Abstract
The presence and role of Archaea in artificial, human-controlled environments is still unclear. The search for Archaea has been focused on natural biotopes where they have been found in overwhelming numbers, and with amazing properties. However, they are considered as one of the major group of microorganisms that might be able to survive a space flight, or even to thrive on other planets. Although still concentrating on aerobic, bacterial spores as a proxy for spacecraft cleanliness, space agencies are beginning to consider Archaea as a possible contamination source that could affect future searches for life on other planets. This study reports on the discovery of archaeal 16S rRNA gene signatures not only in US American spacecraft assembly clean rooms but also in facilities in Europe and South America. Molecular methods revealed the presence of Crenarchaeota in all clean rooms sampled, while signatures derived from methanogens and a halophile appeared only sporadically. Although no Archaeon was successfully enriched in our multiassay cultivation approach thus far, samples from a European clean room revealed positive archaeal fluorescence in situ hybridization (FISH) signals of rod-shaped microorganisms, representing the first visualization of Archaea in clean room environments. The molecular and visual detection of Archaea was supported by the first quantitative PCR studies of clean rooms, estimating the overall quantity of Archaea therein. The significant presence of Archaea in these extreme environments in distinct geographical locations suggests a larger role for these microorganisms not only in natural biotopes, but also in human controlled and rigorously cleaned environments.
Keywords: Archaea, artificial environment, clean room, microbial diversity, planetary protection
Introduction
The study of the biology and ecology of Archaea, the third domain of life on Earth, has undergone significant changes and great discoveries during recent years. For more than 20 years Archaea were considered to be extremophiles that were ecologically restricted, and highly adapted to specific and often hostile biotopes (for example, Pyrodictium, Picrophilus; Woese and Fox, 1977; Moissl et al., 2008). However, new molecular methodologies have revealed an omnipresence of Archaea in almost any ‘normal' natural biotope, like marine and freshwater environments, or soil (Bintrim et al., 1997; Karner et al., 2001; Rudolph et al., 2004). Nevertheless, the successful enrichment or even isolation of cold loving or mesophilic Archaea (besides methanogens) has only rarely been reported (Konneke et al., 2005). By using indirect techniques such as metagenomics, fluorescence in situ hybridization (FISH)-based methods and submicronscale ion imaging, great insights were obtained into the ecological and physiological properties of Archaea in their diverse habitats, and their role as a significant motor in chemical conversions (Teira et al., 2004; Cavicchioli et al., 2007; Dekas et al., 2009). These methodologies also revealed an astonishing quantitative presence: It is estimated that about 1/3 of all the marine microbes are constituted by archaeal cells (Karner et al., 2001).
So far, most studies searching for Archaea still focus on the natural habitats. Almost nothing is known about the presence of Archaea in artificial systems and human maintained environments. Although very rare studies point to a possible pathogenic role of some methanogens (Vianna et al., 2006), Archaea are considered harmless to human health, and therefore are not, unlike bacteria, in the focus of medical surveys. Humans can be carriers of certain methanogens in their intestine, however it is not certain whether humans are hosts for other groups of Archaea, and/or have immediate contact with Archaea in their external environments. Thus far, no surveys have been carried out looking for Archaea in, for instance, human households, offices, airplanes, clinical environments or other restricted environments like pharmaceutical and industrial clean rooms.
Clean rooms are unusual and extreme environments for microorganisms. Low nutrient levels, low humidity, frequent cleaning and sterilization procedures pose a threat for all organisms thriving or living in these environments (Venkateswaran et al., 2001). Challenging the survival of microbes is one of the major aims of spacecraft-associated clean rooms: In terms of planetary protection, biological contamination on spacecraft flying to extraterrestrial points of interest has to be controlled (understood and reduced) in order to avoid compromising the integrity of the future life-detection missions on Mars, Titan or other solar bodies (see also: Horneck et al., 2010). As nowadays spacecraft cannot be sterilized (‘baked') as a whole, a microbial quality control of each hardware item is tremendously important. Standard procedures for the detection of microbial contamination are still based on cultivation, specifically by counting the colonies of robust, spore-forming bacteria, surviving a heat shock at 80 °C (National Aeronautics and Space Administration (NASA) standard procedure, Anonymous, 1999; European Space Agency (ESA) standard procedure, Anonymous, 2008). The number of colonies serves as a proxy for the overall contamination of the spacecraft. Nevertheless, recent studies have shown that a broad variety of bacteria is present on or around the spacecraft, out of which many showing special adaptations and extreme tolerances (LaDuc et al., 2007). Molecular studies have revealed an even broader bacterial diversity than that assessable through cultivation procedures, and even the estimated cell numbers likely needs to be corrected upwards (LaDuc et al., 2004). These findings clearly show that diverse bacterial communities are able to resist conventional sterilization and cleaning procedures, and emphasize the need for novel sterilization methodologies and improvement of the detection methods (Moissl et al., 2007).
Being aware of a specialized, adapted bacterial community in such clean rooms, the main detection is still directed towards bacteria, but Archaea (and Eukarya) are further considered as a possible contamination source in the field of planetary protection. Archaea were discussed as one of the most likely life forms on Earth to survive a space flight, and survive or even thrive on Mars (mainly methanogens and halophiles; Kendrick and Kral, 2006; Landis, 2001), but it was unclear, if Archaea even could be found in spacecraft-associated clean rooms.
Very recently, we reported the molecular detection of Archaea in two US American NASA spacecraft assembly facilities (Moissl et al., 2008). Overall, 30 different crenarchaeal and euryarchaeal 16S rRNA gene sequences were derived from these American clean rooms, however the physiological status and quantity of associated cells were unknown.
In this study, the presence of Archaea in spacecraft assembly clean rooms around the globe was confirmed by the analysis of three distinct spacecraft assembly clean rooms in Europe and South America. In addition to molecular 16S rRNA gene sequence analysis, data were supported through FISH, and quantitative real-time PCR (qPCR). This study reveals that Archaea are a significant part of the microbial community in spacecraft assembly clean rooms all over the world, and that this group of microorganisms is able to persist in these artificial, human maintained environments.
Materials and methods
Sampling
Samples were collected from selected surface areas within the two European spacecraft-associated clean rooms, as well as one clean room (final assembly building) in French Guiana, South America. All sampling locations and their characteristics are summarized in Table 1. During sampling, the European clean rooms were in full operation, and harbored the Herschel Space Observatory. Further details concerning these samplings are given in Stieglmeier et al. (2009). Within final assembly building at Centre Spatial Guyanais in French Guiana, different clean room surfaces were sampled. The final assembly building clean room is nominally operated as ISO 8 (according ISO 14644-1), but was opened 1 day before sampling to move in a container, harboring Ariane 5 fairings to be launched with the Herschel observatorium.
Table 1. Sampling locations and characteristics.
| Sampling location | EADS Friedrichshafen (FR) | ESTEC Noordwijk (ES) | CSG Kourou (KO) |
| Sampling date | November 2007 | March 2008 | April 2009 |
| Country | Germany | The Netherlands | French Guiana |
| Clean room facility | Hall 6, room 6101–04 | Hydra | BAF |
| Clean room specificsa | ISO 5 | ISO 8 | ISO 8 |
Abbreviations: BAF, final assembly building; CSG, Centre Spatial Guyanais; EADS, European Aeronautic Defence and Space Company; ESTEC, European Space Research and Technology Centre; ISO, International Organization for Standardization.
Clean room classification according ISO 14644–1.
All samples for molecular analysis were taken by using BiSKits (QuickSilver Analytics, Abingdon, MD, USA) as recommended by the manufacturer (sample area up to 1 m2). The selected area was wiped in three different directions while rotating the sampling device (this protocol was adapted from the ESA ‘Microbial examination of flight hardware and cleanrooms' standard for wipe sampling; Anonymous, 2008). Whereas clean room floors were sampled from European facilities, at Centre Spatial Guyanais the inside floor of the fairing container was sampled for comparison.
Samples for cultivation were taken using SpongeSicles (Biotrace (3 M), St Paul, MN, USA) as described in Stieglmeier et al. (2009). SpongeSicles were premoistened with water (10 ml) before sampling. Also here, the sampling area was wiped thrice in different directions. After sampling, 10 ml of sterile phosphate-buffered saline (PBS) was added. In case of anaerobic sample treatment, SpongeSicles were stored under anoxic conditions immediately after sampling (see Stieglmeier et al., 2009). Within the final assembly building at Centre Spatial Guyanais, samples were taken from various clean room surfaces. In addition, platform 12 was sampled, located at the upper end of Ariane 5, where the integration of the Herschel observatorium took place. All samples and their characteristics are given in Table 2.
Table 2. Summary of all study-relevant samples and their characteristics.
| Sample ID | Sampling location | Sampled area (in m2) | Sampling tool | Total volume | Analysis (used volume in ml)a |
|---|---|---|---|---|---|
| FR | |||||
| WA1, WA2, WA3b | Clean room floor | 0.36 | SpongeSicle | 80.0 | Cultivation (1.0) |
| W4 | Clean room floor | 0.36 | SpongeSicle | 12.5 | Cultivation (0.5), FISH (0.5) |
| W5 | Stairs, GSE | 0.07 | SpongeSicle | 12.5 | Cultivation (0.5), FISH (0.5) |
| B3 | Clean room floor | 0.54 | BiSKit | 4.5 | DNA extraction (1.5 ml), pooled with B5, B4/B5 |
| B4 | Clean room floor | 0.45 | BiSKit | 4.5 | DNA extraction (1.5 ml), pooled with B3/B5 |
| B5 | Floor ramp, GSE | 0.63 | BiSKit | 4.5 | DNA extraction (1.5 ml), pooled with B3, B3/B4 |
| ES | |||||
| W1 | Clean room floor | 0.36 | SpongeSicle | 11.0 | Cultivation (0.4) |
| W2 | Clean room floor | 0.36 | SpongeSicle | 7.5 | Cultivation (0.2) |
| W3 | Clean room floor | 0.28 | SpongeSicle | 9.3 | Cultivation (0.4) |
| WA1b | Stairs, GSE | 0.11 | SpongeSicle | 80.0 | Cultivation (1.0) |
| B1 | Clean room floor | 0.36 | BiSKit | 3.6 | DNA extraction (1.0), pooled with B2; FISH (0.5) |
| B2 | Clean room floor | 0.36 | BiSKit | 3.0 | DNA extraction (1.0), pooled with B1; FISH (0.5) |
| B3 | Clean room floor | 0.16 | BiSKit | 3.5 | FISH (0.5) |
| B4 | Clean room floor | 0.36 | BiSKit | 0.9 | FISH (0.5) |
| KO | |||||
| W1 | Clean room floor | 1.0 | SpongeSicle | 12.0 | Cultivation (0.2), FISH (0.5) |
| W3 | Clean room floor (p12) | 1.0 | SpongeSicle | 12.0 | Cultivation (0.2), FISH (0.5) |
| W4 | Container floor | 1.0 | SpongeSicle | 12.0 | Cultivation (0.2), FISH (0.5) |
| WA1b | Clean room floor | 1.0 | SpongeSicle | 80.0 | Cultivation (1.0) |
| WA2b | Clean room floor (p12) | 1.0 | SpongeSicle | 80.0 | Cultivation (1.0) |
| B1 | Clean room floor | 1.0 | BiSKit | 0.7 | FISH (0.5) |
| B2 | Clean room floor (p12) | 1.0 | BiSKit | 4.0 | FISH (0.5) |
| B4 (B) | Container floor | 1.0 | BiSKit | 2.9 | DNA extraction (1.0), FISH (0.5) |
Abbreviations: ES, European Space Research and Technology Centre; FISH, fluorescence in situ hybridization; FR, Friedrichshafen; GSE, ground support equipment; KO, Kourou; p12, platform 12.
For cultivation studies, the inoculation volume was given for one assay.
Samples were taken anaerobically. See also Stieglmeier et al. (2009).
Sample treatment
BiSKit samples for molecular analysis were stored chilled (4–8 °C) during transportation, and frozen immediately after arrival at the laboratory.
SpongeSicle samples were stored chilled during transportation. To extract the samples, SpongeSicles were squeezed and dispersed within their plastic transportation bag. The liquid was removed under a sterile hood and inoculated immediately. Anaerobic samples were shaken rigorously and samples were removed by using syringes and needles (Stieglmeier et al., 2009). Smaller fractions from SpongeSicle and BiSKit samples for FISH were fixed immediately (see below), or as soon as possible after return to the Regensburg laboratory.
Cultivation
The following liquid media were used for the cultivation of Archaea (see also Stieglmeier et al. (2009); the recipes are given for 1 l medium to be prepared with distilled water): TG (thioglycolate liquid medium): peptone from casein (Becton Dickinson (BD), Franklin Lakes, NJ, USA) 15.0 g; yeast extract (BD) 5.0 g; D(+) glucose 5.5 g; NaCl 2.5 g; sodium acetate 3.0 g; cysteine HCl 0.5 g; sodium thioglycolate 0.5 g; sodium resazurin 0.001 g; gas phase: N2, pH 7.1. TS (trypticase soy liquid medium): Trypticase soy broth (BD) 30.0 g; sodium resazurin 0.001 g; sodium thioglycolate 0.5 g; cysteine·HCl 1.0 g; gas phase N2 (80%) and CO2 (20%). MM (methanogenic Archaea liquid medium): NH4Cl 0.5 g; KH2PO4 0.4 g; MgCl2·6 H2O 0.15 g; CaCl2·2 H2O 0.05 g; trace element solution (10 × ) 1 ml; vitamin solution (10 × ) 1 ml; sodium resazurin 0.001 g; Na2S 0.5 g; cysteine HCl 0.5 g; gas phase H2 (80%) and CO2 (20%), supplemented with either acetate, methanol or no addition). BM (basal medium): NH4Cl 0.5 g; KH2PO4 0.4 g; MgCl2·6 H2O 0.15 g; CaCl2·2 H2O 0.05 g; NaHCO3 1.0 g; trace element solution (10 × ) 1 ml; vitamin solution (10 × ) 1 ml; sodium resazurin 0.001 g; Na2S 0.25 g; cysteine·HCl 0.25 g; pH 7.0. ASM (Archaea supporting liquid medium): per 20 ml BM, add 0.1 ml of an antibiotics mixture (carbenicillin (0.2%, w/v), streptomycin (0.2%, w/v), rifampicin (0.4%, w/v) and cephalosporin (0.2%, w/v); gas phases either aerobic, microaerobic (3% oxygen) or anaerobic (N2), pH 7.0, supplemented with either yeast extract or ammonium chloride). AAM (autotrophic all-rounder liquid medium): KH2PO4 0.4 g; CaCl2·2 H2O 0.05 g; MgCl2·6 H2O 0.15 g; NaHCO3 1.5 g; Fe2O3·9 H2O 0.25 g; NaNO3 0.5 g; Na2S2O3·5 H2O 1.56 g; trace element solution (10 × ) 1 ml; vitamin solution (10 × ) 1 ml; sodium resazurin 0.001 g; Na2S 0.5 g; gas phase: N2 (80%) and CO2 (20%) For the composition of the trace element solution and vitamin solution: see DSMZ medium 141 (http://www.dsmz.de). For the preparation of the media see Stieglmeier et al. (2009).
All cultivations were done at 32 °C for 5 to 6 months, and examined weekly through microscopy for growth. If growth occurred, phylogenetic affiliation of the microbes was determined using FISH, or 16S rRNA gene sequencing.
Field blanks and media blanks were included as negative controls for the cultivation attempts.
FISH
SpongeSicle samples from Friedrichshafen (FR) sampling were used for FISH (see Table 2). Because of problems with background fluorescence in SpongeSicle samples, for the other samplings, mainly BiSKit samples were used for FISH (Table 2). For each analysis, 500 μl of the original sample was fixed using 3% (w/v) paraformaldehyde, either directly after sampling (FR and ES), or at arrival at the laboratory (Kourou). Because of very low biomass in the samples, they were concentrated by centrifugation to 15 μl. Bacteria- and Archaea-specific FISH was carried out as described previously (Moissl et al., 2002). For the detection of bacteria, Cy3-labeled EUB 338 probe (Amann et al., 1990) was used. For Archaea-specific hybridization, three probes (rhodamine green-labeled) were used as a mixture (ARCH344, ARCH915, ARCH1060; Moissl et al., 2003). Samples, field blanks and positive controls (SM1 Euryarchaeon (Moissl et al., 2003) and Escherichia coli) were stained in parallel, but on different slides to avoid cross-contamination.
DNA extraction
Sampling liquid from BiSKits (up to 4.5 ml for each individual DNA extraction) was concentrated using UV sterilized Amicon Ultra-15 filters (cutoff 50 kDa; Millipore GmbH, Schwalbach, Germany). The resulting suspension was subjected to DNA extraction using the XS buffer method (Tillett and Neilan, 2000, modified): XS buffer (2 × ) was freshly prepared as follows: (20 ml stock solution): 1 M Tris/HCl (pH 7.4) (4 ml); 7 M ammonium acetate (4.56 ml); 250 m ethylene diamine tetraacetic acid (3.2 ml); 10% sodium dodecyl sulfate (w/v) (4 ml); potassium ethyl xanthogenate (0.4 g); PCR-grade water (4.99 ml). For completely dissolving the xanthogenate, the buffer was incubated at 65 °C for 15 min.
Starting with for example 1 ml sample, in total, 1 ml of 2 × XS buffer was added, and the mixture was stirred gently (short vortex). After an incubation of 2 h at 65 °C, and mixing by hand for about every 30 min, the suspension was vortexed for 10 s. The tube was placed on ice for 10 min and centrifuged afterwards (100 g, 5 min, 4 °C). The supernatant was transferred into a PhaseLock Gel tube (Eppendorf, Hamburg, Germany), and an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added. The suspension was mixed gently and centrifuged (2000 g, 5 min, 15 °C). The aqueous layer was transferred into a new tube. To precipitate DNA, the same volume of cold 100% isopropanol and 1/10 volume of 4 M ammonium acetate was added and gently mixed. After incubation at −20 °C overnight, the suspension was centrifuged at 13600 g at 4 °C for 30 min. The (invisible) pellet was washed with 1 ml 70% ethanol (ice cold) and centrifuged (13600 g, 30 min, 4 °C). The pellet was dried completely and afterwards dissolved in 15 μl PCR-grade water.
PCR and cloning of archaeal 16S rRNA genes
For the analyses in this study, nested PCR was carried out. Primers 8aF (Burggraf et al., 1992) and 1406uR (Lane, 1991) served as a primer pair for the first amplification, for which 1 μl of the DNA extract was used. The PCR was carried out using the Pure Taq system (Peqlab, Erlangen, Germany) under the following conditions: 95 °C for 4 min; 33 cycles of 95 °C for 50 s, 60 °C for 50 s and 72 °C for 1 min 30 s; and final incubation at 72 °C for 10 min. Afterwards, 1 μl of the amplicon was used for the second PCR reaction with primer combination 345aF and 1119aR (Burggraf et al., 1997). The length and quantity of the amplicons were visualized on a 1% agarose gel. Amplified PCR fragments were subjected to cloning using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The presence of inserts of the expected size was verified by direct PCR screening of up to 100 transformants without plasmid extraction. For restriction fragment length polymorphism analyses, the PCR products were digested with a mixture of two different restriction endonucleases (BsuRI and HinfI; Fermentas GmbH, St Leon-Rot, Germany). The resulting restriction patterns were compared, and representative transformants were selected and their plasmid DNA extracted (Qiaprep kit, Qiagen, Valencia, CA, USA) as recommended by the manufacturer. Sequencing of the 16S rRNA gene sequences was carried out by the companies Geneart or Entelechon (Regensburg, Germany).
Controls
In each step of the sampling, cultivation, FISH, DNA extraction and PCR, suitable negative controls were used. Field blanks were taken (sampling tool was opened on sampling site and waved through the air), and served as an additional negative control, besides media controls (cultivation), extraction blanks (molecular studies) and water blanks (molecular studies).
In addition to water controls an unopened BiSKit buffer was included as a negative control.
All PCR reactions were carried out with the following controls: a positive control (DNA of Methanococcus aeolicus), and three negative controls (field blank, extraction blank and water blank). In order to check whether any inhibitory molecules were present in the extract, all samples that showed negative PCR amplification for Archaea were spiked with a known amount of M. aeolicus DNA.
Quantitative PCR
Quantitative PCR was performed using the RotorGene 6000 Real-Time PCR system (Corbett Life Science, Concorde, NSW, Australia). The qPCR reactions were carried out in 20 μl with 1 × SYBR Green Taq Premix (Quantitect SYBR Green PCR kit, Qiagen, Hilden, Germany). In total, 1 μl of extracted DNA was used for the amplification with the Archaea-specific primer combination 345aF-517uR (Lane, 1991; Burggraf et al., 1997). The amplification protocol was as follows: hold at 95 °C, 15 min; cycles (40 repeats): 94 °C (15 s), 60 °C (30 s) and 72 °C (30 s). Fluorescence measurements were taken at the end of each cycle. Standards were developed from PCR products of the 16S rRNA gene from M. aeolicus, and 10-fold serial dilutions thereof. For this, PCR amplicons were purified and the concentration was determined using Nanodrop (ND-1000, Thermo Scientific, Wilmington, USA); on the basis of the 16S rRNA gene sequence of M. aeolicus, its nucleotide composition and weight was determined, and the copy number was calculated subsequently.
Coverages and phylogenetic calculations
These analyses (calculation of coverage of clone libraries, chimera check, ARB) were carried out as given previously (Moissl et al., 2008).
Nucleotide sequence accession numbers
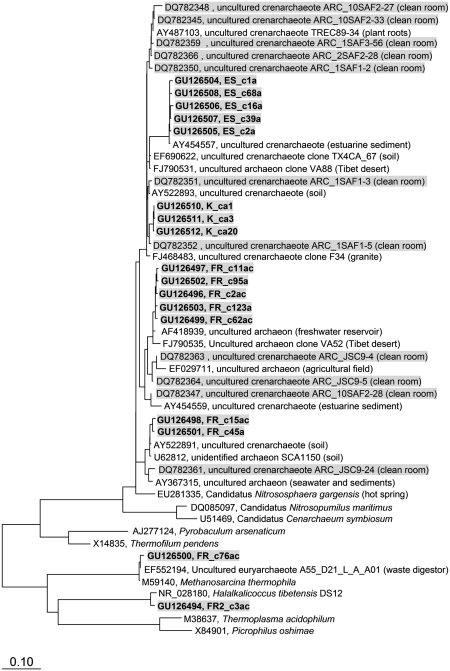
The 16S rRNA gene sequences of the clones were deposited in the NCBI nucleotide sequence database. The accession numbers are given in Figure 1.
Figure 1.
Phylogenetic tree, showing selected archaeal 16S rRNA genes from US American, European and South American spacecraft-associated clean rooms. The tree was calculated on the basis of the maximum likelihood methodology, implemented in the ARB software package (Ludwig et al., 2004). Sequences obtained from this study are shown in bold and highlighted (gray background). Sequences from previous studies are shown with gray background. Abbreviations: FR, Friedrichshafen; ES, ESTEC; KO, Kourou; SAF, spacecraft assembly facility at the Jet Propulsion Laboratory (Pasadena, CA, USA); JSC, Johnson Space Center (Houston, TX, USA).
Results
During a biodiversity study connected to the Herschel mission, launched in May 2009, samples were taken from three different locations in Europe and South America (Table 1, see also Stieglmeier et al., 2009). Focusing on the archaeal diversity, samples were taken for cultivation attempts, molecular analysis (qPCR and 16S rRNA gene screening) and FISH assays. Overall, 24 different clean room surface samples (and three pooled samples) were analyzed for the presence of archaeal signatures and cultivable cells.
For cultivation, samples were taken both aerobically or anaerobically (see also Stieglmeier et al., 2009). In total, 32 different cultivation strategies were applied, 9 of them focusing mainly on Archaea: media specific for methanogenic Archaea (acetotrophs, methylotrophs and hydrogenotrophs), as well as media for a broad variety of metabolic pathways supplied with a combination of four different antibiotics were applied. This mixture of antibiotics has been used successfully for the cultivation of mesophilic Crenarchaeota by suppressing the concomitant bacteria (Konneke et al., 2005). Nevertheless, none of our cultivation assays revealed positive (microscopically visible) archaeal growth in the time frame of 5 to 6 months.
In general, the microbial density in samples taken from clean rooms is very low (LaDuc et al., 2004). For FISH, samples had to be concentrated through centrifugation by a factor of 33 in order to obtain visible cell densities on hybridization slides. The majority of detectable microorganisms showed strong 4′,6-diamidino-2-phenylindole (DAPI) signals, and a low to bright signal with a bacteria-specific probe. These results confirmed that the hybridization conditions used, allowed for the detection of a broad variety of bacterial cells, even though many of them were most likely not in an actively growing status. Bacteria of different morphological types were detected, in particular diplorods (FR), rods (all locations), diplococci (all locations), single cocci (ES and KO) and tetrads (ES).
Samples from Kourou gave comparatively low fluorescence signals, probably because of the longer transportation time from South America to the laboratory in Regensburg, Germany.
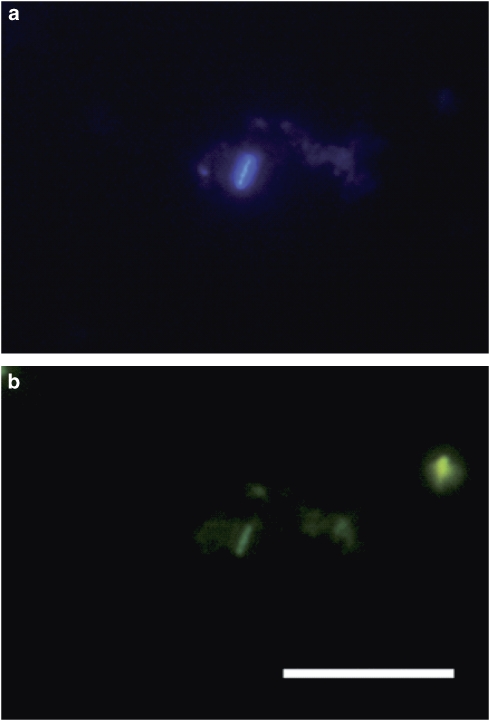
Notably, some cells from a European Space Research and Technology Centre sample (B1, clean room floor, right entrance) gave positive signals with Archaea-specific probes on duplicate slides. These archaeal cells, estimated 1 cell per 100 bacteria, and were brightly DAPI-stained, and did not hybridize with the bacteria specific-probe (Figure 2). The cells detected were rod-shaped, revealing a length of about 2 μm and a width of 0.5 μm. Because of the brightness of the hybridization signal, which depends on the number of ribosomes in the cells, the microbes appeared to be physiologically active.
Figure 2.
Fluorescence in situ hybridization (FISH) of an ESTEC clean room sample, showing a rod shaped microorganism stained with archaeal probes. (a) Dapi stain. (b) Same section. Rod shows a positive archaeal signal, staining with an archaeal probe mixture (rhodamine green labeled). Bar: 10 μm. No signal was obtained with bacteria-specific probe mix staining in parallel (Cy3 labeled, not shown).
A previous study showed that because of low microbial biomass observed in spacecraft-assembly clean rooms, and the even lower abundance of Archaea, parallel samples taken in one clean room led to 2/3 negative results when screened for archaeal 16S rRNA genes (Moissl et al., 2008). Therefore, samples from larger surface areas were taken and pooled in order to detect archaeal signatures more effectively. BiSKit samples were pooled before DNA extraction as given in Table 2. Extracted DNA from BiSKit clean room samples was subjected to nested-archaeal PCR. None of the negative controls resulted in the amplification of archaeal 16S rRNA genes, but all clean room samples revealed the presence of signatures from Archaea (Table 3). The ∼800-bp PCR amplicons obtained were subjected to cloning, and more than 100 clones per sampling location were analyzed. One FR sequence was judged as a chimera, and therefore removed from all calculations.
Table 3. Summary of results: Clone libraries, FISH and qPCR.
| Friedrichshafena | ESTEC | Kourou | |
|---|---|---|---|
| Clone libraries | |||
| Samples | B3/5 and B3/4/5 | B1/2 | B |
| Euryarchaeotab | 2 | − | − |
| Crenarchaeotab | 9 | 6 | 3 |
| Clones analyzed | 100 and 86 | 109 | 110 |
| Coverage | 100 and 96% | 98.2% | 99.1% |
| FISH | |||
| Bacteria | + | + | + |
| Archaea | − | + | − |
| Archaeal qPCR | |||
| Samples | B3/5 | B1/2 | B |
| 16S rRNA gene copies per m2b | 13461 | 85388 | 18760 |
| Estimated cell number per m2b | 7.6 × 103 | 4.8 × 104 | 1.1 × 104 |
Abbreviations: ESTEC, European Space Research and Technology Centre; FISH, fluorescence in situ hybridization, qPCR, quantitative polymerase chain reaction.
Two clone libraries were carried out: One with sample B3/5 and other with sample B3/4/5.
Number of different 16S rRNA gene sequences detected.
The clone coverage values of the 16S rRNA recombinant libraries were fairly high (96–100%), indicating an almost complete accession of the 16S rRNA genes present, on the basis of the methods applied. The archaeal clean room sequences were affiliated with Euryarchaeota and Crenarchaeota, whereas Euryarchaeota were detected in FR samples only. The majority of sequences obtained, clustered with mainly uncultivated Crenarchaeota from very diverse biotopes like soil, sediments, deserts or plant roots (Figure 1). Interestingly, the US American clean room sequences (Moissl et al., 2008) clustered within this group of Archaea as well (crenarchaeal soil group 1b; DeLong, 1998), and were closely related to the newly retrieved clone sequences. The closest cultivated neighbor to these sequences was the thermophilic ammonia oxidizer candidatus Nitrososphaera gargensis (Hatzenpichler et al., 2008), showing up to 96% similarity in the 16S rRNA gene with the sequences from this study. A phylogenetic tree with representative sequences is given in Figure 1.
The crenarchaeal clean room sequences from this study spanned a difference of up to ∼5% from each other within the ∼800 bp fragment of the 16S rRNA gene. The sequences obtained from locations ESTEC and Kourou formed small clusters with up to 1.2% difference in the single sequences. On the other hand, the broadest diversity of Archaea was observed in FR samples: Besides Crenarchaeota, one sequence from a methanogen was obtained (Methanosarcina). Additionally, a sequence from a halophilic and maybe even alkaliphilic Archaeon has been amplified from these samples (Halalkalicoccus, Figure 1).
All samples were subjected to archaeal quantitative PCR, in order to estimate the quantity of archaeal cells per m2. The average 16S rRNA gene copy number of Archaea was measured to be around 4 × 104 per m2, which corresponds to an estimated cell number of about 2.2 × 104 (the calculation was on the basis of the average number of 16S rRNA gene copies for Archaea (1.77), as available at rrnDB (Klappenbach et al., 2001; Lee et al., 2009; Table 3). The typical bacterial cell number was estimated to be around 105–107 per m2, approximately two logs higher than the archaeal cell number. The highest number of Archaea was detected in the ES-pooled sample, in agreement with the appearance of the positive archaeal FISH signal.
Discussion
This study is a completion, continuation and deepening of a previous study, that reported the finding of archaeal 16S rRNA gene signatures in US American spacecraft assembly facilities (Moissl et al., 2008). Until now, the significance of the finding, in terms of the abundance of these archaeal cells, their physical and physiological state, and their distribution throughout the globally distributed clean rooms, was unknown.
In this study we addressed these crucial open questions, and were able to confirm the presence of a particular crenarchaeal community in European and South American clean rooms: Again, the majority of the archaeal 16S rRNA genes detected, clustered within the Crenarchaeota group 1b (300 of the 400 clones). The presence of these Crenarchaeota is significant as different procedures, and setups were used in our two studies (Moissl et al., 2008): Different sampling methods (wipe sampling, BiSKit sampling), extraction methods (phenol–chloroform extraction, XS buffer method), DNA amplification procedures (different Taq systems, primer sets) were applied. Additionally, the experiments were conducted in two different laboratories (Jet Propulsion Laboratory, USA and Institute for Microbiology, Regensburg, Germany). To date, 48 different crenarchaeal 16S rRNA gene sequences were obtained from five global clean rooms.
Using a sensitive qPCR method, the number of Archaea m−2 clean room surface was estimated to be around 2.2 × 104, which is approximately two to three logs lower than the estimated number of bacteria. Even though this archaeal abundance appears low, the clean room Crenarchaeota were detected in every facility analyzed, so that the presence of these Archaea appears to be as consistent as clean room-associated bacterial genera such as Staphylococcus, Micrococcus, Bacillus and other typical bacterial contaminants (Moissl et al., 2007).
Our study is the first report of the application of a qPCR method for the detection and quantification of Archaea in clean room environments. Future qPCR studies of other clean rooms and human-controlled environments will be necessary to confirm this low, but constant abundance of Archaea.
This study also reports the first visualization of archaeal cells in spacecraft-assembly clean rooms. Rod-shaped microorganisms were observed, that showed a bright signal with Archaea-specific probes, which hints towards the presence of intact and potentially active cells in these environments. Nevertheless, because of the low biomass available for FISH, the Archaea-specific hybridization result could not be analyzed further to identify Euryarchaeota or Crenarchaeota signals. As ES samples (from which the positive FISH signal was obtained) resulted in the detection of crenarchaeal 16S rRNA gene sequences only, it can be assumed that the FISH signal probably belonged to crenarchaeal representatives.
Even though archaeal 16S rRNA gene sequences have now been amplified from a number of different clean rooms in geographically distinct locations, the cultivation of Archaea from these artificial environments has not been successful thus far. It is possible, that the selected temperature and media have not been suitable for their successful isolation, although a broad variety of possible metabolic pathway specialists were targeted (for example, methanogens, sulfate reducers, iron oxidizers, iron reducers, ammonia oxidizers, nitrate reducers, organotrophs). These negative results show again that the cultivation of Archaea existing in mesophilic environments still remains very difficult. At present, nothing is known about archaeal ‘viable, but nonculturable' (VBNC) states, which have been reported for bacteria (Oliver, 2009), and might also affect the cultivability of Archaea. This problematic background has also been discussed previously for the bacterial diversity in clean rooms (LaDuc et al., 2004).
Because of the fact that most bacteria found in clean room environments are human symbionts, commensals or pathogens (for example, Staphylococcus, Clostridia, Propionibacteria, Moissl et al., 2007; Stieglmeier et al., 2009), the detection of a typical archaeal clean room population might also point towards a human involvement. Humans are the only omnipresent, flexible, not sterilizable or cleanable items, causing traffic and air turbulences within the clean rooms. It appears plausible that the Archaea detected in these clean rooms were introduced by human activity, and they could therefore be envisaged as human associated.
So far, Archaea appear to be harmless to humans, but they are not harmless in the context of space travel. Detailed resistance studies of vegetative thermophilic and hyperthermophilic Archaea have unexpectedly revealed tolerances against desiccation, vacuum and radiation (UV and γ, Beblo et al., 2009). It is unclear however, whether these organisms could cope with the problematic and extremely harsh conditions during space travel, or lack of nutrients and low temperatures in extraterrestrial environments.
Methanogens in particular are considered as one of the most likely life forms on Earth to be able to survive on Mars or other planets (McCollom, 1999; Krasnopolsky et al., 2004). Many methanogens are primary producers, adapted to anaerobic environments with very low or no organic compounds making them perfect candidates for the colonization of extraterrestrial environments. In particular the presence of methanogens on Mars has already been discussed, as the Martian atmosphere contains traces of methane, and an imbalance of efflux and production was observed. Although the origin of the detected methane is unclear, a microbiological methane source is one possibility (Lefevre and Forget, 2009).
Signatures from methanogens have been found in the US American SAF (Moissl et al., 2008), and also in the FR facility. This shows the possibility that these organisms may be in contact with spacecraft, and could potentially contaminate other planets.
Besides methanogens, halophiles have been discussed as possible survivors on Mars, as the Martian liquid water (if available) would contain high concentrations of different salts (Landis, 2001). Signatures of a halophilic Archaeon has been obtained from the clean room in FR (Halalkalicoccus), an Archaeon that appears to be adapted to salty, alkalic environments (Xue et al., 2005).
The cultivability and availability of closely related strains allow a certain speculation and discussion about the possible properties of the detected methanogens and halophile. However, it is unclear at present whether the signatures obtained from the clean rooms were extracted from living, intact cells that could possibly pose a threat to planetary protection.
In contrast, much less information is available about the possible role of the particular crenarchaeal clean room populations that have been detected in all spacecraft assembly facilities analyzed. Closer relatives thereof were detected in molecular studies of natural biotopes only, the closest cultivated neighbor (Nitrososphaera gargensis) shows more than 4% difference in the 16S rRNA gene sequence. Nevertheless, many of the uncultivated relatives, and also Nitrososphaera were described to have the appropriate genes for ammonia oxidation (Hatzenpichler et al., 2008; Erguder et al., 2009). The focussed search for functional genes in clean room samples, such as archaeal amo A genes, could possibly shed light onto the physiological properties of the clean room Crenarchaeota. The ability for ammonia oxidation is not necessarily an advantage for the survival on the Red Planet, but could be beneficial when introduced in the environments of the Saturn's moons Titan or Enceladus. Although the temperature on both moons is quite low, the presence of ammonia, H2O and other prerequisites for a possible growth of specialized microbes has been discussed (Israel et al., 2005).
For sure, the affect of the finding of Archaea in spacecraft-associated clean rooms on planetary protection requires further discussion. Besides including Archaea in biodiversity studies of spacecraft assembly clean rooms, an expansion and adaptation of the standard microbial detection procedures to include Archaea is definitely recommended.
In summary, the presence of Archaea in highly specific, human maintained and controlled environments can no longer be denied. Their detection emphasizes the ability of this domain of life to exist in the most unexpected biotopes, including in habitats that are of human origin. Other studies attempting to detect Archaea on artificial surfaces and in human environments are very rare and have not yet resulted in positive signals. Because of their lower abundance, Archaea require different, more sensitive methods for identification and quantification, and maybe also variation in primers and amplifying systems. Looking at other human-maintained habitats might also increase awareness of potential interactions between Archaea and humans, including symbiotic or even pathogenic associations.
Acknowledgments
I thank Michaela Stieglmeier and Petra Schwendner for technical support and cultivation attempts. I am grateful to Gerhard Kminek (ESA) and Reinhard Wirth (Regensburg) for support and discussion. I thank Ruth Henneberger, Ariane Briegel and Anne Dekas for critically reading the paper. Funding by ESA is gratefully acknowledged (contract no. 20508/07/NL/EK).
The author declares no conflict of interest.
References
- Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous 2008Microbial examination of flight hardware and cleanrooms ECSS-Q-ST-70-55C European Cooperation for Space StandardizationESA-ESTEC.
- Anonymous 1999NASA standard procedures for the microbiological examination of space hardware, NPG 5340.1D Jet Propulsion Laboratory communication, National Aeronautics and Space Administration.
- Beblo K, Rabbow E, Rachel R, Huber H, Rettberg P. Tolerance of thermophilic and hyperthermophilic microorganisms to desiccation. Extremophiles. 2009;13:521–531. doi: 10.1007/s00792-009-0239-1. [DOI] [PubMed] [Google Scholar]
- Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraf S, Huber H, Stetter KO. Reclassification of the crenarchael orders and families in accordance with 16S rRNA sequence data. Int J Syst Bacteriol. 1997;47:657–660. doi: 10.1099/00207713-47-3-657. [DOI] [PubMed] [Google Scholar]
- Burggraf S, Olsen GJ, Stetter KO, Woese CR. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R, Demaere MZ, Thomas T. Metagenomic studies reveal the critical and wide-ranging ecological importance of uncultivated Archaea: the role of ammonia oxidizers. Bioessays. 2007;29:11–14. doi: 10.1002/bies.20519. [DOI] [PubMed] [Google Scholar]
- Dekas AE, Poretsky RS, Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science. 2009;326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- DeLong EF. Everything in moderation: archaea as ′non-extremophiles′. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horneck G, Klaus DM, Mancinelli RL. Space Microbiology. Microbiol Mol Biol Rev. 2010;74:121–156. doi: 10.1128/MMBR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel G, Szopa C, Raulin F, Cabane M, Niemann HB, Atreya SK, et al. Complex organic matter in Titan′s atmospheric aerosols from in situ pyrolysis and analysis. Nature. 2005;438:796–799. doi: 10.1038/nature04349. [DOI] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Kendrick MG, Kral TA. Survival of methanogens during desiccation: implications for life on Mars. Astrobiology. 2006;6:546–551. doi: 10.1089/ast.2006.6.546. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. Rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konneke M, Bernhard AE, de lTorre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Krasnopolsky VA, Maillard JP, Owen TC. Detection of methane in the martian atmosphere: evidence for life. Icarus. 2004;172:537–547. [Google Scholar]
- LaDuc MT, Dekas A, Osman S, Moissl C, Newcombe D, Venkateswaran K. Isolation and characterization of bacteria capable of tolerating the extreme. Appl Environ Microbiol. 2007;73:2600–2611. doi: 10.1128/AEM.03007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaDuc MT, Kern R, Venkateswaran K. Microbial monitoring of spacecraft and associated environments. Microb Ecol. 2004;47:150–158. doi: 10.1007/s00248-003-1012-0. [DOI] [PubMed] [Google Scholar]
- Landis GA. Martian water: are there extant halobacteria on Mars. Astrobiology. 2001;1:161–164. doi: 10.1089/153110701753198927. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M. (eds).Nucleic acid techniques in bacterial systematics John Wiley & Sons Chichester: England; 115–175. [Google Scholar]
- Lee ZM, Bussema C, Schmidt TM.2009RrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea Nucleic Acids Res 37Database issueD489–D493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre F, Forget F. Observed variations of methane on Mars unexplained by known atmospheric chemistry and physics. Nature. 2009;460:720–723. doi: 10.1038/nature08228. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Research. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM. Methanogenesis as a potential source of chemical energy for primary biomass production by autotrophic organisms in hydrothermal systems on Europa. J Geophys Res. 1999;104:30729–30742. [Google Scholar]
- Moissl C, Bruckner JC, Venkateswaran K. Archaeal diversity analysis of spacecraft assembly clean rooms. ISME J. 2008;2:115–119. doi: 10.1038/ismej.2007.98. [DOI] [PubMed] [Google Scholar]
- Moissl C, Osman S, LaDuc MT, Dekas A, Brodie E, DeSantis T, et al. Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol Ecol. 2007;61:509–521. doi: 10.1111/j.1574-6941.2007.00360.x. [DOI] [PubMed] [Google Scholar]
- Moissl C, Rudolph C, Rachel R, Koch M, Huber R. In situ growth of the novel SM1 euryarchaeon from a string-of-pearls-like microbial community in its cold biotope, its physical separation and insights into its structure and physiology. Arch Microbiol. 2003;180:211–217. doi: 10.1007/s00203-003-0580-1. [DOI] [PubMed] [Google Scholar]
- Moissl C, Rudolph C, Huber R. Natural communities of novel archaea and bacteria with a string-of-pearls-like morphology: molecular analysis of the bacterial partners. Appl Environ Microbiol. 2002;68:933–937. doi: 10.1128/AEM.68.2.933-937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2009;34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- Rudolph C, Moissl C, Henneberger R, Huber R. Ecology and microbial structures of archaeal/bacterial strings-of-pearls communities and archaeal relatives thriving in cold sulfidic springs. FEMS Microbiol Ecol. 2004;50:1–11. doi: 10.1016/j.femsec.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Stieglmeier M, Wirth R, Kminek G, Moissl-Eichinger C. Cultivation of anaerobic and facultatively anaerobic bacteria from spacecraft-associated clean rooms. Appl Environ Microbiol. 2009;75:3484–3491. doi: 10.1128/AEM.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and Archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett D, Neilan BA. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol. 2000;35:1–8. [Google Scholar]
- Vianna ME, Conrads G, Gomes BP, Horz HP. Identification and quantification of archaea involved in primary endodontic infections. J Clin Microbiol. 2006;44:1274–1282. doi: 10.1128/JCM.44.4.1274-1282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran K, Satomi M, Chung S, Kern R, Koukol R, Basic C, et al. Molecular microbial diversity of a spacecraft assembly facility. System Appl Microbiol. 2001;24:311–320. doi: 10.1078/0723-2020-00018. [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Fan H, Ventosa A, Grant WD, Jones BE, Cowan DA, et al. Halalkalicoccus tibetensis gen. nov., sp. nov., representing a novel genus of haloalkaliphilic archaea. Int J Syst Evol Microbiol. 2005;55:2501–2505. doi: 10.1099/ijs.0.63916-0. [DOI] [PubMed] [Google Scholar]