter phosphatis'-enriched enhanced biological phosphorus removal sludge Environ Microbiol 121205–1217.Epub 2010 Feb 9. [DOI] [PubMed] [Google Scholar]
- Hesselmann RPX, Werlen C, Hahn D, van der Meer JR, Zehnder AJB. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst Appl Microbiol. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- Hesselmann RPX, von Rummell R, Resnick SM, Hany R, Zehnder AJB. Anaerobic metabolism of bacteria performing enhanced biological phosphate removal. Water Res. 2000;34:3487–3494. [Google Scholar]
- Kato J, Yamamoto T, Yamada K, Ohtake H. Cloning, sequence and characterization of the polyphosphate kinase-encoding gene (ppk) of Klebsiella aerogenes. Gene (Amsterdam) 1993;137:237–242. doi: 10.1016/0378-1119(93)90013-s. [DOI] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol. 2004;70:5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg HL. Role and control of glyoxylate cycle in Escherichia coli - The first Colworth Medal Lecture. Biochem J. 1966;99:1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner H, Zumft WG. Expression of denitrification enzymes in response to the dissolved-oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl Environ Microbiol. 1989;55:1670–1676. doi: 10.1128/aem.55.7.1670-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BK, Bishop PL. Micro-profiles of activated sludge floc determined using microelectrodes. Water Res. 2004;38:1248–1258. doi: 10.1016/j.watres.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Louie TM, Mah TJ, Oldham W, Ramey WD. Use of metabolic inhibitors end gas chromatography/mass spectrometry to study poly-beta-hydroxyalkanoates metabolism involving cryptic nutrients in enhanced biological phosphorus removal systems. Water Res. 2000;34:1507–1514. [Google Scholar]
- Lynch AS, Lin ECC.1996Response to molecular oxygenIn: Neidhardt FC, Curtis R, Ingraham JL, Lin ECC, Low KB, Magasanik B et al (eds).Escherichia coli and Salmonella: Cellular and molecular biology American Society for Microbiology: Washington DC; 1526–1538. [Google Scholar]
- Maurer M, Gujer W, Hany R, Bachmann S. Intracellular carbon flow in phosphorus accumulating organisms from activated sludge systems. Water Res. 1997;31:907–917. [Google Scholar]
- McMahon KD, Dojka MA, Pace NR, Jenkins D, Keasling JD. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl Environ Microbiol. 2002;68:4971–4978. doi: 10.1128/AEM.68.10.4971-4978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Ragsdale SW. Mechanism of the Clostridium thermoaceticum pyruvate:ferredoxin oxidoreductase: evidence for the common catalytic intermediacy of the hydroxyethylthiamine pyropyrosphate radical. Biochemistry. 1997;36:8484–8494. doi: 10.1021/bi970403k. [DOI] [PubMed] [Google Scholar]
- Mino T, Arun V, Yoshiaki T, Matsuo T.1987Effect of phosphorus accumulation on acetate metabolism in the biological phosphorus removal processIn: Ramadori R. (edn).Advances in Water Pollution Control -Biological Phosphate Removal from Wastewaters Pergamon Press: Oxford, UK; 27–38. [Google Scholar]
- Mulkerrins D, Dobson ADW, Colleran E. Parameters affecting biological phosphate removal from wastewaters. Environ Int. 2004;30:249–259. doi: 10.1016/S0160-4120(03)00177-6. [DOI] [PubMed] [Google Scholar]
- Oehmen A, Lemos PC, Carvalho G, Yuan ZG, Keller J, Blackall LL, et al. Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res. 2007;41:2271–2300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Partridge JD, Scott C, Tang Y, Poole RK, Green J. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J Biol Chem. 2006;281:27806–27815. doi: 10.1074/jbc.M603450200. [DOI] [PubMed] [Google Scholar]
- Peterson SB, Warnecke F, Madejska J, McMahon KD, Hugenholtz P. Environmental distribution and population biology of Candidatus Accumulibacter, a primary agent of biological phosphorus removal. Environ Microbiol. 2008;10:2692–2703. doi: 10.1111/j.1462-2920.2008.01690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan M, Guisasola A, Baeza JA, Carrera J, Casas C, Lafuente J. Aerobic phosphorus release linked to acetate uptake: influence of PAO intracellular storage compounds. Biochem Eng J. 2005;6:184–190. [Google Scholar]
- Pijuan M, Guisasola A, Baeza JA, Carrera J, Casas C, Lafuente J. Net P-removal deterioration in enriched PAO sludge subjected to permanent aerobic conditions. J Biotechnol. 2006;123:117. doi: 10.1016/j.jbiotec.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Pijuan M, Saunders AM, Guisasola A, Baeza JA, Casa C, Blackall LL. Enhanced biological phosphorus removal in a sequencing batch reactor using propionate as the sole carbon source. Biotechnol Bioeng. 2004;85:56–67. doi: 10.1002/bit.10813. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria; 2008. [Google Scholar]
- Richardson DJ. Bacterial respiration: a flexible process for a changing environment. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- Shehab O, Deininger R, Porta F, Wojewski T. Optimizing phosphorus removal at the ann arbor wastewater treatment plant. Water Sci Technol. 1996;34:493–499. [Google Scholar]
- Wanner BL.1996Phosphorus assimilation and control of the phosphate regulonIn: Neidhardt FC, Curtis R, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al. (eds).Escherichia coli and Salmonella: Cellular and molecular biology American Society for Microbiology: Washington DC; 1357–1381. [Google Scholar]
- Wentzel MC, Lotter LH, Loewenthal RE, Marais GVR. Metabolic behavior of Acinetobacter spp. in enhanced biological phosphorus removal - a biochemical model. Water SA. 1986;12:209–224. [Google Scholar]
- Wexler M, Richardson DJ, Bond PL. Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal. Environ Microbiol. 2009;11:3029–3044. doi: 10.1111/j.1462-2920.2009.02007.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Pijuan M, Zeng RJ, Yuan Z. Involvement of the TCA cycle in the anaerobic metabolism of polyphosphate accumulating organisms (PAOs) Water Res. 2009;43:1330. doi: 10.1016/j.watres.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Zilles JL, Peccia J, Kim M-W, Hung C-H, Noguera DR. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl Environ Microbiol. 2002;68:2763–2769. doi: 10.1128/AEM.68.6.2763-2769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.