Abstract
The potential of metatranscriptomic sequencing to provide insights into the environmental factors that regulate microbial activities depends on how fully the sequence libraries capture community expression (that is, sample-sequencing depth and coverage depth), and the sensitivity with which expression differences between communities can be detected (that is, statistical power for hypothesis testing). In this study, we use an internal standard approach to make absolute (per liter) estimates of transcript numbers, a significant advantage over proportional estimates that can be biased by expression changes in unrelated genes. Coastal waters of the southeastern United States contain 1 × 1012 bacterioplankton mRNA molecules per liter of seawater (∼200 mRNA molecules per bacterial cell). Even for the large bacterioplankton libraries obtained in this study (∼500 000 possible protein-encoding sequences in each of two libraries after discarding rRNAs and small RNAs from >1 million 454 FLX pyrosequencing reads), sample-sequencing depth was only 0.00001%. Expression levels of 82 genes diagnostic for transformations in the marine nitrogen, phosphorus and sulfur cycles ranged from below detection (<1 × 106 transcripts per liter) for 36 genes (for example, phosphonate metabolism gene phnH, dissimilatory nitrate reductase subunit napA) to >2.7 × 109 transcripts per liter (ammonia transporter amt and ammonia monooxygenase subunit amoC). Half of the categories for which expression was detected, however, had too few copy numbers for robust statistical resolution, as would be required for comparative (experimental or time-series) expression studies. By representing whole community gene abundance and expression in absolute units (per volume or mass of environment), ‘omics' data can be better leveraged to improve understanding of microbially mediated processes in the ocean.
Keywords: metatranscriptomics, marine, bacterioplankton, gene expression, biogeochemistry
Introduction
Metatranscriptomics is a powerful tool for capturing gene expression patterns in natural microbial communities without previous assumptions as to the ongoing activities or dominant taxa (Poretsky et al., 2005; Frias-Lopez et al., 2008). In contrast to metagenomics, which provides an inventory of the community gene pool, metatranscriptomics identifies which of those genes are being transcribed in a given ecological context, including under experimentally manipulated conditions (Gilbert et al., 2008; Poretsky et al., 2010).
The advent of second-generation sequencing has increased metatranscriptome library sizes by orders of magnitude (Poretsky et al., 2005, 2009b; Frias-Lopez et al., 2008; Urich et al., 2008; Hewson et al., 2009a), yet how deeply a community transcriptome is ‘covered' by the sequence library remains a critical issue. If too shallow, libraries will be dominated by transcripts from metabolic pathways shared by most cells and poor in those representing specialized biogeochemical pathways (Hewson et al., 2009b; Poretsky et al., 2009b). As a consequence, unique expression patterns within a community may be missed, and comparative analyses between communities can be insensitive.
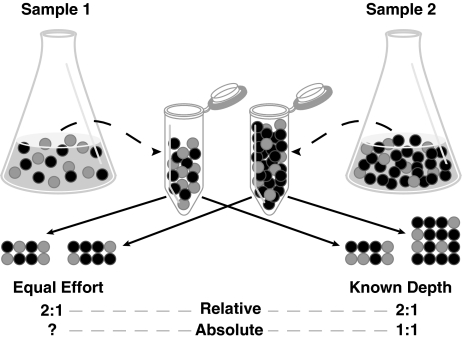
Variability in sample-sequencing depth between community metatranscriptomes, regardless of coverage level, further limits the power of comparative analyses by restricting assessments to relative data (that is, as a proportion of the transcriptome; Figure 1). This is problematic because changes occurring in the abundance of some mRNAs in response to shifting conditions (van de Peppel et al., 2003; Hannah et al., 2008; Robinson and Oshlack, 2010) leads to changes in the percent representation of other mRNAs whose absolute abundance has not changed (Figure 1). Comparative analyses based on ratios of mRNA copies to DNA copies (that is, relative abundance in metatranscriptomes vs relative abundance in metagenomes; Frias-Lopez et al., 2008) does not solve this problem, as both are similarly affected by unknown and possibly different sample-sequencing depths. Thus as currently obtained, metatranscriptomic data provide information on enrichment or depletion of a transcript category in the community transcriptome, but not on the absolute abundance of these transcript categories per volume or mass of environment (Figure 1), which is the most relevant comparison for biogeochemical studies and ecosystem modeling. Metagenomic-, metaproteomic- and environmental microarray-based studies suffer these same proportional data constraints.
Figure 1.
Effect of sample-sequencing depth on quantification of transcripts (or genes) in environmental samples. ‘Equal-effort' sequences the same number of reads per sample volume, regardless of the size of the mRNA pool, and therefore conveys only relative abundance. ‘Known-depth' sequences a known proportion of the transcript pool (50% for both, in this example), and therefore also conveys absolute copy numbers per sample volume. The latter is more relevant to biogeochemical rate measurements, as mRNAs of biogeochemical interest (gray dots) can make up different proportions in community transcriptomes yet have identical numbers in the environment.
In this study, we report the deep sequencing of two replicate metatranscriptomes from southeastern United States coastal seawater to characterize microbial gene expression and address three critical questions about sequencing effort: (1) What was the sample-sequencing depth for the bacterioplankton community transcriptome? We used an internal mRNA standard to estimate the number of transcripts in the natural sample compared with the number of transcripts sequenced. (2) What was the abundance of transcripts representing key bacterial transformations in the nitrogen (N), phosphorus (P) and sulfur (S) cycles in coastal seawater? We used BLAST analysis normalized to internal standard recovery to estimate absolute transcript numbers for over 80 diagnostic steps in marine elemental cycles. (3) Was the sequencing strategy sufficient to detect differences in gene transcription between samples? We examined the effect of coverage depth on statistical comparisons of biogeochemically diagnostic transcripts in the metatranscriptomes.
Materials and methods
Sample collection
Two replicate seawater samples (FN56 and FN57) were collected at Marsh Landing, Sapelo Island, Georgia, USA (31°25′4.08N, 81°17′43.26W; http://www.simo.marsci.uga.edu) on 6 August 2008 at 2330 hours local time, 1 h before high tide and 3 h after sunset. These samples are part of a multiyear time series of the Sapelo Island Microbial Observatory (http://www.simo.marsci.uga.edu), in which collections are made every 3 months and each collection set consists of duplicate samples from four consecutive high tides (2 days and 2 nights). Surface water (5.75 l from a depth of 0.5 m) was pumped directly through 3 and 0.22 μm filters. The 0.22 μm filter was placed in a Whirl-Pak bag (Nasco, Fort Atkinson, WI, USA) and immediately flash frozen in liquid N2. Total time from the start of filtration to freezing was 10 min. Although the samples are considered biological replicates within the larger time series, we note that there was 8 min between the end of the first collection (sample FN56) and the start of the second (sample FN57). Nutrient data are collected monthly at station GCE6 (∼3 km from Marsh Landing) as part of the Georgia Coastal Ecosystems Long Term Ecological Research program (http://gce-lter.marsci.uga.edu).
RNA processing and sequencing
RNA processing in preparation for pyrosequencing was carried out as previously described (Poretsky et al., 2009a, 2009b), with the exception of the addition of an in vitro transcribed standard to the extraction tube before beginning the extraction. The standard was constructed by linearizing a pGem-3Z plasmid (Promega, Madison, WI, USA) with ScaI restriction enzyme (Roche, Penzberg, Germany) and cleaned with a phenol/chloroform/isoamyl alcohol extraction. Complete digestion of the plasmid was confirmed with a 1% agarose gel. The DNA fragment was then in vitro transcribed using the Riboprobe in vitro Transcription System (Promega) according to the manufacturer's protocol; an SP6 RNA polymerase was used to create a 994 nt long RNA fragment. The pGem plasmid had another internal T7 promoter region, but it was present in the reverse complement sequence during in vitro transcription and aRNA amplification (see below), and did not interfere. Residual DNA was removed with RQ1 RNase-Free DNase, and the RNA was cleaned with a phenol/chloroform/isoamyl alcohol extraction. The RNA standard was quantified with a Nanodrop Spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and correct fragment size was confirmed with an Experion automated electrophoresis system (Bio-Rad, Hercules, CA, USA).
RNA standard (25 ng; 4.7 × 1010 copies) was added to a 50 ml conical tube containing 8 ml RLT lysis buffer (Qiagen, Valencia, CA, USA) and 3 g of RNA PowerSoil beads (Mo-Bio, Carlsbad, CA, USA). The sample filters were removed from −80 °C storage, shattered and added to the extraction tubes. RNA was then extracted using an RNEasy kit (Qiagen), and any residual DNA was removed using the Turbo DNA-free kit (Applied Biosystems, Austin, TX, USA). To reduce the number of rRNAs in the pyrosequencing reads, total RNA was treated in two ways to enrich for mRNA. Epicentre's mRNA-Only isolation kit (Madison, WI, USA) was first used to decrease rRNA contamination enzymatically. The samples were then treated with MICROBExpress and MICROBEnrich kits (both from Applied Biosystems), which couple an oligonucleotide rRNA probe with magnetic separation to enrich for mRNA. Successful reduction of rRNA was confirmed by running both pre- and post-treated samples on an Experion automated electrophoresis system (Bio-Rad). To obtain enough mRNA for pyrosequencing, the samples were linearly amplified using the MessageAmp II-Bacteria kit (Applied Biosystems). The amplified RNA was then converted to cDNA using the Universal RioboClone cDNA synthesis system with random primers (Promega), which produced cDNAs primarily in the size range of 200–600 bp. Residual reactants and nucleotides from cDNA synthesis were removed from the sample using the QIAquick PCR purification kit (Qiagen), and gel-based size selection was used to select fragments in the 250–500 bp range. cDNAs from each replicate sample were loaded into 1/2 of each of four GS-FLX plates for 454 pyrosequencing. Sequences are deposited in the CAMERA database (http://camera.calit2.net/about-camera/full-datasets) under accession name ‘CAM_PROJ_Sapelo2008'.
Read annotation
Duplicate clusters were identified using an online program (Gomez-Alvarez et al., 2009). Ribosomal RNA sequences were identified with a BLASTn search against the small and large subunit SILVA database (http://www.arb-silva.de) with a bit score cutoff ⩾50; sequences identified as rRNA were then removed from further consideration. To identify small, nonprotein encoding RNAs (Shi et al., 2009), all nonribosomal reads were compared with the RFam database (http://rfam.janelia.org) using BLASTn with a bitscore ⩾40, and hits were considered putative small RNAs if the best hit in the RefSeq database was a hypothetical protein or if the RFam alignment was ⩾95 nt (Supplementary Figure S1).
Remaining reads were annotated using BLASTx searches against the NCBI RefSeq and Clusters of Orthologous Genes (COG) databases (Tatusov et al., 2003) with a bit score cutoff ⩾40. Taxonomic binning was based on RefSeq hit. Collector's curves were produced from a custom script in the R environment (R Development Core Team, 2009). Read coverage of proteorhodopsin PU1002_03206 gene bin and the internal standard was assessed by assembling reads against the reference sequence using Geneious version 4.8 (Biomatters, Auckland, New Zealand) with gaps and default scoring (word length=18, maximum gap size=1, maximum gaps per read=20, maximum mismatches=20 and maximum ambiguities=4) and the consensus sequence representing the majority nucleotide at each position. Sequence variation at each nucleotide position was determined using a custom script in R with the BioStrings package (Pages et al., 2009).
Elemental cycle transcripts
Reference diagnostic genes representing transformations in the N, P and S cycles were selected from marine alphaproteobacteria, gammaproteobacteria and bacteriodetes genomes (the three most common taxa in marine metagenomic libraries), or from other taxa if these three groups did not contain an ortholog to the gene of interest. These reference sequences were used as query sequences in BLASTX analysis against the metatranscriptomic data (bitscore ⩾40, E-value <10−3) and redundant hits were removed. Remaining hits were manually checked with BLASTX against the RefSeq database and discarded if the top three hits were not to a similar annotation as the original reference gene.
Statistics
Pairwise statistical comparisons were carried out with Xipe, a bootstrapped difference of means calculation developed by Rodriguez-Brito et al. (2006), using 20 000 bootstrap iterations and 95% confidence intervals, or with 2 × 2 contingency tables and the Fisher's exact test (White et al., 2009) using P<0.05. Subsampled libraries for Xipe analyses were created by sampling without replacement using R (R Development Core Team, 2009). The Benjamini–Hochberg correction was used to adjust the Fisher's exact test P-values as a control for the false discovery rate using the R package ‘multtest' (Strimmer, 2008), and only those genes with an adjusted P-value <0.05 were considered significant. A simulation analysis of Fisher's exact test significance threshold as a function of count number was carried out using an R script that ran 2 × 2 contingency tables at incrementing count values for library sizes of 125 000 reads.
Results and discussion
Sequence libraries
cDNAs derived from two replicate coastal bacterioplankton samples (samples FN56 and FN57 in the Sapelo Island Microbial Observatory series; http://simo.marsci.uga.edu) were sequenced in four GS-FLX 454 runs (Margulies et al., 2005), with four technical sequencing replicates per biological replicate. Over a million reads averaging 210 nt in length were obtained per sample (Table 1). After removal of rRNAs and putative small RNAs (Shi et al., 2009), there were ∼500 000 possible protein encoding reads in each library (Table 1).
Table 1. Summary statistics for coastal ocean metatranscriptome datasets.
| FN56 | FN57 | Combined | |
|---|---|---|---|
| Total reads | 1 067 363 | 1 114 536 | 2 181 899 |
| rRNA | 466 834 (44%) | 623 804 (56%) | 1 090 638 (50%) |
| psRNA | 100 437 (9%) | 25 213 (2%) | 125 650 (6%) |
| Possible proteins | 500 092 (47%) | 465 519 (42%) | 965 611 (44%) |
| RefSeq Hits | 255 280 | 260 739 | 516 019 |
| RefSeq Genes | 96 573 | 109 395 | 168 669 |
| RefSeq Taxa | 1707 | 1761 | 1909 |
| COG hits | 162 925 | 170 593 | 333 518 |
| Unassigned | 244 812 | 204 780 | 449 592 |
Abbreviation: COG, Clusters of Orthologous Groups.
Percentages are of total reads.
RefSeq Hits, number of reads with significant homology to the RefSeq database; RefSeq Genes, number of unique accession numbers within those hits; Unassigned, =number of possible proteins that did not have a significant hit to either the RefSeq or COG databases (47% of possible proteins).
Sample-sequencing depth
Sample-sequencing depth is defined here as the percent of mRNA molecules present in a sample that is represented in the sequence library. The greater the sequencing depth of an mRNA pool, the more thorough the representation of microbial gene transcription. Further, if the volume or weight of the sample is also known, information on the sample-sequencing depth allows absolute transcript abundance to be calculated for a given quantity of the environment, not just proportional abundance in the community transcriptome. To estimate sample-sequencing depth, a known number of artificial RNA sequences serving as an internal standard was added immediately before cell lysis at the initiation of nucleic acid extraction. This approach may have some biases, for example if the internal standard is more susceptible to degradation than natural mRNA or if the efficiency of release of natural mRNA from cells is <100%, but it provides a consistent accounting across samples through extraction, processing and sequencing steps. Similar approaches have been successfully applied to qPCR (Coyne et al., 2005) and microarray studies (Hannah et al., 2008).
A total of 4014 internal standards were identified in the FN56 sequence library out of 4.7 × 1010 copies added before cell lysis, leading to an estimate of 1.0 × 1012 bacterioplankton mRNA molecules per liter of coastal seawater (Table 2). Two other estimates of the size of the community transcriptome were derived using literature values for mRNA content of marine bacterioplankton (Table 2), and these were in reasonable agreement with the internal standard method. The sample-sequencing depth was therefore ∼0.00001%, or 1 in 107 transcripts, with FN57 sequenced slightly deeper than FN56 (Table 2).
Table 2. Estimation of the number of bacterioplankton mRNA molecules in coastal seawater and sequencing depth of the metatranscriptomic libraries.
| Calculation method | Sample | mRNA molecules per liter | mRNA molecules per cellb | Sequencing depth (%) |
|---|---|---|---|---|
| Internal standarda | FN56 | 1.0 × 1012 | 238 | 0.000009 |
| FN57 | 0.6 × 1012 | 142 | 0.000015 | |
| Extracted RNA massc | FN56 | 0.2 × 1012 | 48 | 0.000043 |
| FN57 | 0.4 × 1012 | 95 | 0.000020 | |
| Per cell RNA contentd | FN56 | 2.6 × 1012 | 619 | 0.000003 |
| FN57 | 2.6 × 1012 | 619 | 0.000003 |
The libraries contained 4014 (FN56) and 6865 (FN57) copies of the internal standard out of a total of 500 092 (FN56) and 465 519 (FN57) potential protein encoding sequences. The standard was added at 4.7 × 1010 copies per 5.75 liter of seawater just before cell lysis for total RNA extraction (see Materials and methods for details).
Cell numbers in the 3 μ filtrate averaged 4.2 × 109 per liter based on epifluorescence microscopy.
Extraction yields were 14.4 (FN56) and 32.9 (FN57) μg total RNA from 5.75 liter of seawater. Total RNA is assumed to contain 4% mRNA by mass (Neidhardt and Umbarger, 1996) and bacterial mRNAs are assumed to average 924 nt (Xu et al. (2006)).
Marine bacterial cells are assumed to contain 5.7 fg total RNA per cell (mid point of 1.9–9.5 fg range reported by Simon and Azam (1989)). See footnote c for estimate of percent mRNA by mass and footnote b for cell counts per litre.
Direct cell counts indicated 4.2 × 109 bacterioplankton cells per liter in the seawater samples, and therefore an average of 190 mRNA transcripts per cell (Table 2). Laboratory cultures of Escherichia coli in exponential growth phase have ∼1400 transcripts per cell (Neidhardt and Umbarger, 1996). The sevenfold lower estimate for coastal bacterioplankton was not unexpected, however, because the cells are considerably smaller in size (Azam and Hodson, 1977) and have much lower growth rates (Ducklow, 2000) than laboratory-grown E. coli. On the basis of this per cell abundance estimate, it can be deduced that transcript copy number was lower than gene copy number for most of the bacterial and archaeal genes present in this coastal ocean.
Coverage depth
Coverage depth is defined here as the percent of the unique mRNAs present in a sample that is represented in the sequence library. Sample-sequencing depth and coverage depth are not strictly coupled, as a low richness/high evenness community transcriptome will be well covered even with shallow sample sequencing.
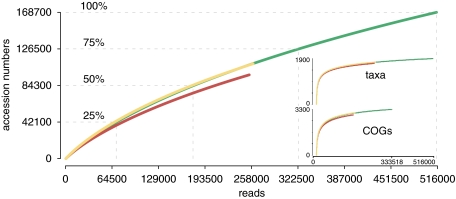
We evaluated coverage depth for the coastal metatranscriptomes in terms of taxa, functional gene categories and genes. Taxonomic coverage, as assessed by a collector's curve of NCBI taxonomy bins at the species or strain level, was approaching saturation for the library (Figure 2, inset); indeed, 75% of the total taxonomic richness emerging from this analysis would have been discovered with <15% of the sequencing effort. Saturating coverage was also found for functional gene assignments based on best hits to the COGs database (Table 1); 75% of the total richness would have been found with <10% of the sequencing effort (Figure 2, inset). However, these coverage assessments are constrained by the composition of the reference database, as apparent richness can be no higher than the number of reference bins available for transcript assignment. We found 1909 taxon bins represented in the metatranscriptomic libraries out of 8054 entries in the NCBI taxonomy database, and 3298 COGs out of 5666 entries in the COG database.
Figure 2.
Collector's curve of gene richness as a function of reads analyzed. Light gray: FN56; dark gray: FN57; medium gray: combined libraries. Dashed lines indicate the number of reads needed to reach quarter percentiles of the total richness of the combined library. Inset: collector's curves for taxonomic and functional gene category (COG) richness, with the y axis corresponding to the number of unique reference organisms or COG numbers.
When coverage was assessed based on gene assignments in the RefSeq database (>6 million accession numbers), transcripts binned to over 168 000 genes (Table 1), and the collector's curve indicated that the metatranscriptome library was far from saturating (Figure 2). Singletons made up 59% of the sequences (Supplementary Figure S2), and abundant transcripts (>10 hits accession number−1) and highly abundant transcripts (>100 hits accession per number) composed only 3% and 0.5% of the library, respectively. Although RefSeq binning could overestimate transcript richness (if identical transcripts bin to different reference genes because of differences in the region sequenced or because of sequencing errors), it also underestimates richness (if a variety of sequence variants bin to the same reference gene; Figure 3). In any event, despite efforts to sequence more deeply than typical, our libraries exhibited the same low coverage that has been reported in previous metatranscriptomic analyses of marine bacterioplankton communities (Frias-Lopez et al., 2008; Poretsky et al., 2010; Stewart et al., 2010).
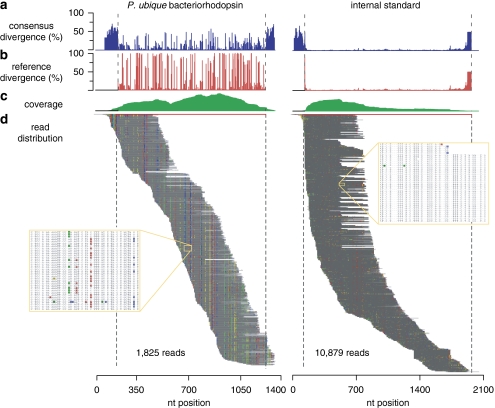
Figure 3.
Assembly of 1825 reads (out of 2259 total) binning to the P. ubique HTCC1002 proteorhodopsin gene PU1002_03206 (left), and of 10 879 reads (out of 10 879 total) binning to the internal transcript standard (right). (a) Percent nucleotide divergence from the consensus sequence. (b) Percent nucleotide divergence from the reference sequence. (c) Coverage by nucleotide position. (d) Read assembly to the reference gene (shown in red), with dashed lines indicating start and end positions of the reference. Note that the reference gene lengths are extended by assembly gaps. Divergence from the consensus sequence (that is, the majority nucleotide at a given position) is indicated as follows: A= red, T= green, C= blue and G= yellow. Insets show close-up regions of assemblies.
Metatranscriptomes might be expected to have lower richness compared with metagenomes if expression is limited to a small fraction of the bacterial genome at any one time. In this case, they would also have higher coverage than metagenomes for the same size sequence library (Gilbert et al., 2008). However, for this coastal metatranscriptome, the distribution of hits per gene (Supplementary Figure S2) did not indicate dominance by a limited number of highly transcribed genes (Figure 2). Similarly, a synchronized clonal population of Bacillus anthracis expressed 40–80% of genes under all growth conditions tested (Passalacqua et al., 2009), suggesting that the population's transcriptome was only slightly less rich than its genome. Even allowing for significant advances in sequencing technology, the extremely low sample-sequencing depth found in this study suggests that most natural community transcriptomes will continue to be undersampled.
Microdiversity
Assembly of transcripts from the most highly expressed genes (>1000 reads for some) revealed significant variation within reference bins (Hollibaugh et al., in press). For example, the 2259 reads that binned to the Pelagibacter ubique HTCC1002 proteorhodopsin gene (PU1002_03206; 1 of 28 proteorhodopsin bins in the libraries) had high sequence diversity (Figure 3). That this observed diversity was in fact real biological variation was substantiated by an assembly of the internal standard reads (Figure 3), which indicated a mean sequencing error rate in this study of 3.7±7.4 per 1000 bp compared with a mean sequence variation rate of 97.6 (±28.0) per 1000 bp for transcripts binning to PU1002_03206. Although high diversity in proteorhodopsin genes has been found previously in the ocean (Rusch et al., 2007; Campbell et al., 2008), the metatranscriptomic data revealed simultaneous expression of scores of microdiverse sequence variants. Transcriptome coverage estimates based on gene binning to the RefSeq database are considerable underestimates of the true sequence richness.
Detection of biogeochemically informative transcripts
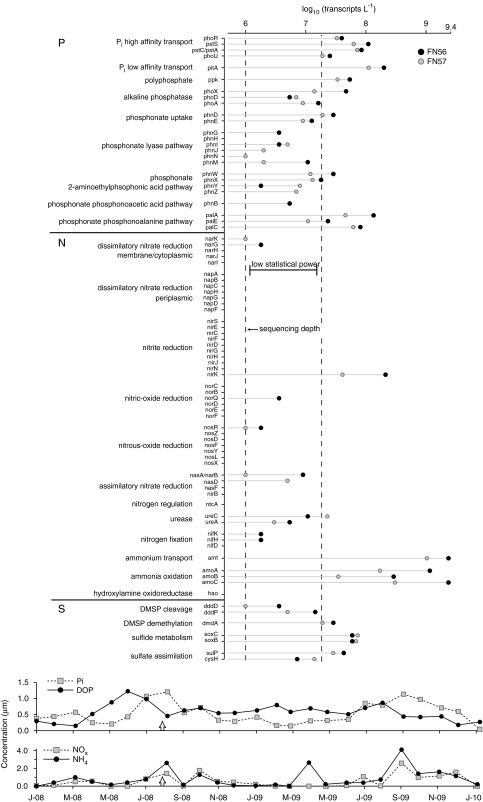
We determined the absolute abundance of transcripts for key genes representing the P cycle (25 diagnostic genes), N cycle (50 genes) and S cycle (7 genes) (Figure 4). Transcripts were found for 56% of genes surveyed. Most P and sulfur cycle transformations were represented by at least one transcript. N cycle expression was dominated by ammonia transporter and ammonia monooxygenase transcripts, which had the highest copy numbers of any gene category (2.7 × 109 transcripts per liter; Figure 4); many other N cycle genes were not detected at all.
Figure 4.
Copy numbers of phosphorus, nitrogen and sulfur cycle transcripts in a coastal ocean microbial community. The left line represents the limit of detection for this study, and together with the right line defines the region where copy numbers are too low for robust statistical analysis (that is, where the fold-difference requirement is >2). Symbols indicate copy numbers in biological duplicates. Bottom graphs show monthly nutrient concentrations for GCE-LTER station six. The arrows mark the date of sample collection.
To examine detection of biogeochemically diagnostic mRNAs in theoretically smaller libraries, the full metagenomic libraries were randomly subsampled in silico to generate subsets. The majority of the elemental cycle transcripts detected in the full libraries were still evident in smaller libraries. For example, >80% of the P-cycle related genes would have had at least one hit in a library one-fourth the size (Supplementary Figure S3).
Statistical resolution
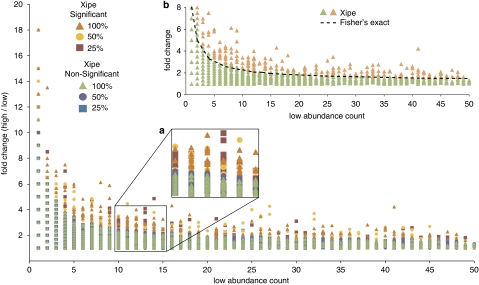
Comparative metatranscriptomics seeks to differentiate transcript abundance between samples, for example, across natural environmental gradients or in response to experimental manipulations. We examined the statistical power of comparative analyses as a function of library size, starting first with broad categories of gene function as represented by COG assignments. Subsets of each of the replicate libraries were generated in silico and the fold-difference criteria (high abundance count/low abundance count) needed for statistically significant differences were compared using a resampling method based on difference of medians (Xipe; Rodriguez-Brito et al., 2006). Even for libraries one-fourth of the original size, there was little effect on the fold-difference threshold required for a COG category to be considered significantly different between samples (Figure 5). This was true as well for an alternate statistical approach using contingency tables and Fisher's exact test (White et al., 2009) (Figure 5, inset b), and also when analyzing libraries much smaller than the original (for example, the average fold-difference threshold for significance in a 10 000 read library was <1% greater than in a 500 000 read library).
Figure 5.
Minimum fold difference required for statistical significance (Xipe, P<0.05) as a function of both the count in the lower abundance sample and the library size. Samples and subsamples were from the combined libraries (FN56 and FN57). Marker color is based on the statistical outcome (significant or nonsignificant) and library size (percent of full library). (a) Zoom of region in the main figure. Note that the minimum fold-difference for significance is independent of the three library sizes analyzed. (b) An alternative analysis of the significance threshold using contingency tables and Fisher's exact test. The minimum fold-difference threshold at which a low abundance count is significant by the Fisher's exact test is plotted as a dotted black line. The results from the Xipe analysis (main figure) at the 100% library size are also shown in inset B for direct comparison with the Fisher's exact test.
Library size, however, had a direct impact on the number of counts in a transcript category, thereby affecting the power of statistical comparisons. Transcript categories with low copy numbers (defined here as ⩽15 hits in the lower abundance sample) required from 2- to 8-fold difference between the two samples for statistical significance (Figure 5). For smaller in silico subsets of the libraries or for more specific transcript annotation categories (for example, RefSeq gene bins), both of which result in lower counts per category, the power to detect statistical differences between two samples decreased. For example, 17 out of the 25 genes that mediate key steps in the marine P cycle fell into a low-count category even with the full-size library (Figure 4), and nearly all would do so if the library was one-fourth of the original size. For metatranscriptomic libraries of the magnitude obtained in this study (>1 000 000 454 FLX reads), only those transcripts present at concentrations >1 × 106 per liter had a good probability of being detected, and only those present at concentrations >1.5 × 107 per liter (which would exclude all singletons and other low-count transcript categories) could be compared across samples with good statistical power.
Replication
The need to improve sample-sequencing depth competes with the need for replication in comparative metatranscriptomic analyses. Two important sources of variability that can be quantified through replication include technical variation during sample processing/sequencing, and natural biological variation within the environment sampled.
For the first type, 454 pyrosequencing is prone to artifacts in which single DNA fragments are sequenced more than once (‘duplicate sequences'). Although artifactual duplicates are recognized in metagenomes as sequences with identical 5′-sequence and high identity throughout (Dinsdale et al., 2008; Gomez-Alvarez et al., 2009), true duplicate sequences can arise in metatranscriptomes from discrete mRNAs from highly expressed genes. In this study, 24% of RefSeq reads were ‘duplicates' (same start site and ⩾90% identity). As each replicate biological sample was sequenced as four technical replicates (independent emulsion PCRs and sequencing runs), and assuming that artifactual duplicates arise during the emulsion PCR step (Gomez-Alvarez et al., 2009; Stewart et al., 2010), artifactual duplicates should have uneven distributions across the four 454 runs, whereas natural duplicates should be evenly distributed. We found that most duplicate clusters averaged ∼25% per technical sequencing replicate (Supplementary Figure S4), and a statistical comparison of COG assignments for all six within-sample pairwise combinations of the technical sequencing replicates indicated that only 0.2% fit the pattern for artifactual duplicates (significantly higher in one technical replicate compared to the other three). For the transcript with the highest copy number in the combined library (Rac prophage; ZP_03400590), removal of duplicate reads would have decreased the count by 98% (from 6235 to 111 hits) despite evidence from technical replicates that many of these are natural (Supplementary Figure S5). Duplicate removal from metatranscriptomic libraries based on sequence start position and percent identity (Gomez-Alvarez et al., 2009; Stewart et al., 2010) may therefore produce systematic underestimates of abundance for the most highly transcribed genes in the community, and statistical analysis of technical replicates is a recommended alternative.
For the second type of variation, within-treatment biological variability sets the false-positive rate against which differences in gene expression patterns across treatments or environments can be evaluated (Poretsky et al., 2010). In this study, patchiness in community gene transcription patterns was detectable in paired coastal seawater samples separated by ∼300 m (based on tidal flushing rates past a fixed collection point). At the level of functional gene categories, pairwise comparisons indicated significant differences between the samples for 461 of 3298 COGs (14%) (Xipe, P<0.05). Only nine significant COGs contained sequences from a putative artifactual duplicate cluster (see above), highlighting the benefit of technical replicate averaging for reducing spurious differences from sequencing artifacts. In accordance with other studies of environmental sequence libraries (Rodriguez-Brito et al., 2006), as well as our observations above, decreasing the library size had a major influence on the number of significant differences that were detectable (Supplementary Table S1).
Differences between replicate samples at the individual gene level (that is, transcripts binned by RefSeq hits) were also examined, using Fisher's exact test coupled with a correction for the false discovery rate (Strimmer, 2008) to control for type I errors arising when simultaneously conducting large numbers of statistical tests (in this case, for >186 000 different RefSeq bins). Eighty-three (0.05%) of the gene bins were statistically different between the two samples (P<0.05 with Benjamini–Hochberg correction), including those representing phage genes, ammonia oxidation genes and various genes for light-driven energy acquisition (Supplementary Figure S6). The replicate samples therefore established within-treatment variability (Figure 4) for future between-treatment comparisons.
Microbial gene expression in a coastal ocean
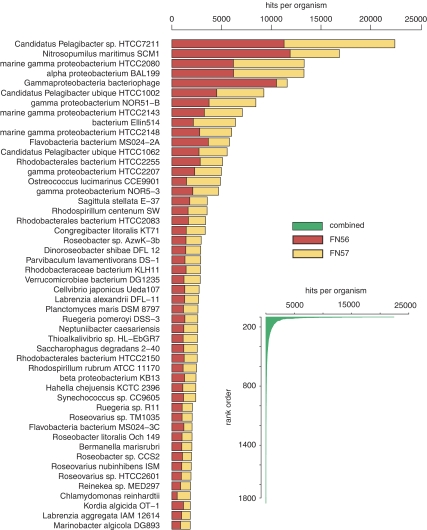
Transcripts from the combined library binned to genes from 1909 reference organisms. Thirty-three percent of the sequences had best hits to alphaproteobacteria genes (with roseobacters accounting for 11% and P. ubique for 7%) and 27% had best hits to gammaproteobacteria genes (Figure 6). Unexpectedly, 4% of the transcripts binned to the two archaeal genomes of Nitrosopumilus maritimus (3.3%) and Cenarchaeum symbiosum (0.1%). Although Archaea are often abundant and active in deep ocean environments, they were not expected to contribute significantly to gene expression in this shallow coastal water system; however, the N. maritimus taxonomic bin was the second largest in the metatranscriptome (Hollibaugh et al., in press). The oligotrophic marine gammaproteobacteria clades, which are usually in low abundance in oceanic 16S rRNA libraries (<3% Cho and Giovannoni, 2004), were also unexpectedly well represented in the metatranscriptome (Figure 6; 8% of transcripts), and transcripts binning to three genes from a gammaproteobacteria prophage (3% of transcripts) may indicate an ongoing infection of these oligotrophic marine gammaproteobacteria populations. Eukaryotic transcripts composed 6% of the total, with those binning to Ostreococcus spp. particularly well represented (20% of eukaryotic hits).
Figure 6.
Rank-order abundance of taxonomic bins (species or strain level). Main figure: top 50 taxonomic annotation bins; inset: all 1909 taxonomic annotation bins.
Copy numbers of transcripts representing 82 genes diagnostic for P, N and S cycling were determined simultaneously from the metatranscriptomic data (Figure 4). For P transformations, annotations suggest bacterioplankton were transporting phosphate by both high- and low-affinity transporters. The expression of low-affinity transporters, along with polyphosphate storage genes, is consistent with elevated phosphate concentrations (1.2 μ) at the time of sampling, which is typical of late summer in this coastal ocean (Figure 4). Expression patterns also indicated ongoing use of organic P, including phosphoesters (by phoX, phoD and phoA) and phosphonates (although transcripts for the canonical C–P lyase pathway were near the limit of detection). For N transformations, sampling occurred during a local ammonia peak (2.6 μ; Figure 4), and transcripts related to the uptake and oxidation of ammonia (amt, amoA, B and C) were orders of magnitude higher in abundance than genes mediating nitrate or nitrite processing (for example, nar, nap and nir genes) (Figure 4). Transcripts for urea metabolism, the only representative of dissolved organic N use included in the analysis, made up the second most abundant group of N-related sequences (Figure 4). Nitrogen is often the limiting nutrient (or co-limiting with carbon; Pomeroy et al., 2000) to microbial activity in this ecosystem, and dissolved organic N is 2- to 200-fold higher in concentration than inorganic N. For S transformations, gene expression suggested substantial use of reduced S compounds typically found in high concentrations in marsh-dominated coastal systems (Kiene and Capone, 1988; Pakulski and Kiene, 1992). Transcripts were found for metabolism of dimethylsulfoniopropionate (dmdA, dddP and dddD), as well as oxidation of sulfide/thiosulfate (sox genes) (Figure 4). This broad inventory of P, N and S cycle transcripts represents an absolute benchmark against which time-series and experimentally manipulated transcriptomes in this ecosystem can be compared.
Conclusions
Addition of an internal mRNA standard provides a significant advantage in metatranscriptomics protocols as it allows estimation of the fraction of the microbial transcriptome captured in the sequence library, as well as the absolute quantification of transcript copy number in the environment (Figures 1 and 4). Although RT-qPCR approaches can also provide absolute transcript numbers, often with greater sensitivity (Church et al., 2010), they are currently limited to a handful of functional genes at a time. Furthermore, the high microdiversity found in many natural gene populations (Figure 3) makes primer design challenging (Varaljay et al., 2010), and likely results in RT-qPCR only quantifying a subset of the total functional gene population. Multiple internal standards that vary in length and concentration (van de Peppel et al., 2003) will allow for more robust calculations of sequencing depth in future studies, and better position ‘omics' data for integration with biogeochemical rate measurements.
As low-count transcript categories are difficult to resolve statistically, library size had a critical effect on comparative metatranscriptomic analyses. Many of the biogeochemically diagnostic transcripts detected in our libraries would have been detected in ones that were one-fourth or one-tenth the size, but these theoretically smaller libraries resulted in a decreased ability to statistically differentiate between samples. Typical library sizes for metatranscriptomes (105–106 sequence reads) are therefore sufficient for descriptive studies, but significant gains in comparative analyses of biogeochemically informative gene expression patterns will require a greater sequencing investment. Indeed, 54% of the 46 detected steps in the marine N, P and S cycles would require at least a twofold difference in copy number between samples to meet statistical criteria for hypothesis testing (that is, P<0.05). Although a twofold change in transcript abundance is an appropriate minimum criterion for expression studies of clonal bacterial cultures in synchronized growth (Bürgmann et al., 2007), it may fail to catch smaller expression differences among complex microbial communities that are ecologically relevant.
Despite a sample-sequencing depth of only 1 in 107 transcripts, the libraries provided remarkable insights into gene expression in a marine microbial community, including evidence for active microbes not known previously to have a major role in the ecosystem and quantification of transcripts for scores of steps in marine elemental cycles. These data establish the foundation for comparative assessments of diel, seasonal and annual changes in microbial gene expression that will provide insights into the regulation of biogeochemical processes in the coastal ocean.
Acknowledgments
We thank R Newton for assistance with sample collection and bioinformatics analysis, L Tomsho and S Schuster at Penn State University for 454 sequencing expertise, JT Hollibaugh for comments and discussion on the article and S Rathbun for helpful discussions on statistical methods. Nutrient data were provided by K Hunter and S Joye through the Georgia Coastal Ecosystems Long Term Ecological Research program (OCE-0620959). This project was supported by funding from the Gordon and Betty Moore Foundation and the National Science Foundation Microbial Observatories Program (MCB-0702125).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Azam F, Hodson RE. Size distribution and activity of marine microheterotrophs. Limnol Oceanogr. 1977;22:492–501. [Google Scholar]
- Bürgmann H, Howard EC, Ye WY, Sun F, Sun SL, Napierala S, et al. Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP) Environ Microbiol. 2007;9:2742–2755. doi: 10.1111/j.1462-2920.2007.01386.x. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Waidner LA, Cottrell MT, Kirchman DL. Abundant proteorhodopsin genes in the North Atlantic Ocean. Environ Microbiol. 2008;10:99–109. doi: 10.1111/j.1462-2920.2007.01436.x. [DOI] [PubMed] [Google Scholar]
- Cho J-C, Giovannoni SJ. Cultivation and growth characteristics of a diverse group of oligotrophic marine gammaproteobacteria. Appl Environ Microbiol. 2004;70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne KJ, Handy SM, Demir E, Whereat EB, Hutchins DA, Portune KJ, et al. Improved quantitative real-time PCR assays for enumeration of harmful algal species in field samples using an exogenous DNA reference standard. Limnol Oceanogr Meth. 2005;3:381–391. [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Ducklow HW.2000Bacterial production and biomass in the oceansIn: Kirchman (ed).Microbial Ecology of the Ocean, 1st edn.Wiley-Liss: New York, NY [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, et al. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS ONE. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Hannah MA, Redestig H, Leisse A, Willmitzer L. Global mRNA changes in microarray experiments. Nat Biotechnol. 2008;26:741–742. doi: 10.1038/nbt0708-741. [DOI] [PubMed] [Google Scholar]
- Hewson I, Poretsky RS, Beinart RA, White AE, Shi T, Bench SR, et al. In situ transcriptomic analysis of the globally important keystone N2-fixing taxon Crocosphaera watsonii. ISME J. 2009a;3:618–631. doi: 10.1038/ismej.2009.8. [DOI] [PubMed] [Google Scholar]
- Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ, et al. Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J. 2009b;3:1286–1300. doi: 10.1038/ismej.2009.75. [DOI] [PubMed] [Google Scholar]
- Hollibaugh JT, Gifford SM, Sharma S, Bano N, Moran MA.2010Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage ISME JIn press. [DOI] [PMC free article] [PubMed]
- Kiene RP, Capone DG. Microbial transformations of methylated sulfur-compounds in anoxic salt-marsh sediments. Microb Ecol. 1988;15:275–291. doi: 10.1007/BF02012642. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Umbarger HE.1996Chemical composition of Escherichia coliIn: Böck A, Curtiss III R, Kaper JB, Karp PD, Neidhardt FC, Nyström T et al (eds).EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology2nd edn.ASM Press: Washington, DC [Google Scholar]
- Pages H, Aboyoun P, Gentleman R, DebRoy S.2009Biostrings: string objects representing biological sequences, and matching algorithmsR package version 2.14.8.
- Pakulski JD, Kiene RP. Foliar release of dimethylsulfoniopropionate from Spartina alterniflora. Mar Ecol-Prog Ser. 1992;81:277–287. [Google Scholar]
- Passalacqua KD, Varadarajan A, Ondov BD, Okou DT, Zwick ME, Bergman NH. Structure and complexity of a bacterial transcriptome. J Bacteriol. 2009;191:3203–3211. doi: 10.1128/JB.00122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy LR, Sheldon JE, Sheldon WM, Blanton JO, Amft J, Peters F. Seasonal changes in microbial processes in estuarine and continental shelf waters of the south-eastern USA. Estuar Coast Shelf S. 2000;51:415–428. [Google Scholar]
- Poretsky RS, Bano N, Buchan A, LeCleir G, Kleikemper J, Pickering M, et al. Analysis of microbial gene transcripts in environmental samples. Appl Environ Microbiol. 2005;71:4121–4126. doi: 10.1128/AEM.71.7.4121-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretsky RS, Gifford SM, Rinta-Kanto J, Vila-Costa M, Moran MA.2009aAnalyzing gene expression from marine microbial communities using environmental transcriptomics JoVE 2 . ( http://www.jove.com/index/details.stp?ID=1086 ). [DOI] [PMC free article] [PubMed]
- Poretsky RS, Hewson I, Sun SL, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009b;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Poretsky RS, Sun S, Mou X, Moran MA. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol. 2010;12:616–627. doi: 10.1111/j.1462-2920.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2009R: A Language and Environment for Statistical Computing v210.0 R Foundation for Statistical Computing: Vienna, Austria; , ( http://www.R-project.org ). [Google Scholar]
- Robinson M, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest atlantic through eastern tropical pacific. PLoS Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YM, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- Simon M, Azam F. Protein-content and protein-synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9:303. doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich TA, Lanzen J, Qi DH, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE. 2008;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, Holstege FCP. Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep. 2003;4:387–393. doi: 10.1038/sj.embor.embor798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaljay VA, Howard EC, Sun SL, Moran MA. Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl Environ Microbiol. 2010;76:609–617. doi: 10.1128/AEM.01258-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Chen H, Hu XH, Zhang RM, Zhang Z, Luo ZW. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between the two kingdoms. Mol Biol Evol. 2006;23:1107–1108. doi: 10.1093/molbev/msk019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.