Abstract
The populations of dominant species within the human colonic microbiota can potentially be modified by dietary intake with consequences for health. Here we examined the influence of precisely controlled diets in 14 overweight men. Volunteers were provided successively with a control diet, diets high in resistant starch (RS) or non-starch polysaccharides (NSPs) and a reduced carbohydrate weight loss (WL) diet, over 10 weeks. Analysis of 16S rRNA sequences in stool samples of six volunteers detected 320 phylotypes (defined at >98% identity) of which 26, including 19 cultured species, each accounted for >1% of sequences. Although samples clustered more strongly by individual than by diet, time courses obtained by targeted qPCR revealed that ‘blooms' in specific bacterial groups occurred rapidly after a dietary change. These were rapidly reversed by the subsequent diet. Relatives of Ruminococcus bromii (R-ruminococci) increased in most volunteers on the RS diet, accounting for a mean of 17% of total bacteria compared with 3.8% on the NSP diet, whereas the uncultured Oscillibacter group increased on the RS and WL diets. Relatives of Eubacterium rectale increased on RS (to mean 10.1%) but decreased, along with Collinsella aerofaciens, on WL. Inter-individual variation was marked, however, with >60% of RS remaining unfermented in two volunteers on the RS diet, compared to <4% in the other 12 volunteers; these two individuals also showed low numbers of R-ruminococci (<1%). Dietary non-digestible carbohydrate can produce marked changes in the gut microbiota, but these depend on the initial composition of an individual's gut microbiota.
Keywords: human colon, resistant starch, 16S rRNA, phylotypes, Ruminococcus, temporal change
Introduction
The remarkable diversity of the human colonic microbiota at the level of bacterial species and phylotypes has become apparent from 16S rRNA-based analyses. Not only are hundreds of phylotypes typically estimated to be present in the human colonic microbiota from a given faecal sample (Suau et al., 1999; Eckburg et al., 2005), but samples from different individuals have been reported to show limited overlap in the phylotypes present (Ley et al., 2006; Turnbaugh et al., 2008). A recent study (Tap et al., 2009), however, has indicated that certain phylotypes occur more commonly than others among the dominant faecal bacteria of different individuals. Identifying the dominant bacterial species that colonize the large intestine and the extent to which these are influenced by diet and host factors is of key importance in uncovering the impact of the colonic microbiota upon human health (Flint et al., 2007; Sokol et al., 2008).
A few studies to date have examined temporal changes, and these suggest a degree of stability in the colonic microbiota of individuals consuming their normal diets (Franks et al., 1998; Zoetendal et al., 1998; Costello et al., 2009). In contrast, little is known about the impact of dietary change upon microbial community composition. There is evidence that dietary supplementation with prebiotics such as fructo-oligosaccharides and inulin can promote specific groups of bacteria, including bifidobacteria (Bouhnik et al., 2004; Ramirez-Farias et al., 2009). It has also been shown that reductions in total carbohydrate content, in weight loss (WL) diets for obese subjects, have major effects upon the composition and metabolic outputs of the bacterial community in the colon (Duncan et al., 2007, 2008; Brinkworth et al., 2009). These changes are assumed to reflect the fermentation of non-digestible (ND) carbohydrate components (mainly non-starch polysaccharides (NSPs), resistant starch (RS) and certain oligosaccharides) that reach the large intestine. The impact upon the colonic microbiota of controlled changes in the main types of ND carbohydrate normally present in the diet (RS and NSP) has not however been examined in any detail. The ND carbohydrate content of the human diet is considered to influence health. For example, diets high in RS have been shown to benefit insulin sensitivity, possibly mediated by bacterial fermentative activity in the colon (Robertson et al., 2005). Diets containing RS and NSP offer potential benefits in prevention of colorectal cancer through the delivery of fermentation acids, in particular butyrate, to the distal colon (McIntyre et al., 1993; Duncan et al., 2007). Microbial breakdown of NSP also releases bound phytochemicals into the colon (Gill and Rowland, 2002). These health benefits may be particularly important in obese and overweight subjects who are at increased risk of developing colorectal cancer and diabetes (Polednak, 2003; Cani et al., 2007).
This study asks to what extent changes in the major type of ND carbohydrate in the diet influence the dominant bacterial phylotypes present in an individual. It also explores the dynamics of such changes, including their reversibility, for the first time through the use of detailed time courses and a cross-over design. A necessary supplementary question is whether the same dietary change elicits similar changes in the gut microbiota of different individuals. Our results reveal rapid and marked changes in the colonic microbiota of a group of overweight volunteers after a shift in the main type of ingested fermentable carbohydrate (RS or NSP). We also find that these changes can be highly specific to the individual, with potentially important implications for the design of fibre-enriched diets.
Materials and methods
Volunteer recruitment
Volunteers were initially recruited by newspaper advertisement. Inclusion criteria for men were a body mass index >27 kg m−2, waist circumference >102 cm and fasting plasma glucose concentration >6.0 mmol l−1, indicating metabolic syndrome. They underwent a medical examination and their general practitioner was contacted to confirm medical and medication history. None of the subjects had a history of gastrointestinal disease or disturbance, or took antibiotics, either in the 6 months leading up to the study or during the study. Two of the original 16 subjects left the study for reasons unconnected with the protocol. The remaining 14 volunteers had a mean age of 54 years (range 27–73 years) with a mean body mass index (kg m−2) of 39.4 (range 27.9–51.3) (Supplementary Table S1). The study was approved by the North of Scotland Research Ethics Service and all volunteers provided informed, written consent.
Experimental design and dietary regime
The volunteers were weight stable (<3 kg change in the past 4 months) before entry on the trial. Over the first 7 weeks, intakes were provided at energy maintenance as 1.4–1.5 × measured resting metabolic rate for each individual. The final intervention period consisted of high protein WL diet for an additional 3 weeks (Supplementary Table S2, Supplementary Figure S1). The initial maintenance diet (M diet) comprised protein/carbohydrate/fat % as 13:52:35 of metabolizable energy and 27.7 g per day NSP, provided for 7 days. Subjects were then provided with fixed intakes of two diets, which consisted of either a high RS diet or a high NSP diet each supplied for 3 weeks in a randomized cross-over design (Supplementary Figure S1). The RS diet contained added type III RS whereas the NSP diet intake contained added wheat bran. All meals were of the same energy density (5.5 MJ kg−1) and daily intakes were recorded by weigh-back after each meal (Supplementary Table S2). Daily macronutrient intakes were calculated using the Windiet software program (Robert Gordon University, Aberdeen, UK), based on the type and quantity of each ingredient consumed and published food composition tables (Food Standards Agency 2002). Faecal samples were collected on average twice each week. In addition, one 24 h collection from the final week of each dietary period was used for chemical analysis of digestibility.
Chemical analysis of diet composition and digestibility
Diets were analysed for total gross energy, RS and insoluble and soluble NSPs (Supplementary Table S2). Estimation of RS content of diets and 24 h faecal sample collections was as described by Englyst et al. (1992) whereas NSPs were quantified as described by Englyst and Cummings (1988).
DNA extraction from faecal samples
Faecal samples were kept at 4 °C and processed within 5 h of collection. This short period of storage is not expected to influence molecular estimation of microbial community composition (Lauber et al., 2010). Each sample was mixed and 5 g dispersed (3500 r.p.m. for 1 min using a Dispomix Drive, Medic Tools, Lussiwag, Switzerland) in 10 ml sterile phosphate-buffered saline buffer before aliquoting. One aliquot was used immediately for DNA extraction using the FastDNA Spin for soil kit following manufacturer's (Qbiogene, MP Biomedicals, Illkirch, France) instructions.
Phylogenetic analysis of sequences derived from 16S rRNA clone libraries
PCR amplification of 16S rRNA genes from the extracted DNA involved initial denaturation at 94 °C for 5 min; 20 cycles of denaturation (30 s at 95 °C), annealing (30 s at 52 °C), extension at 72 °C for 2 min with a final cycle at 72 °C for 8 min. Taq1 polymerase from Promega (Southampton, UK) was used at 0.025 U μl−1 in the presence of 2.5 m MgCl2. The forward primer comprised a 4:1:1:1:1 mixture of 7f, 27fChl, 27fBor, 27fBif and 27fAto and the reverse primer was 1510r (Supplementary Table S3). PCR products were cleaned using the Wizard PCR product purification kit (Promega) and were then cloned and sequenced as described (Lawley et al., 2009). The sequences (spanning variable regions V2–V5) were aligned using the SILVA-derived reference alignment in the mothur software package (Schloss et al., 2009) and were tested for the presence of chimeras by Mallard (Ashelford et al., 2006), Pintail (Ashelford et al., 2005) and BLAST (Johnson et al., 2008). After removal of chimeric sequences, the alignment was subjected to extensive manual curation using the editor function in the ARB package (Ludwig et al., 2004). Using the curated alignment, a distance matrix, with Felsenstein correction, was created using ARB. This matrix was then used as an input for DOTUR (Schloss and Handelsman, 2005) using a 98% identity cut-off under the default furthest-neighbour setting. Sequences with >98% phylogenetic similarity were regarded as belonging to the same phylotype. This generated 320 phylotypes (Supplementary Table S4), which were assigned an identity from the phylum to genus level by the RDP Classifier (Cole et al., 2009) and to the closest match in the NCBI database using MegaBLAST (Johnson et al., 2008). Rarefaction analysis, Shannon and Simpson diversity index calculations, community structure comparisons with ∫-Libshuff, parsimony and weighted and unweighted versions of UniFrac, Jaccard and Yue and Clayton theta tree clustering analysis were all performed using the mothur software package (Schloss et al., 2009).
The final 16S rRNA clone library data set from six volunteers (v16, v19, v20, v22, v23 and v24) contained 5915 sequences (median length of 678 bases) (GenBank accession numbers GU238434–GU244348). These sequences were from one faecal sample from each of the four diets per individual, except for volunteer v16 when two samples were analysed from the M and NSP diets (see Supplementary Table S4). An additional 768 sequences were subsequently obtained from the RS and NSP diets only of v14 and v25 (Supplementary Table S5) (GenBank accession numbers HM191774–HM192541).
qPCR and DGGE analysis of 16S rRNA genes
Quantitative real-time PCR (qPCR) was performed on all available samples from all 14 volunteers. Primer sequences and conditions are reported in Supplementary Table S3, based largely on the study by Ramirez-Farias et al. (2009). Primers for Oscillibacter-related bacteria were designed in this study.
For denaturing gradient gel electrophoresis (DGGE), bacterial DNA was amplified by PCR using primers for the hyper-variable V3 region of the 16S gene (Supplementary Table S3). Reaction mixtures contained 1.5 m MgCl2 and 0.025 U Taq1 polymerase per μl. Products were separated in an 8% acrylamide denaturing gradient gel containing urea and formamide (35–60% gradient) at 80 V for 16 h at 60 °C and visualized by staining with SybrGold dye (Invitrogen, Paisley, UK).
Statistical analysis
For phylotypes with large abundance (more than 1% and found in 5 or 6 out of 6 volunteers, which was the case for 8 out of 320 phylotypes) the Sanger sequences (clone counts) were analysed as Hierarchical Generalized Linear Models using a binomial model with logistic link and estimated overdispersion. This was carried out for one phylotype at a time, with the total number of clones per sample regarded as the total count. Volunteer and diet were regarded as random (normally distributed) and fixed effects, respectively. Similarities between faecal samples were displayed graphically by means of principal coordinate analysis, where the Canberra distance matrix was applied to the numbers of clones assigned to each phylotype (based on all 320 phylotypes) for each faecal sample. Principal coordinate analysis was also used to obtain a graphical representation of the phylogenetic distances (obtained from the DOTUR software package) between the 320 phylotypes.
The effect of diet was tested by means of analysis of variance with volunteer regarded as random and diet regarded as fixed effect. When the effect of diet was significant, diet means were compared by post hoc t-test. Data containing many zeros (that is, below limit of detection) were analysed by Friedman's non-parametric analysis of variance instead. For all analyses, the effect of order was included initially as a fixed effect but it and its interaction with diet were found not to be significant and was removed from subsequent analyses. All statistical analyses were performed by using Genstat 11th Edition Release 11.1 (VSN International Ltd, Hemel Hempstead, Hertfordshire, UK). Significance was set at P-value of <0.05.
Results
Study design
Fourteen overweight male volunteers completed this study (see Materials and Methods section) (Supplementary Table S1). Complete diets were provided daily resulting in precise control over all dietary intake throughout the 10-week study period (Supplementary Table S2). Each volunteer received a control weight maintenance (M) diet for the first week, followed by three weeks each of diets in high type III resistant starch (RS diet) or high in wheat bran (NSP) and a WL diet reduced in carbohydrate but high in protein (WL diet). The first three diets were provided at energy maintenance and were closely matched for total carbohydrate, protein and fat content; the order of the RS and NSP diets was reversed for half of the volunteers (Supplementary Figure S1, Supplementary Table S2). Faecal samples were processed as soon as possible after collection, without prior freezing, for the extraction of nucleic acids (see Materials and Methods section).
Dominant bacterial phylotypes within the intestinal bacterial community
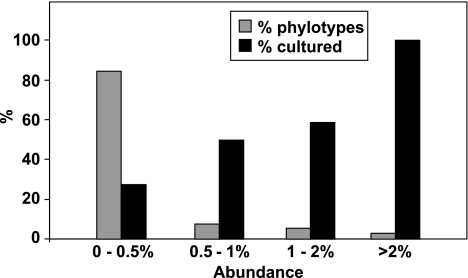
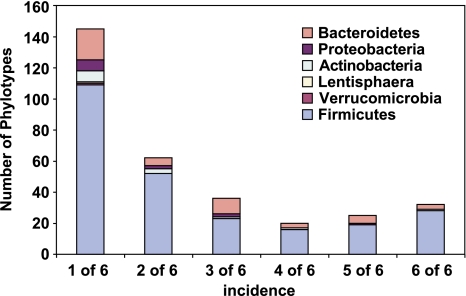
16S rRNA profiles from all available faecal samples were analysed first using DGGE, which revealed that profiles were consistent over time within an individual for a given diet (Supplementary Figure S2). A switch between the RS and NSP diets, however, altered the profile substantially, indicating a shift in bacterial community composition. Sequences of amplified 16S rRNA genes were analysed for 6 of the 14 volunteers (3 from each diet order) using a sample from the final week of each dietary regime. This yielded 5915 non-chimeric sequences (averaging 227 sequences per sample). Phylogenetic analysis using a 98% sequence identity cut-off revealed 320 phylotypes across all samples (Supplementary Table S4). The proportion of cultured strains increased with increasing abundance of phylotypes, with all phylotypes present at >2% of the total being identified with cultured species (Figure 1). None of the 320 phylotypes was detected in every sample, but 32 phylotypes were present in all six volunteers accounting for 47.1% of all sequences, whereas 77 phylotypes occurred in at least four of the six individuals (Figure 2, Supplementary Figure S3). Figure 2 suggests a bimodal frequency distribution for phylotype occurrence, indicating that there may be a group of widespread and highly abundant phylotypes within the human gut microbiota (Tap et al., 2009). The most abundant phylotype was Eubacterium rectale although three phylotypes corresponding to three divergent strains of Faecalibacterium prausnitzii together comprised 8.0% of total sequences (Table 1).
Figure 1.
Culturability of 16S rRNA phylotypes in relation to their abundance. 16S rRNA sequences (5915) obtained from faecal samples of six volunteers (for all four diets) were classified into 320 phylotypes (defined at >98% sequence identity) (Supplementary Table S4). The % of phylotypes showing >98% sequence identity to a cultured bacterium is seen to increase with increasing phylotype abundance. The phylotype frequency distribution is shown as a percentage; actual numbers of phylotypes were 9 (>2% of all sequences), 17 (>1%<2%), 24 (>0.5%<1%) and 270 (<0.5%) (total 320). Data refer to volunteers 16, 20, 22 (diet order RS-NSP) and 19, 23, 24 (diet order NSP-RS).
Figure 2.
Incidence of phylotypes in different individuals. The distribution of all 320 16S rRNA phylotypes was determined across the six volunteers (see Figure 1, Supplementary Table S4); the numbers of phylotypes found in all six individuals (32) and in five (25), four (20), three (36), two (62) or one (145) of the six are shown here according to bacterial phylum.
Table 1. Impact of diet upon the 10 most abundant 16S rRNA phylotypes detected in six overweight male volunteers.
| Phylotype, speciesa | Phylum | Mean % clones | Diets |
P-valueb | |||
|---|---|---|---|---|---|---|---|
| % total clones | M | NSP | RS | WL | |||
| Eubacterium rectale | F (L) | 4.43 | 2.9x | 4.1x | 8.0y | 1.7x | <0.001 |
| Collinsella aerofaciens | A | 3.67 | 4.3x | 3.4x | 5.2x | 0.6y | 0.032 |
| Clostridium clostridioforme | F (L) | 3.83 | 4.8 | 2.6 | 2.5 | 3.1 | 0.217 |
| Bacteroides vulgatus | B | 3.21 | 2.6 | 4.1 | 1.3 | 4.9 | 0.057 |
| Faecalibacterium prausnitzii L2-6 | F (R) | 2.96 | 2.5 | 3.4 | 1.6 | 3.8 | 0.224 |
| Faecalibacterium prausnitzii A2-165 | F (R) | 2.55 | 1.8 | 2.8 | 2.9 | 3.4 | 0.338 |
| Faecalibacterium prausnitzii M21/2 | F (R) | 2.47 | 2.1 | 1.8 | 2.2 | 3.1 | 0.771 |
| Anaerostipes colic SSC/2 | F (L) | 2.25 | 3.0 | 2.5 | 1.1 | 1.8 | 0.091 |
| Ruminococcus bromii | F (R) | 2.11 | 1.5x | 0.4x | 5.0y | 1.5x | <0.001 |
| Eubacterium hallii | F (L) | 2.00 | 2.4 | 2.5 | 1.2 | 1.4 | 0.251 |
Abbreviations: F, Firmicutes (L, Lachnospiraceae; R, Ruminococcaceae); B, Bacteroidetes; A, Actinobacteria.
Detected in all six volunteers, except for R. bromii (four of six volunteers).
Within a row values not sharing superscripts (x and y) differ significantly (P< 0.05). Clone counts were analysed with Hierarchical Generalized Linear Model with Volunteer as random and Diet as fixed effect.
New species name soon to be formally proposed (Allen-Vercoe et al., in preparation).
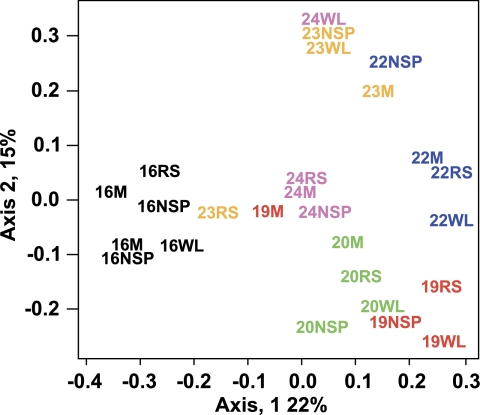
Overall community structure was analysed for each sample using the mothur software package (Schloss et al., 2009). ∫-Libshuff, parsimony and UniFrac (weighted and unweighted) analysis revealed a high degree of variation between individuals and showed that each sample was significantly different from all others. Similarity in community membership and structure was estimated using the Jaccard and the Yue and Clayton theta similarity coefficients (Yue and Clayton, 2005) (Supplementary Figure S4). These results and those from principal coordinate analysis (Figure 3) indicate that samples predominantly clustered by volunteer, suggesting that responses to dietary change are influenced by the initial species composition of the individual's colonic microbiota. Rarefaction analysis revealed that the observed species richness was similar for all four diets (Supplementary Figure S5). Simpson (1/D) and Shannon diversity indices were slightly lower on the RS and NSP diets compared with the M and WL diets, but did not differ significantly (Supplementary Figure S5; see also Supplementary Table S4 for individual values).
Figure 3.
Impact of diet and individual variation upon faecal microbiota composition. A principal coordinates analysis (using Canberra distance matrix) based on 16S rRNA clone libraries from 26 faecal samples (obtained from six donors under four dietary conditions) (Supplementary Table S4). Colour code is based on donor (v16, v19, v20, v22, v23 and v24). Diets are indicated as M, maintenance; NSP, non-starch polysaccharide; RS, resistant starch; WL, weight loss. Two samples were analysed for v16 from the M and NSP diets.
Response of the colonic microbial community to dietary change
At the phylum level there was no significant effect of diet upon the proportions of Bacteroidetes (mean 21.5%), Firmicutes (mean 70.6%), Actinobacteria (mean 4.9%) or Proteobacteria (mean 3.0%) within the faecal microbiota. At finer taxonomic levels, however, two individual phylotypes, E. rectale and Ruminococcus bromii, showed increased proportions on the RS diet whereas Collinsella aerofaciens showed decreased proportions on the WL diet (P<0.001) (Table 1). Because the sequence data also suggested that groups of related phylotypes might be affected by diet, we decided to use qPCR for further analysis.
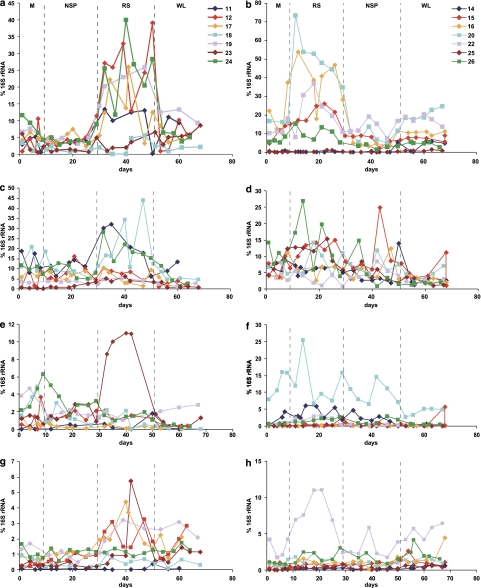
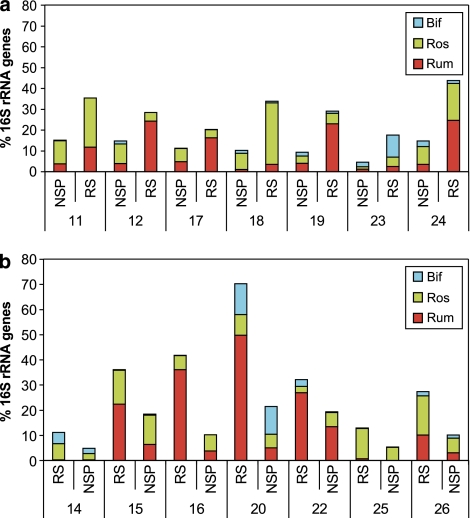
PCR primer sets (Supplementary Table S3) were used to target groups for which sequence analysis indicated a response to the RS diet (Rum, Ros, Osc), or for which previous in vitro evidence has shown the ability to use starch (Bac, Bif, Fprau) (Table 2). Methanogens were also monitored because of their potential impact on the polysaccharide-utilizing community (Robert and Bernalier-Donadille, 2003). All available time points were analysed. Mean data for the targeted groups are shown in Table 2 and complete time courses for all 14 subjects are shown in Figure 4 for those bacterial groups that showed responses to the RS diet. The Rum group includes relatives of R. bromii, R. flavefaciens and R. albus that belong to the Ruminococcaceae, and will be referred to here as ‘R-ruminococci'. There was a significant increase of around 4.5-fold (P<0.001) in this group on the RS diet, compared to the NSP diet (Table 2, Figure 4). A new primer set was designed to recognize another group of Ruminococcaceae, related to Oscillibacter valericigenes (Iino et al., 2007) that has not been cultured from the human gut. This group increased significantly on average on the RS and WL relative to the M and NSP diets (P<0.001) (Table 2, Figure 4). A previously reported primer set (Mackie et al., 2003) targeting relatives of this group from the rumen was also used here, but detected <0.1% of human faecal bacteria (results not shown). Populations of a third abundant group of Ruminococcaceae, F. prausnitzii, showed no significant response to changes of diet (Table 2).
Table 2. Populations estimated by qPCR of bacteria and methanogenic archaea expressed as % relative to total bacteria.
| Bacterial group (%) |
Diet |
P dieta | ||||
|---|---|---|---|---|---|---|
| M | NSP | RS | WL | SED | ||
| Bac | 27.8 | 25.7 | 20.2 | 24.0 | 3.58 | 0.387 |
| Ros | 7.3x | 6.5x | 10.1y | 3.3z | 1.381 | <0.001 |
| Fprau | 11.2 | 14.4 | 12.1 | 12.5 | 1.622 | 0.278 |
| Rum | 6.5x | 3.8x | 17.0y | 7.5x | 2.253 | <0.001 |
| Osc | 0.74x | 0.77x | 2.0y | 1.6y | 0.34 | <0.001 |
| Bif | 1.9 | 1.8 | 2.4 | 0.8 | 0.632 | 0.059 |
| Met | 0.080 | 0.034 | 0.124 | 0.123 | 0.042 | 0.155b |
Abbreviations: Bac, Bacteroides/Prevotella; Ros, E. rectale/Roseburia spp.; Fprau, F. prausnitzii; Rum, R-ruminococci; Osc, Oscillibacter relatives; Bif, Bifidobacterium spp.; Met, methanogens.
Significance based on analysis of variance (ANOVA). Within each row, values not superscripts (x, y and z) differ significantly (P<0.05, post hoc t-test).
Friedman non-parametric ANOVA.
Primer sequences and amplification conditions are shown in Supplementary Table S3. One mean value is shown per diet for the 14 volunteers. For the M diet, means are for all samples from the 1 week of this diet, whereas for the NSP, RS and WL diets means are for all samples from the last 2 weeks on each diet.
Figure 4.
Diet-driven changes in four groups of human colonic bacteria detected by qPCR. Abundance for each targeted group is expressed as a percentage of the signal obtained with a general bacterial primer set (see Supplementary Table S3 for primers and conditions used). All available time points are shown for the 14 volunteers: left-hand panels show the diet order M-NSP-RS-WL and right-hand panels M-RS-NSP-WL, (a and b) R-ruminococci (relatives of R. bromii); (c and d) relatives of E. rectale and Roseburia spp.; (e and f) Bifidobacterium spp.; (g and h) relatives of O. valericigenes.
Firmicutes bacteria related to Roseburia and E. rectale also increased significantly (P<0.048) on the RS diet, and decreased (approximately threefold compared to RS) (P<0.029) on the reduced carbohydrate WL diet. (Ros, Table 2). Individual differences were evident for all groups shown in Figure 4. Notably, Bifidobacterium spp. did not show a significant mean response to RS, but their population increased markedly on RS in one volunteer (v23) (Figures 4 and 5). Bifidobacteria are often poorly represented in 16S rRNA clone libraries (Eckburg et al., 2005; Hold et al., 2002) and this was also the case in our analysis in spite of using a broad bacterial primer set reported to include this group (Frank et al., 2008). There was no significant change in the % Bacteroides in response to diet (Table 2).
Figure 5.
Populations of three groups of potentially amylolytic bacteria on the RS (high-resistant starch) and NSP (low-resistant starch) diets. Populations, estimated by qPCR, are shown for R-ruminococci, E. rectale/Roseburia spp. and Bifidobacterium spp. for all 14 volunteers, according to diet order. Data are the mean values for each volunteer during the second and third week of the RS and NSP diets (overall means are given in Table 2).
Methanogenic archaea were detected by qPCR in >90% of samples from nine volunteers, but were detected in fewer than 10% of samples from v11, 14, 17, 24 and 25. This group showed no significant effect of diet (Table 2).
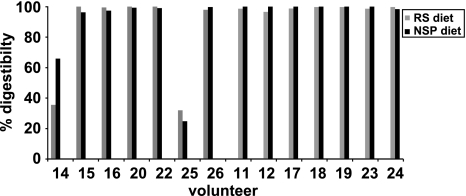
Digestibility of RS and NSP
Whole tract digestibility of RS and NSP was estimated from chemical analysis of the diet and of 24 h faecal collections. For 12 of the 14 volunteers <4% of ingested RS was recovered in faecal samples for all four diets, reflecting almost complete microbial fermentation. For two individuals (v14 and v25), however, faecal RS recovery was 69% and 65%, respectively (31% and 35% digestibility) on the RS diet (Figure 6). These two individuals both harboured low R-ruminococci numbers as assessed by qPCR, although one of them showed an increase in the Roseburia and E. rectale group on the RS diet (Figures 4 and 5). Additional clone library analyses were subsequently performed on the final samples from the RS and NSP dietary periods for these two individuals. No R-ruminococci were detected whereas 36% and 46.5% of clones belonged to the Bacteroidetes among the 381 and 387 clones analysed for v14 and v25, respectively (Supplementary Table S5).
Figure 6.
Starch digestibilities. The whole tract % digestibility of resistant starch was determined after estimating resistant starch present in the supplied diet and in 24 h faecal samples (see Materials and Methods section). Results are shown only for the NSP and RS diet periods. The markedly reduced resistant starch digestibilities in v14 and v25 correspond with low R-ruminococcal numbers in these individuals (see text and Supplementary Table S5).
Soluble NSP digestibility averaged 90% and was slightly lower (P<0.05) for RS and WL diets (88%) than for the M diet (92%). Insoluble NSP digestibility averaged 66% and showed no difference between diets (mean range 58–71%).
Discussion
This is the first study to combine precise control and monitoring of human dietary intake and digestibility with detailed analysis of changes in the faecal microbiota at the level of bacterial phylotypes. As a result it has been possible to reveal the interplay between diet and inter-individual differences that influence the human colonic community. Analysis of amplified 16S rRNA sequences suggested that samples from six overweight male volunteers clustered more strongly by individual than by diet. This is in line with previously reported evidence of inter-individual variation in the faecal microbiota (Franks et al., 1998; Ley et al., 2006). In contrast, diet composition had very substantial effects on specific groups of bacteria that were detected both through clone libraries and extensive qPCR analysis on 279 faecal samples. The time courses show that most diet-driven changes occurred rapidly, being detectable within 3–4 days, and were reversed equally rapidly. These kinetics appear consistent with immediate effects of dietary residue upon relative bacterial growth rates in the colon that are subsequently reflected in faecal samples as colonic contents turnover, assuming mean transit colonic times of around 60 h (Stephen et al., 1987).
Firmicutes bacteria related to R. bromii (R-ruminococci) and E. rectale were commonly stimulated by the RS diet. In most individuals, qPCR analysis revealed a surge in the population of R-ruminococci with values exceeding 25% of total bacteria in some samples. R-ruminococci were the only group of human gut bacteria previously found to be preferentially associated with particulate material in human faecal samples (Walker et al., 2008), suggesting that they have an important role in the breakdown of particulate substrates. R. bromii isolates show amylolytic activity (Salyers et al., 1977) but our results suggest that many related, but uncultured, bacteria may also possess this activity. One previous study, using non-quantitative DGGE analysis, reported that R. bromii-related bacteria were prominent in faecal samples from humans on a diet high in RS and NSP (Abell et al., 2008). The E. rectale and Roseburia group is also known to contain amylolytic species (Ramsay et al., 2006) but has not previously been shown to respond to RS in vivo. An earlier study failed to detect an increase in specific Eubacterium spp. in human subjects on a diet high in RS type III but, importantly, detection methods were not available for E. rectale (Schwiertz et al., 2002). In this work we also detected a dietary response for an uncultured group of Ruminococcaceae related to Oscillibacter. It is not known whether these bacteria are starch degraders, and their increase both on the RS and WL diets suggests that other factors must be involved in their response to diet.
Early studies indicated a broad distribution of amylase activity among cultured human gut bacteria with >50% of strains, including many Bacteroides, able to grow on amylose or amylopectin starch (Salyers et al., 1977). The potential role of Bifidobacterium spp. has been emphasized particularly in the degradation of high amylose RS (Macfarlane and Englyst, 1986; Wang et al., 1999). In this study, bifidobacterial numbers showed a strong response to RS only in one individual (v23), whereas there was no evidence for a response of the Bacteroides group to RS. Bacteroides spp. may perhaps be better adapted to the use of solubilized starch molecules (Flint et al., 2008). These findings in vivo however correspond better with more recent work in vitro that identified R. bromii, E. rectale and Bifidobacterium spp. as the major species in human faeces that colonize starch particles (Leitch et al., 2007) and that use 13C-labelled starch (Kovatcheva-Datchary et al., 2009).
Different groups of amylolytic bacteria differ markedly in their metabolic products and potential host interactions. Bifidobacterium spp., which produce lactate and acetate, are widely used as probiotics and as targets for prebiosis (Furrie et al., 2005). Members of the E. rectale group are flagellated bacteria that are major producers of butyrate in the large intestine, and may therefore contribute to the butyrogenic effect of RS (Aminov et al., 2006; Duncan et al., 2007; Louis et al., 2010). R. bromii is a producer of acetate, ethanol and hydrogen that is likely to contribute to gas production, but otherwise little is currently known about the impact of this group upon the host. It will be important in future to establish whether different types of RS select for different groups of amylolytic bacteria in view of their differing effects on fermentation and host responses (Le Leu et al., 2009).
Starch is considered to show high digestibility across the whole gastrointestinal tract, with most RSs being completely fermented in the large intestine (Bird et al., 2000). Remarkably, however, significant amounts of starch survived fermentation to be recovered in the faeces in 2 of the 14 individuals studied here. This difference in fermentation cannot be ascribed to dietary intake, which was standardized for all 14 subjects, and seems likely to lie with the strain composition of each individual's colonic microbiota. These two individuals showed very low numbers of R-ruminococci, and the relationship between community composition and starch fermentation will clearly warrant further investigation. Although these two individuals were also non-methanogenic, no simple relationship was evident between methanogens and R-ruminococci. Methanogen populations are known to vary between individuals and are influenced by a variety of factors (Florin et al., 2000).
In contrast to these responses to RS, there was little evidence that the high NSP diet resulted in major alterations in the composition of the faecal microbiota. In part, however, this may reflect the fact that a smaller increase was achieved in NSP intake (1.5-fold) than with RS intake (4.8-fold) when compared with the M diet. It is possible that larger changes in specific NSP components would affect the populations of specific groups of colonic bacteria, as was observed with RS. Significant decreases were observed for C. aerofaciens and for the E. rectale group on the WL diet. The WL changes do not show a simple relationship with RS and NSP intakes, and it is possible that the increased dietary protein content of this diet might have a role in altering microbiota composition.
The distribution of major bacterial phyla observed here did not depart dramatically from that reported in non-obese subjects (Eckburg et al., 2005; Walker et al., 2008). The data that we obtained by qPCR and clone library analysis are consistent with recent reports on obese subjects using fluorescence in situ hybridization microscopy (Duncan et al., 2007, 2008; Schwiertz et al., 2010) or 16S rRNA sequencing (Zhang et al., 2008) although lower % Bacteroides have been reported in another study (Ley et al., 2006). More subtle differences may occur between the gut microbiota of obese and non-obese individuals at the species level, and indeed the dietary responses reported here make this likely. Nevertheless 5 of the 10 most abundant phylotypes identified in this study group were also among the 10 most abundant phylotypes described by Tap et al. (2009) in non-obese volunteers.
A high proportion of the most abundant 16S rRNA phylotypes that we detected here corresponded to cultured bacteria, with 66% of the 50 phylotypes that accounted for >0.5% of sequences having close cultured relatives. This suggests that the limited coverage of the human gut microbiota through cultivation may be because of insufficient anaerobic isolation work, rather than to intrinsic non-culturability of human colonic bacteria. Four of the five most abundant phylotypes detected here (E. rectale, F. prausnitzii, C. aerofaciens and B. vulgatus) corresponded with the most abundant species reported by anaerobic cultivation in Japanese–Hawaiians, North American Caucasians and polyp patients (Moore and Moore, 1995). Overall, however, 33.4% of phylotypes showed <98% identity with cultured bacteria in this study, reflecting the relatively poor coverage of the less abundant bacterial groups by cultured strains. Many abundant phylotypes were found to be widespread, with 32 of the 320 phylotypes detected being present in all six individuals surveyed and accounting for almost half (47%) of all sequences.
In conclusion, we find that increased intake of RS, an important ND carbohydrate in the human diet, can substantially alter the species composition of the colonic microbiota. Such responses are likely to occur also with other fermentable ND dietary components. It follows that the colonic microbial community must typically be in a state of continuous change over time, driven by short-term changes in dietary intake. Thus, only the most successful and versatile organisms will be found commonly among the dominant microbiota at different sampling times and in different individuals. In addition, however, our evidence shows that the bacterial strain composition of the colonic microbiota is subject to inter-individual variation. Furthermore, we show that this can be associated with profound inter-individual differences in the response of the microbial community to dietary change, and in microbial fermentation of dietary substrates in the colon. This suggests that dietary advice on the consumption of ND carbohydrates might need to be personalized in the future.
Acknowledgments
We acknowledge support for this project from the World Cancer Research Fund. The Rowett Institute of Nutrition and Health (University of Aberdeen) and Biomathematics and Statistics Scotland received support from the Scottish Government Rural Environment Research and Analysis Directorate. We thank Claire Fyfe and the RINH Human Studies Unit. We also thank David Harris and the rest of his team at the Wellcome Trust Sanger Institute for generating the 16S rRNA gene sequences. Funding for AWW, MDS, PS, JP and sequencing was provided by The Wellcome Trust (grant no. WT 76964).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abell GCJ, Cooke CM, Bennett CN, Conlon MA, McOrist AL. Phylotypes related to Ruminococcus bromii are abundant in the large bowel of humans and increase in response to a diet high in resistant starch. FEMS Microbiol Ecol. 2008;66:505–515. doi: 10.1111/j.1574-6941.2008.00527.x. [DOI] [PubMed] [Google Scholar]
- Aminov RI, Walker AW, Duncan SH, Harmsen HJM, Welling GW, Flint HJ. Molecular diversity, cultivation, and improved FISH detection of a dominant group of human gut bacteria related to Roseburia spp. or Eubacteriumrectale. Appl Environ Microbiol. 2006;72:6371–6376. doi: 10.1128/AEM.00701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AR, Brown IL, Topping DL. Starches, resistant starches, the gut microflora and human heath. Curr Issues Intest Microbiol. 2000;1:25–37. [PubMed] [Google Scholar]
- Bouhnik Y, Raskine L, Simoneau G, Vicaut E, Neut C, Flourie B, et al. The capacity of nondigestible carbohydrates to stimulate faecal bifidobacteria in healthy humans: a double blind, randomized, placebo-controlled, parallel-group, dose response relation study. Am J Clin Nutr. 2004;80:1658–1664. doi: 10.1093/ajcn/80.6.1658. [DOI] [PubMed] [Google Scholar]
- Brinkworth GD, Noakes M, Clifton PM, Bird AR. Comparative effects of very low carbohydrate, high fat and high carbohydrate, low-fat weight loss diets on bowel habit and faecal short chain fatty acids and bacterial populations. Brit J Nutr. 2009;101:1493–1502. doi: 10.1017/S0007114508094658. [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across time and space. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obesity. 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englyst HN, Cummings JH. Improved method for measurement of dietary fiber as non-starch polysaccharides in plant foods. J Assoc Off Anal Chem. 1988;1:247–286. [PubMed] [Google Scholar]
- Englyst HN, Kingman SM, Cummings JE. Classification and measurement of nutritionally important starch fractions. Eur J Clin Nutr. 1992;46 (Suppl 2:S33–S50. [PubMed] [Google Scholar]
- Food Standards Agency 2002McCance and Widdowson's The Composition of Foods, 6th summary edn.Royal Society of Chemistry: Cambridge [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- Florin THF, Zhu G, Kirk KM, Martin NG. Shared and unique environmental factors determine the ecology of methanogens in humans and rats. Am J Gastroenterol. 2000;95:2872–2879. doi: 10.1111/j.1572-0241.2000.02319.x. [DOI] [PubMed] [Google Scholar]
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks AH, Harmsen HJM, Raangs GC, Jansen GJ, Schut F, Welling GW. Variations of bacterial populations in human feces measured by fluorescent in situ hybridisation with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrie E, Macfarlane S, Kennedy A, Cummings JH, Walsh SV, O'Neill DA, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54:242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill CIR, Rowland IR. Diet and cancer: assessing the risk. Br J Nutr. 2002;88 (Suppl 1:S73–S87. doi: 10.1079/BJN2002632. [DOI] [PubMed] [Google Scholar]
- Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. 2002;39:33–39. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilic-Stojanovic M, de Graaf AA, Smidt H, et al. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol. 2009;11:914–926. doi: 10.1111/j.1462-2920.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- Iino T, Mori K, Tanaka K, Suzuki K-I, Harayama S. Oscillibacter valericigenes gen nov., a valerate-producing anaerobic bacterium isolated from the alimentary canal of a Japanese corbicula clam. Int J Syst Evol Microbiol. 2007;57:1840–1845. doi: 10.1099/ijs.0.64717-0. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Effect of storage conditions on the assessment of bacterial community structure in soil and human associated samples. FEMS Microbiol Lett. 2010;307:80–85. doi: 10.1111/j.1574-6968.2010.01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch ECM, Walker AW, Duncan SH, Holtrop G, Flint HJ. Selective colonization of insoluble substrates by human colonic bacteria. Environ Microbiol. 2007;72:667–679. doi: 10.1111/j.1462-2920.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- Le Leu RK, Hu Y, Brown IL, Young GP. Effect of high amylose maize starches on colonic fermentation and apoptotic response to DNA-damage in the colon of rats. BMC Nutr Metab. 2009;6:11–19. doi: 10.1186/1743-7075-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology—human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA transferase gene. Environ Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Englyst HN. Starch utilization by the human large intestinal microflora. J Appl Bacteriol. 1986;60:195–201. doi: 10.1111/j.1365-2672.1986.tb01073.x. [DOI] [PubMed] [Google Scholar]
- Mackie RI, Aminov RI, Hu W, Klieve AV, Ouwekerk D, Sundset MA, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol. 2003;69:6808–6815. doi: 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre AP, Gibson P, Young GP. Butyrate production from dietary fibre and protection against large bowel cancer in a gut model. Gut. 1993;34:386–391. doi: 10.1136/gut.34.3.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WEC, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polednak AP. Trends in incidence rates for obesity-associated cancers in the U.S. Cancer Detect Prev. 2003;27:415–421. doi: 10.1016/j.cdp.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- Ramsay AG, Scott KP, Martin CJ, Rincon MT, Flint HJ. Cell-associated α-amylases of butyrate-producing Firmicute bacteria from the human colon. Microbiology. 2006;152:3281–3290. doi: 10.1099/mic.0.29233-0. [DOI] [PubMed] [Google Scholar]
- Robert C, Bernalier-Donadille A. The cellulolytic microflora of the human colon: evidence of microcrystalline cellulose-degrading bacteria in methane-excreting subjects. FEMS Microbiol Ecol. 2003;46:81–89. doi: 10.1016/S0168-6496(03)00207-1. [DOI] [PubMed] [Google Scholar]
- Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN. Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr. 2005;82:559–567. doi: 10.1093/ajcn.82.3.559. [DOI] [PubMed] [Google Scholar]
- Salyers AA, West SEH, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Lehmann U, Jacobasch G, Blaut M. Influence of resistant starch on the SCFA production and cell counts of butyrate-producing Eubacterium spp. in the human intestine. J Appl Microbiol. 2002;93:157–162. doi: 10.1046/j.1365-2672.2002.01679.x. [DOI] [PubMed] [Google Scholar]
- Schwiertz A, Tara D, Schafer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn's disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AM, Wiggins HS, Cummings JH. Effect of changing transit time on colonic microbial metabolism in man. Gut. 1987;28:601–609. doi: 10.1136/gut.28.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;24:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Duncan SH, Harmsen HJM, Holtrop G, Welling GW, Flint HJ. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol. 2008;10:3275–3283. doi: 10.1111/j.1462-2920.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Conway PL, Brown IL, Evans AJ. In vitro utilization of amylopectin and high amylose maize (amylomaize) starch granules by human colonic bacteria. Appl Environ Microbiol. 1999;65:4848–4854. doi: 10.1128/aem.65.11.4848-4854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JC, Clayton MK. A similarity measure based on species proportions. Commun Stat Theo Methods. 2005;34:2123–2131. [Google Scholar]
- Zoetendal EG, Akkermans ADL, de Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human faecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, Kudma D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2008;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.