Abstract
Marine sponges are ancient, sessile, filter-feeding metazoans, which represent a significant component of the benthic communities throughout the world. Sponges harbor a remarkable diversity of bacteria, however, little is known about the functional properties of such bacterial symbionts. In this study, we present the genomic and functional characterization of an uncultured δ-proteobacterium associated with the sponge Cymbastela concentrica. We show that this organism represents a novel phylogenetic clade and propose that it lives in association with a cyanobacterium. We also provide an overview of the predicted functional and ecological properties of this δ-proteobacterium, and discuss its complex interactions with surrounding cells and milieu, including traits of cell attachment, nutrient transport and protein–protein interactions.
Keywords: δ-proteobacterium, sponge, symbiosis, cyanobacterium, genome
Introduction
Sponges are ancient, sessile, filter-feeding metazoans, which represent a significant component of marine, benthic communities throughout the world. Despite their simple body plan, they are remarkably efficient in obtaining food by pumping surrounding seawater through a specialized canal system. Studies have estimated that as much as 24 000 l of water can be pumped though a kilogram of sponge on a daily basis and that the filtering system used by the sponge leaves the expelled water essentially sterile (Reiswig, 1971, 1974; Turon et al., 1997). Despite this process, sponges also host permanently a large number of microorganisms in their body, which can contribute up to 60% of the total sponge biomass (Wilkinson, 1978; Hentschel et al., 2006). These sponge-associated bacteria cover a remarkable diversity across major bacterial and archaeal phyla, and some clades found in sponges represent host-specific groups of bacteria (Taylor et al., 2007; Webster et al., 2008; Zhu et al., 2008). The interactions between sponges and microorganisms have been postulated to range from mutualism to commensalism and parasitism, however, little is understood regarding the exact nature of the symbiosis or the functional role played by certain groups of bacteria within the sponge (Webster and Blackall, 2008). One of the major limitations for our understanding of sponge–microorganism interactions is the inherent difficulty of culturing symbiotic bacteria. For most sponges the dominant or representative members are not available for further analysis.
We have recently applied whole-environment shotgun sequencing to characterize the bacterial community of the sponge Cymbastela concentrica, an abundant sponge species in shallow, temperate waters along the Australian east coast (Thomas et al., 2010). This metagenomic study has revealed previously unrecognized genomic signatures and functions of sponge bacteria including defense mechanisms against the introduction of foreign DNA, metabolic interactions with the host through vitamin production, nutrient transport and utilization, redox sensing and response, as well as protein–protein interactions mediated through ankyrin and tetratricopeptide repeat (TPR) proteins. In our metagenomic data set, we also discovered a divergent 16S ribosomal RNA (rRNA) gene sequence that belongs to the δ-proteobacteria class. To understand the functional characteristics of this novel organism and how these relate to sponge–bacteria interactions, a detailed genomic analysis and determination of cellular localization are presented here. We show that this organism represents a novel clade and that it lives within the sponge in association with a putative cyanobacterium. The predicted properties of the partial δ-proteobacterial genome indicate complex interactions with surrounding cells and their environment as reflected in traits for cell attachment, nutrient transport and protein–protein interactions.
Materials and methods
Phylogenetic analysis
The initial taxonomic classification of the δ-proteobacterial 16S rRNA gene sequence was carried out using the Bayesian classifier algorithm with default parameters at the Ribosomal Database Project (Wang et al., 2007; Cole et al., 2009). The nearest-neighbor sequences (both for uncultured and isolated representatives) of the δ-proteobacterial 16S rRNA gene sequence were extracted from the Ribosomal Database Project version 10 using the seqmatch function and then aligned using the SINA web aligner (Pruesse et al., 2007). A phylogenetic tree was constructed using the maximum likelihood algorithm with near full-length 16S rRNA gene sequences (>1200 nt) in the ARB software package (Ludwig et al., 2004). To avoid the use of highly variable regions for comparisons, a positional variability-by-parsimony mask (pos_var_Bacteria_94) and a user-specific end mask were used resulting in the comparison of 1178 nucleotides in the tree construction. An uncultured euryarchaeote sp. (Genbank Accession: AB077227) was used as an outgroup in the tree construction.
Genomic reconstruction and comparative genomics
The partial genome of the δ-proteobacterial species was re-constructed by binning of shotgun-sequencing data from the bacterial community of the sponge C. concentrica (Thomas et al., 2010). Binning of scaffolds with >20 kb sequence was done according to a modification of the strategy outlined by Woyke et al. (2006) and Thomas et al. (2010) and is further described in the Supplementary Information.
For comparative analysis, the complete and annotated genome sequences of the species Bdellovibrio bacteriovorus strain HD100 (3587 open reading frames (ORFs)) (Rendulic et al., 2004) and Desulfurmonas acetoxidans DSM 684 (3234 ORFs) (unpublished) were downloaded from National Center for Biotechnology Information. The complete, predicted protein sets of these two organisms and the partial protein data set for the sponge δ-proteobacterium were searched with blastP (E-value <10e−5) against the Cluster of Orthologous Groups of proteins (COG) database (Tatusov et al., 2003). Proteins with at least 30% identity to an entry were then classified into specific COGs and COG categories. Statistical, pairwise, comparison of the COG profile between the sponge δ-proteobacterium, B. bacteriovorus and D. acetoxidans was performed by resampling (n=1000) of COG subsamples (n=300) as outlined in Lauro et al. (2009). MEGAN (Huson et al., 2007) was used to examine the taxonomic relationship of proteins within the δ-proteobacterial partial genome. MEGAN uses a lowest common ancestor algorithm to assign ORFs into taxonomical groups based on Basic Local Alignment Search Tool bit scores (Altschul et al., 1990). Parameters were set with ‘min support' at 1, ‘min score' at 30 and ‘top percentage' set at 10%.
Localization of δ-proteobacterium
Specimens of the marine sponge C. concentrica were collected from Botany Bay near Bare Island, Sydney, Australia (S 33.59.461; E 151.13.946) during August 2009. Following collection, C. concentrica tissue was rinsed with calcium and magnesium-free seawater (25 g NaCl, 0.8 g KCl, 1 g Na2SO4, 0.04 g NaHCO3 per 1 l) and immediately transferred to a 15% sucrose solution. The specimens were then transferred back to the laboratory on ice within 1 h. Fluorescent in-situ hybridization (FISH) was performed as described in the Supplementary Information with validated probes targeting bacteria in general and the δ-proteobacterium specifically.
Results and Discussions
A novel δ-proteobacterial clade of sponge-associated bacteria
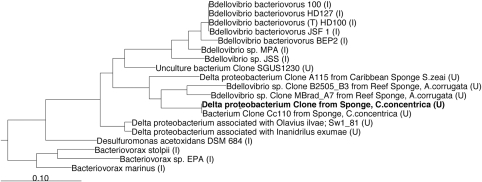
In the phylogenetic survey of the bacterial community associated with the sponge C. concentrica, we discovered a 16S rRNA gene sequence (Thomas et al., 2010) that was assigned by the Ribosomal Database Project classifier to the class δ-proteobacteria (87% confidence) and the order of Bdellovibrionales (81% confidence). Phylogenetic analysis and comparison of the sponge δ-proteobacterial sequence with sequences from closely related isolates and uncultured organisms (Figure 1) showed that it falls into a cluster of uncultured bacteria that were all found in marine sponges. Specifically, the sponge δ-proteobacterium sequence was identical to a previously described sequence of an uncultured bacterium (Cc110, AY942772) from C. concentrica, indicating a permanent association of this bacterium with this sponge. Other sequences in this cluster were from the reef sponge Axinella corrugata (clones B2505_B3 (EF092213) and MBrad_A7 (EF092260)) and the Caribbean sponge Svenzea zeai (clone A115 (FJ529308)) (Lee et al., 2009). These five sponge-associated organisms were only distantly related (>15% sequence divergence) to the nearest cluster of cultured organisms that was defined by B. bacteriovorus strains. The latter strains were isolated from soil (100T, AF084850), animal gut (HD100, AJ292759 and HD127, AJ292760) and an aquaculture farm (JSF1, EU884925) and had high levels of 16S rRNA gene sequence identity (>99%). In addition, three other B. bacteriovorus isolates from soil (MPA, AY294215), root extract (BEP2, AF148938) and sewage (JSS, EF687743) were found in the same cluster, but with lower levels of identity.
Figure 1.
Maximum likelihood tree of the δ-proteobacterium's 16S rRNA gene sequence from C. concentrica and related organisms. The tree was built on 1178 nucleotides of the 16S rRNA gene. An uncultured euryarchaeote sp. (AB077227) was used as an outgroup for the analysis (not shown in the tree). The scale bar indicates 0.1 nucleotide changes (10%) per nucleotide position. U, uncultured; I, isolates.
The sponge-associated δ-proteobacteria cluster was also distinct from the genera Desulfuromonas and Bacteriovorax, with divergence levels of 24% and 30%, respectively. The genus Desulfuromonas contains aquatic species that are known to grow anaerobically by oxidizing acetate with concomitant reduction of elemental sulfur or Fe (III) (Pfennig and Biebl, 1976). Bacteriovorax species parasitizes on other gram-negative bacteria and have a highly motile, free-living, extracellular phase, during which they seek out new hosts. The phenotypic characteristics of this genus are identical to that of Bdellovibrio, and Bacteriovorax was only recently reclassified as a distinct genus based on 16S rRNA gene-based phylogeny (Baer et al., 2000, 2004).
The phylogenetic analysis conducted here showed that the δ-proteobacterium from C. concentrica possibly represents a novel genus or family and that members of this group of yet-to-be-cultured bacteria are associated with sponges.
Functional genomic comparison of the sponge δ-proteobacterium with related bacteria
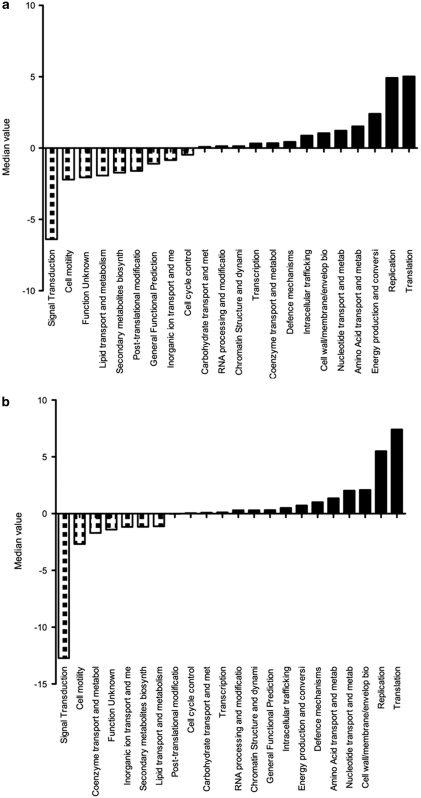
To gain insight into the functional genomic properties of the new δ-proteobacterium, we reconstructed a partial genome for the organism from data of our recent metagenomic study (Thomas et al., 2010). The assembled data represent a composite genome of many related individuals as seen in other partial genome reconstructions from metagenomic shotgun data (for example, Tyson et al., 2004; Hallam et al., 2006) and within population variation may exist at the single-nucleotide or gene level. Through binning based on nucleotide signatures, we could confidently identify 32 genomic scaffolds belonging to the organism spanning an estimated total of 1.1 Mbp with 582 622 bp of unique sequence. The longest scaffold was 116 573 bp and the average span of scaffold was 35 695 bp. We identified 670 open reading frames in this reconstruction and compared their functional annotation to the completed genomes of B. bacterivorus HD100 and D. acetoxidans DSM68, the nearest related organisms with available genome sequences. The sponge δ-proteobacterium showed an overrepresentation in the functional COG categories of translation and replication, recombination and repair, and an underrepresentation in signal transduction when compared with B. bacteriovorus (Figure 2a). On the individual COG level, specific functions associated with signal transduction mechanisms (COG0642, COG0840, COG0784, COG0644 and COG0745) were underrepresented, whereas functions associated with ankyrin repeat (AR) proteins (COG0666), TPR proteins (COG0790), the DNA-directed RNA polymerase, beta subunit per 140 kD subunit (COG0085), the 2-oxoglutarate dehydrogenase complex, dehydrogenase (E1) component (COG0567), restriction endonuclease related to defense mechanism (COG1403) and -alanyl--alanine carboxypeptidase (penicillin-binding protein 4, COG2027) were overrepresented (Supplementary Table S2). A similar pattern of functional characteristics was observed when the sponge δ-proteobacterium was compared with D. acetoxidans DSM684 (Figure 2b). The overall functional comparison again indicated that signal transduction was underrepresented in the sponge δ-proteobacterium and translation and replication, recombination and repair were overrepresented. On the individual COG level, specific functions associated with signal transduction mechanisms (COG2199, COG0642, COG2200, COG0840, COG3706, COG0784, COG2204 and COG2203) were underrepresented, whereas functions associated with AR proteins (COG0666) and TPR proteins (COG0790) are overrepresented (Supplementary Table S3). This analysis highlights the clear functional differences between the sponge δ-proteobacterium and B. bacterivorux and D. acetoxidans, respectively, and in particular within the function of signal transduction.
Figure 2.
Functional genome comparison based on COG categories of δ-proteobacterium partial genome and the genomes of B. bacteriovorus HD100 genome (a) and D. acetoxidans DSM684 (b). The Y axis displays the median value with a significant cut-off value of −2 and 2.
To understand whether certain functions of the δ-proteobacterium genome have a shared evolutionary history with related genomes, we classified all its proteins taxonomically with a lowest common ancestor algorithm implemented in MEGAN (Huson et al., 2007). Taxonomic analysis of 663 proteins showed that 249 could be assigned to B. bacteriovorus HD100, 4 assigned to D. acetoxidans DSM684, 185 had no hits in the non-redundant GenBank database and the remaining proteins were assigned, at various levels, within the bacteria or just cellular organisms (Supplementary Table S4). These results show that the δ-proteobacterium genome shared more conserved proteins with B. bacteriovorus HD100 genome than any other available genome.
Specific functions enriched within the 249 proteins included translation, transcription, replication and cell wall biogenesis, which reflect the evolutionary conserved nature of these functions across phylogenetically related organisms. The sponge δ-proteobacterium and B. bacteriovorus also share an adventurous gliding motility protein R, which is associated with microorganisms that travel in environments with a low water availability, such as biofilms, microbial mats and soil (Spormann, 1999), and which facilitates host cell infection in some Apicomplexan parasites (Kappe et al., 1999). In addition, there is a shared and conserved Flp pilus assembly protein and a cluster of four proteins that are involved in type IV pili assembly and translocation (assembly protein TapB, two twitching motility proteins and a pilin assembly protein PilC). Type IV pili are associated with twitching motility, which has been proposed to be important in cell attachment and invasion by Bdellovibrio species, wherein the force of pili retraction generated by depolymerization could pull the Bdellovibrio cell through its attached prey cell (Lambert et al., 2009). Together these observations support the notion that certain aspects of attachment and motility might be shared and conserved between the sponge δ-proteobacterium and B. bacteriovorus.
A number of functions in cell wall and envelope biogenesis are conserved across the sponge δ-proteobacterium and B. bacteriovorus. These include a murein-degrading enzyme (membrane-bound lytic murein transglycosylase D precursor) that has a role in recycling of muropeptides during cell elongation and/or cell division. Bdellovibrio species can digest up to 20% of their prey's cell wall after invasion, while keeping their newly formed bdelloplast structure from uncontrolled lysis (Thomashow and Rittenberg, 1978). During bacterial predation the amount of digested peptidoglycan would yield enough precursors to form the multiple progeny. Murein biosynthesis requires a crucial disaccharide penta-peptide precursor molecule (UDPMurNAc-pentapeptide) usually formed in the cytoplasm and Bdellovibrio species have therefore retained the six cytoplasmic enzymes (MurA-F) responsible for its de novo biosynthesis and periplasmic transport. The sponge δ-proteobacterium also contains three conserved homologs of these enzymes, namely the UDP-N-acetylmuramoylalanine--glutamate ligase (MurD), the UDP-N-acetylmuramoylalanyl--glutamyl-2, 6-diaminopimelate--alanyl--alanine ligase (MurF), and the UDP-N-acetylmuramoylalanyl--glutamate-2, 6-diaminopimelate ligase (MurE). Clustered with the ORFs of these proteins is another gene (mraY) encoding a phospho-N-acetylmuramoyl-pentapeptide-transferase, which catalyzes the first step of lipid cycle reactions and the biosynthesis of the cell wall peptidoglycan. Immediately downstream of this cluster are two genes that are also associated with cell wall biogenesis. First, there is an ORF encoding for the penicillin-binding protein 2B (PBP2), which has transglycosylase activities to catalyze the formation of the beta 1–4 glycosidic links from the lipid-bound precursors to the nascent end of the peptidoglycan strand, and which is responsible for bulk peptidoglycan synthesis in other bacteria (Vollmer and Bertsche, 2008). The second gene encodes for a peptidoglycan synthetase (FtsI or PBP3), whose homolog in Escherichia coli is essential for the septum formation of the murein sacculus and synthesis of cross-linked peptidoglycan from the lipid intermediates (Nakamura et al., 1983).
This shows that, besides attachment and motility, the process of cell wall biogenesis is also an evolutionary conserved function of the sponge δ-proteobacterium and B. bacteriovorus.
The sponge δ-proteobacterium exhibits a cell-associated life-style
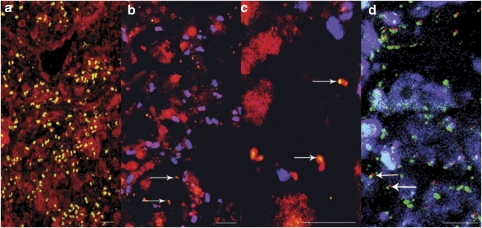
The similarity found in motility, attachment and cell wall biogenesis when compared with B. bacteriovorus led us to the hypothesis that the sponge δ-proteobacterium might also form an association (potentially of predatory nature) with other bacterial cells. We therefore investigated the physical localization of the δ-proteobacterium in the sponge by fluorescence in-situ hybridization. Using a probe for the domain of bacteria (EUB338), we first demonstrated the presence of high numbers of bacteria within the sponge C. concentrica (Figure 3a). The probe directed against the δ-proteobacterium 16S rRNA sequence resulted in the detection of small cells (<1 μm in length) in the sponge tissue. These were generally associated with larger, auto-fluorescent cells (>3 μm in length) and only occasionally found as free-living cells (Figures 3b and c). The relative proportion of the δ-proteobacterium against the total microbiota was estimated to be 6.4%±1.8% (see Supplementary Information), which is similar to 2.2%±0.8% of sequences found in three 16S rRNA gene PCR libraries belonging to the δ-proteobacterium (Thomas et al., 2010). The localization of the δ-proteobacterium was often at the end of the larger, auto-fluorescent cells.
Figure 3.
Detection of sponge-associated bacteria by fluorescence in-situ hybridization (FISH). (a) Section of C. concentrica hybridized with a Cy3-labelled bacteria-specific (EUB322_CY3) probe (yellow) with the auto-fluorescent sponge tissue (red). (b) Section of C. concentrica hybridized with a Cy3-labelled δ-proteobacterium-specific probe (yellow), auto-fluorescence background (red) and larger auto-fluorescence cells (blue-purple). Arrows indicated the sponge δ-proteobacterium associated with auto-fluorescent cells (red). (c) Section of C. concentrica (enlarged) hybridized with a CY3-labelled δ-proteobacterium-specific probe (yellow). Arrows indicated the host-associated nature of the sponge δ-proteobacterium within auto-fluorescent cells (red). (d) Section of C. concentrica (enlarged) hybridized with a Cy3-labelled δ-proteobacterium-specific probe (red) and a fluorescein isothiocyanate (FITC)-labelled bacteria-specific (EUB322_FITC) probe (green). The auto-fluorescent background of sponge tissues (blue) is also shown. Arrows indicated the associated nature of the sponge proteobacterium within a putative cyanobacterial cell (green).
The auto-fluorescent ‘host' cells showed green auto-fluorescence when excited with 488 nm light, which is consistent with the fluorescence reported for Cyanobacteria, diatoms and dinoflagellates (Tang and Dobbs, 2007). To determine, whether the auto-fluorescent cells were eukaryotic (that is, diatoms or dinoflagellets) or bacterial (that is, cyanobacteria), we conducted a further FISH analysis using a two-probe hybridization design with a Cy3-labelled probe specific for the δ-proteobacterium and a fluorescein isothiocyanate-labelled bacteria-specific probe. Figure 3d shows that the auto-fluorescent cells responded to the general bacterial probe. Therefore, based on the particular auto-fluorescence and the binding of a bacteria-specific probe, these cells are most likely cyanobacteria. This finding is consistent with an observation made 30 years ago by Wilkinson (1979), who found ‘Bdellovibrio-like' particles, using electronmicroscopy, within unicellular cyanobacteria in the mesohyl of the two coral sponges, Neofibularia irata and Jaspis stelifera.
Predicted functional properties of the sponge δ-proteobacterium
To gain further insight into the functional properties of the new δ-proteobacterium, we performed a careful manual analysis of its partial genome with respect to physiological and ecological functions.
We identified the subunits of the terminal enzyme in the aerobic respiratory chain, the cytochrome C oxidase, in the partial genome (1108814258778_ORF0001). The sponge δ-proteobacterium is therefore likely able to grow aerobically; further distinguishing it from the related, strictly anaerobic Desulfuromonas species.
A total of 19 proteins had clear functions in membrane transport and could be assigned to four major transport families (Supplementary Table S5) with the adenosine-5′-triphosphate-binding cassette (ABC) superfamily being the largest group. A phosphate transporter (3.A.1.7.2) is encoded by a pstSCAB operon and appears to be homologous to the high-affinity phosphate-specific ABC transport system from Mycobacterium smegmatis (Gebhard and Cook, 2008). A glutathione porter (3.A.1.5.11) is partially encoded by genes similar to yliA, C and D, which encode adenosine-5′-triphosphate-binding cassettes in E. coli K-12 for the import of extracellular glutathione, and which have been shown to be crucial for growth on glutathione as the sole sulfur source (Suzuki et al., 2005). The δ-proteobacterium genome also encodes for a tetracycline resistance protein (1108814258826_ORF0009), which function as a antibiotic efflux transport (metal-tetracycline/H+ antiporter) of the major facilitator superfamily (Yamaguchi et al., 1990; Marger and Saier, 1993), and which can perform a wide variety of processes, including uptake of essential ions, nutrients and removal of toxic compounds (Griffith et al., 1992). An additional tetracycline efflux/multidrug resistance protein (1108814257392_ORF0004) indicates that removal of toxins and antibiotics might be a relevant functional aspect of the δ-proteobacterium.
The δ-proteobacterium also appears to maintain some essential anabolic capacity, despite its host-associated life-style, and has probably not developed an obligate dependence on nutrients provided by the host cell. It possesses two key enzymes for the chorismate biosynthesis (shikimate synthase: 1108814258173_ORF0043 and 3-phosphoshikimate 1-carboxyvinyltransferase: 1108814258173_ORF0042), which provide intermediates for the synthesis of aromatic amino acids and coenzymes such as folic acid, ubiquinone, menaquinone and enterochelin. In addition, proteins involved in menaquinone biosynthesis (menaquinone biosynthesis proteins MenD: 1108814258173_ORF0048; O-succinylbenzoate-CoA ligase MenE: 1108814258778_ORF0042; naphthoate synthase MenB: 1108814258807_ORF0012 and a 1,4-dihydroxy-2-naphthoate octaprenyltransferase: 1108814258807_ORF0011) were also identified and indicate the ability of the sponge δ-proteobacterium to produce this important redox component of the electron transfer chain (Farrand and Taber, 1974). Another cofactor that is possibly synthesized is riboflavin (vitamin B2), a precursor for the essential redox-active cofactors flavin mononucleotide and flavin adenine dinucleotide. Three proteins related to riboflavin biosynthesis were found, including a 6,7-dimethyl-8-ribityllumazine synthase (1108814258173_ORF0015), a riboflavin biosynthesis protein RibAB (1108814258173_ORF0017) and riboflavin biosynthesis protein RibF (1108814258794_ORF0021).
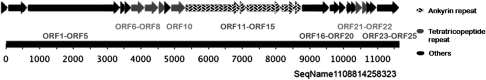
A total of six ORFs encoding multi-functional AR protein (COG0666) were found in the partial δ-proteobacterium, five of which were clustered together (Figure 4). The complete genome of B. bacteriovorus HD100 only contains two AR-containing proteins. AR act as mediators for a diverse range of protein–protein interactions and are found commonly in eukaryotic and viral proteins, but rarely in bacteria (Lux et al., 1990; Bork, 1993; Mosavi et al., 2004). Eukaryotic-like AR have been reported in obligate intracellular bacteria such as the α-proteobacteria Wolbachia pipientis (Iturbe-Ormaetxe et al., 2005) and Ehrlichia canis (Mavromatis et al., 2006), as well as in the γ-proteobacteria Legionella pneumophila (Habyarimana et al., 2008) and Coxiella butnetii (Voth et al., 2009). The involvement of AR proteins in host–bacterial associations and survival of the bacteria inside other cells has been highlighted recently by a mutant study of L. pneumophila, which showed that lack of certain AR proteins cause a significant defect in intracelluar replication (Habyarimana et al., 2008). Seven TPR proteins were also identified, which similarly mediate protein–protein interactions and the assembly of multi-protein complexes (D'Andrea and Regan, 2003). TPR-containing proteins have been found to be involved in a variety of biological processes, such as cell cycle regulation, transcriptional control, mitochondrial and peroxisomal protein transport, neurogenesis and protein folding (Goebl and Yanagida, 1991; Blatch and Lassle, 1999). Interestingly, the genes encoding for the majority of AR and TPR proteins in the δ-proteobacterium are localized closely together on the genome (see Figure 4) and might represent a cluster for the mediation of protein–protein interactions within the cell or between cells.
Figure 4.
Schematic representation of the ORFs encoding AR and TRP proteins in the δ-proteobacterium scaffold 1108814258323. ORFs are shown in arrows with relative nucleotide positions.
Conclusions
We have identified and characterized, through culture-independent approaches, a novel δ-proteobacterium that possibly represents a new genus or family. To our knowledge, this sponge-associated organism represents only the third group of host-associated δ-proteobacteria. The sponge δ-proteobacterium is proposed to live in association with cyanobacteria and likely impacts on the physiology of its photosynthetic host. This in turn might have a consequence for the sponge host, which often harbors photosynthetic symbionts and thereby acquires photosynthetically fixed carbon (Venn et al., 2008). The functional genomic characterization of the uncultured δ-proteobacterium reported here also indicates complex interactions with surrounding cells and environmental milieu. This study offers an insight into multi-level interactions between a sponge, which serves as a host to a cyanobacterium, which in turn is host to a δ-proteobacterium.
Acknowledgments
This work was supported by funding from the Gordon and Betty Moore Foundation and the Australian Research Council (LP0668235). The shotgun sequencing and 16S rRNA gene data of this project have been deposited at GenBank under GenomeProject ID 34751 and are also available through the Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis (CAMERA) website (http://camera.calit2.net/).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baer ML, Ravel J, Chun J, Hill RT, Williams HN. A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int J Syst Evol Microbiol. 2000;50 (Pt 1:219–224. doi: 10.1099/00207713-50-1-219. [DOI] [PubMed] [Google Scholar]
- Baer ML, Ravel J, Pineiro SA, Guether-Borg D, Williams HN. Reclassification of salt-water Bdellovibrio sp. as Bacteriovorax marinus sp. nov. and Bacteriovorax litoralis sp. nov. Int J Syst Evol Microbiol. 2004;54:1011–1016. doi: 10.1099/ijs.0.02458-0. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally. Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Farrand SK, Taber HW. Changes in menaquinone concentration during growth and early sporulation in Bacillus subtilis. J Bacteriol. 1974;117:324–326. doi: 10.1128/jb.117.1.324-326.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard S, Cook GM. Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon. J Bacteriol. 2008;190:1335–1343. doi: 10.1128/JB.01764-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Griffith JK, Baker ME, Rouch DA, Page MG, Skurray RA, Paulsen IT, et al. Membrane transport proteins: implications of sequence comparisons. Curr Opin Cell Biol. 1992;4:684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- Habyarimana F, Al-Khodor S, Kalia A, Graham JE, Price CT, Garcia MT, et al. Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol. 2008;10:1460–1474. doi: 10.1111/j.1462-2920.2007.01560.x. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Usher KM, Taylor MW. Marine sponges as microbial fermenters. FEMS Microbiol Ecol. 2006;55:167–177. doi: 10.1111/j.1574-6941.2005.00046.x. [DOI] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Burke GR, Riegler M, O'Neill SL. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol. 2005;187:5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe S, Bruderer T, Gantt S, Fujioka H, Nussenzweig V, Menard R. Conservation of a gliding motility and cell invasion machinery in Apicomplexan parasites. J Cell Biol. 1999;147:937–944. doi: 10.1083/jcb.147.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Hobley L, Chang CY, Fenton A, Capeness M, Sockett L. A predatory patchwork: membrane and surface structures of Bdellovibrio bacteriovorus. Adv Microb Physiol. 2009;54:313–361. doi: 10.1016/S0065-2911(08)00005-2. [DOI] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee OO, Chui PY, Wong YH, Pawlik JR, Qian P-Y. Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean sponge Svenzea zeai. Appl Environ Microbiol. 2009;75:6147–6156. doi: 10.1128/AEM.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux SE, John KM, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- Marger MD, Saier MH., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Mavromatis K, Doyle CK, Lykidis A, Ivanova N, Francino MP, Chain P, et al. The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J Bacteriol. 2006;188:4015–4023. doi: 10.1128/JB.01837-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Maruyama IN, Soma M, Kato J, Suzuki H, Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191:1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Pfennig N, Biebl H. Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch Microbiol. 1976;110:3–12. doi: 10.1007/BF00416962. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiswig HM. Particle feeding in natural populations of three marine Demosponges. Biol Bull. 1971;141:568–591. [Google Scholar]
- Reiswig HM. Water transport, respiration and energetics of three tropical marine sponges. J Exp Mar Biol Ecol. 1974;14:231–249. [Google Scholar]
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Spormann AM. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Koyanagi T, Izuka S, Onishi A, Kumagai H. The yliA, -B, -C, and -D genes of Escherichia coli K-12 encode a novel glutathione importer with an ATP-binding cassette. J Bacteriol. 2005;187:5861–5867. doi: 10.1128/JB.187.17.5861-5867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YZ, Dobbs FC. Green autofluorescence in dinoflagellates, diatoms, and other microalgae and its implications for vital staining and morphological studies. Appl Environ Microbiol. 2007;73:2306–2313. doi: 10.1128/AEM.01741-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Rusch D, Demaere MZ, Yung PY, Lewis M, Halpern A, et al. 2010Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis ISME Je-pub ahead print 3 June 2010, doi: 10.1038/ismej.2010.74 [DOI] [PubMed]
- Thomashow MF, Rittenberg SC. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: solubilization of Escherichia coli peptidoglycan. J Bacteriol. 1978;135:998–1007. doi: 10.1128/jb.135.3.998-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turon X, Galera J, Uriz MJ. Clearance rates and aquiferous systems in two sponges with contrasting life-history strategies. J Exp Zool. 1997;278:22–36. [Google Scholar]
- Tyson G, Chapman J, P H, Allen E, Ram R, Richardson P, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Venn A, Loram J, Douglas A. Photosynthetic symbioses in animals. J Exp Bot. 2008;59:1069–1080. doi: 10.1093/jxb/erm328. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, et al. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. 2009;191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Blackall LL. What do we really know about sponge-microbial symbioses. ISME J. 2008;3:1–3. doi: 10.1038/ismej.2008.102. [DOI] [PubMed] [Google Scholar]
- Webster NS, Cobb RE, Negri AP. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2008;2:830–842. doi: 10.1038/ismej.2008.42. [DOI] [PubMed] [Google Scholar]
- Wilkinson C. Bdellovibrio-like parasite of cyanobacteria symbiotic in marine sponges. Arch Microbiol. 1979;123:101–103. [Google Scholar]
- Wilkinson CR. Microbial associations in sponges. II. Numerical analysis of sponge and water bacterial populations. Mar Biol. 1978;49:169–176. [Google Scholar]
- Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Udagawa T, Sawai T. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J Biol Chem. 1990;265:4809–4813. [PubMed] [Google Scholar]
- Zhu P, Li Q, Wang G. Unique microbial signatures of the alien Hawaiian marine sponge Suberites zeteki. Microb Ecol. 2008;55:406–414. doi: 10.1007/s00248-007-9285-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.