Abstract
Factors that facilitate the onset of black band disease (BBD) of corals remain elusive, though anoxic conditions under the complex microbial mat and production of sulfide are implicated in necrosis of underlying coral tissues. This study investigated the diversity and quantitative shifts of sulfate-reducing bacterial (SRB) populations during the onset of BBD using real-time PCR (RT-PCR) and cloning approaches targeting the dissimilatory (bi)sulfite reductase (dsrA) gene. A quantitativePCR (qPCR) assay targeting the 16S rRNA gene also provided an estimate of total bacteria, and allowed the relative percentage of SRB within the lesions to be determined. Three Montipora sp. coral colonies identified with lesions previously termed cyanobacterial patches (CPs) (comprising microbial communities unlike those of BBD lesions), were tagged and followed through time as CP developed into BBD. The dsrA-targeted qPCR detected few copies of the gene in the CP samples (<65 per ng DNA), though copy numbers increased in BBD lesions (>2500 per ng DNA). SRB in CP samples were less than 1% of the bacterial population, though represented up to 7.5% of the BBD population. Clone libraries also demonstrated a shift in the dominant dsrA sequences as lesions shifted from CP into BBD. Results from this study confirm that SRB increase during the onset of BBD, likely increasing sulfide concentrations at the base of the microbial mat and facilitating the pathogenesis of BBD.
Keywords: coral disease, black band disease, dissimilatory sulfite reductase gene (dsrA), quantitative PCR
Introduction
Black band disease (BBD) represents one of the most intensively studied coral diseases as a result of its conspicuous and easy diagnosis in the field, wide-spread prevalence on reefs around the world and the potential destructive impacts of the disease on reef ecosystems (Richardson, 2004). BBD consists of a complex microbial mat that is dominated by phototrophic filamentous cyanobacteria along with other highly diverse heterotrophic bacterial groups, including sulfate-reducing and sulfide-oxidising bacterial populations (Richardson, 1997; Cooney et al., 2002; Frias-Lopez et al., 2004; Viehman et al., 2006). Despite being first identified in 1973 (Antonius, 1973), the underlying mechanisms of BBD pathogenesis still remain elusive. With no ‘primary pathogen' identified, it is currently thought that the microbial consortium as a whole are involved in BBD pathogenicity. A number of factors are implicated as etiological agents, facilitating the progression of the disease across coral colonies (Richardson, 2004). Foremost is the observation that the base of the mat in contact with the coral tissue has been shown to be anoxic and high in sulfide levels, with the oxygen/sulfide interface migrating with varying light intensities (Carlton and Richardson, 1995; Richardson et al., 1997). This combination of anoxia and sulfide is lethal to coral tissue, and, therefore sulfide production is suspected to be a substantial component of BBD pathogenicity (Richardson et al., 1997; Richardson, 2004). Sulfate-reducing bacteria (SRB) of the genus Desulfovibrio have been commonly detected in culture independent 16S ribosomal RNA gene assessments of the BBD microbial mats (Cooney et al., 2002; Frias-Lopez et al., 2002, 2004; Barneah et al., 2007). Desulfovibrio strains have also been isolated from BBD lesions at different locations and from different coral species, and thus proposed as essential for BBD pathogenicity (Viehman et al., 2006).
Investigations of BBD ecology at a site on the Great Barrier Reef off Pelorus Island identified lesions termed ‘cyanobacterial patches (CPs)' that preceded the characteristic signs of BBD on some occasions (Sato et al., 2010). Further microbiological studies characterized the transition in the microbial communities as CP developed into BBD with cyanobacterial species shifting from Blennothrix sp.-affiliated sequences dominating CP lesions, to Oscillatoria sp.-affiliated ribotypes in BBD (Sato et al., 2010). Shifts in other bacterial groups were also observed with Alphaproteobacteria affiliated sequences dominant in CP lesions, whereas gammaproteobacterial ribotypes were more abundant in BBD lesions. It is interesting that the sequences affiliated with organisms identified in sulfur cycling were most commonly retrieved from BBD lesions and it was speculated that increased colonization of bacteria involved in sulfur cycling during the onset of BBD potentially contributes to the higher progression rates for BBD compared with CP lesions (Sato et al., 2010).
This study specifically aimed to provide a quantitative assessment of SRB within the lesions during the development of BBD from CP. The metabolic capability of dissimilatory sulfate reduction is widely distributed within both bacterial and archeal phyla and the enzyme responsible (dissimilatory sulfite reductase; DSR) is highly conserved and ubiquitous in all known sulfate-reducing prokaryotes and, therefore, ideal for assessing the diversity of these populations within environmental samples (Wagner et al., 1998, 2005; Leloup et al., 2007; Kondo et al., 2008). This study modified an established quantitative real-time PCR (RT-PCR) approach, targeting the gene coding for the α-subunit of the DSR gene (dsrA) (Leloup et al., 2007; Kondo et al., 2008) to assess both the diversity and relative abundance of SRB within microbial samples collected from CP, intermediate stages (IM) and BBD coral disease lesions. Owing to the variability in bacterial population diversity and sampled biomass within individual coral lesions, the SRB population was determined as a percentage of the total bacterial population by including a quantitative RT-PCR assay targeting the bacterial 16S rRNA gene (Nadkarni et al., 2002). In this study we demonstrate the increase in abundance and shift in diversity of SRB populations within the microbial lesions during the onset of BBD.
Results and discussion
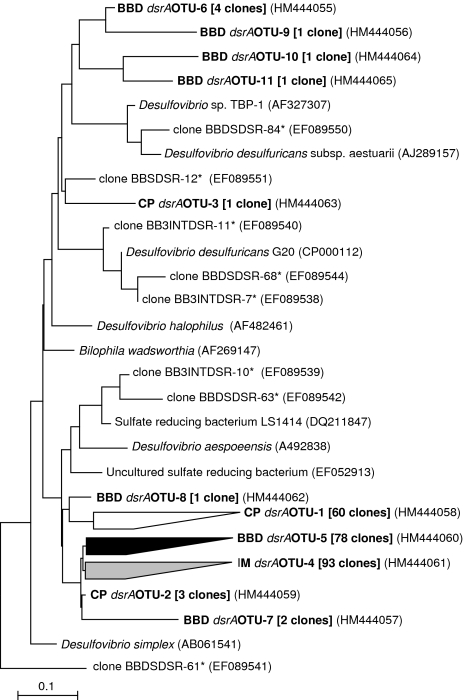
CP lesions on three individually tagged (2.5-3 m depths) Montipora hispida coral colonies (YS-1, YS-2 and YS-3) at Pelorus Island in the central Great Barrier Reef (18°33′S and 146°30′E), were monitored and sampled at approximately 2 week intervals as CP progressed into an IM (slightly darker lesion color) and then developed characteristic BBD signs. For each stage (CP, IM and BBD), biomass from the microbial mat and underlying skeleton was collected (approximately 5 mm in diameter, 2 mm in depth) from the front of the lesion using a sterilized stainless steel chisel and processed for total DNA extraction as outlined previously (Sato et al., 2010). The DNA from the three parallel colonies were pooled according to the disease developmental stages and diversity of dsrA genes from the individual stages was determined through PCR amplification using primers DSR1-F+ (5′-ACSCACTGGAAGCACGGCGG-3′) and DSR-R (5′-GTGGMRCCGTGCAKRTTGG-3′). The primers target a subset of the dsrA sequences identified from SRB (Leloup et al., 2007) and moreover cover sequences from Desulfovibrionaceae that have been implicated as members of the SRB population in BBD microbial mats (Viehman et al., 2006; Barneah et al., 2007; Sato et al., 2010). Amplified products (221 bp in size) were cloned (TOPO TA Kit, Invitrogen, Carlsbad, CA, USA) and sequenced. Subsequent phylogenetic analysis (MEGA; Kumar et al. (2008) see Supplementary Section for detailed methods) of dsrA sequences derived from CP, IM and BBD libraries demonstrated that the majority of identified sequences had less than 90% DNA identity to previously identified dsrA genes present in the public databases (Figure 1), including dsrA genes derived from previous studies of BBD lesions from corals in the Red Sea (Barneah et al., 2007). Sequences within each respective library were grouped into operational taxonomic units (OTUs) using the MOTHUR program (Schloss et al., 2009) and a 90% sequence identity cutoff. The analysis indicated that the majority of sequences within each respective clone library clustered at this 90% gene sequence identity (94%, 100% and 89% of clones within CP, IM and BBD libraries, respectively). It is interesting that the dominant dsrA sequences from the CP sample (CP dsrAOTU-1) displayed <92% sequence similarity (206 over 221 bp) to the dominant OTUs in both the BBD (BBD dsrAOTU-5) and IM (IM dsrAOTU-4) libraries (Figure 1). In contrast the dominant sequences in the IM library (IM dsrAOTU-4) displayed >98% sequence identity (>219 over 221 bp) to the BBD dominant OTU (BBD dsrAOTU-5). These results indicate that the SRB community shifted as CP lesions developed the first initial signs of BBD at the IM stage. The closest related dsrA sequence to the dominant CP, IM and BBD library clones (∼90% over 221 bp identified through BLAST analysis; Altschul et al., 1997) was derived from an uncultured sulfate reducing bacterium derived from waste water (Ben-Dov et al., 2007) (accession number EF052913). The CP library also contained three clones (CP dsrAOTU-2; Figure 1) that were highly similar to the IM and BBD dominant clones indicating that a ‘seed' SRB population is present in these lesions and may become dominant in the IM libraries as CP lesions undergo successional changes in microbial communities (Sato et al., 2010). The BBD dsrAOTU-8 clone, which clustered with the dominant CP cluster, may represent a residual from the CP lesion. A small number of other dsrA sequences recovered from the BBD library (BBD dsrAOTU-6, -9, -10 and -11) and one clone from the CP library (CP dsrAOTU-3), affiliated more closely with previous BBD derived dsrA genes (Figure 1).
Figure 1.
Neighbour-joining phylogenetic tree for the dissimilatory sulfate reductase gene dsrA of SRB showing relationship between sequences obtained from CP, IM and BBD lesion samples and reference sequences of SRBs obtained from GenBank. Sequences were grouped into OTUs on the basis of a 90% sequence identity cutoff. Representative sequences of OTU groups identified in this study are presented in bold text and numbers following in square brackets are indicative of the number of sequences within each OTU group. The * indicates sequences that have been retrieved from previous investigations of SRB in BBD lesions (Barneah et al., 2007). The scale bar is an estimate of sequence divergence (10%).
The diversity of dsrA gene sequences retrieved from this study all clustered within the Desulfovibrionaceae family. In the only other study specifically investigating dsrA genes from BBD lesions, Barneah et al. (2007) similarly observed sequences closely related to Desulfovibrio species and retrieved no representatives from other genera of SRB, implying the dominance of Desulfovibrio in BBD lesions. Studies investigating the microbial diversity of BBD lesions from different geographical regions using 16S rRNA gene profiling techniques have also identified Desulfovibrio affiliated sequences as constituents of the BBD mat (Cooney et al., 2002; Frias-Lopez et al., 2002, 2004), whereas Viehman et al. (2006) isolated a number of Desulfovibrio strains from BBD lesions. From the current study site, Sato et al. (2010) recovered a number of Desulfovibrio affiliated sequences from 16S rRNA gene assessments of BBD lesions, though one Desulfovibrio sp.-affiliated sequence was also observed in one CP library.
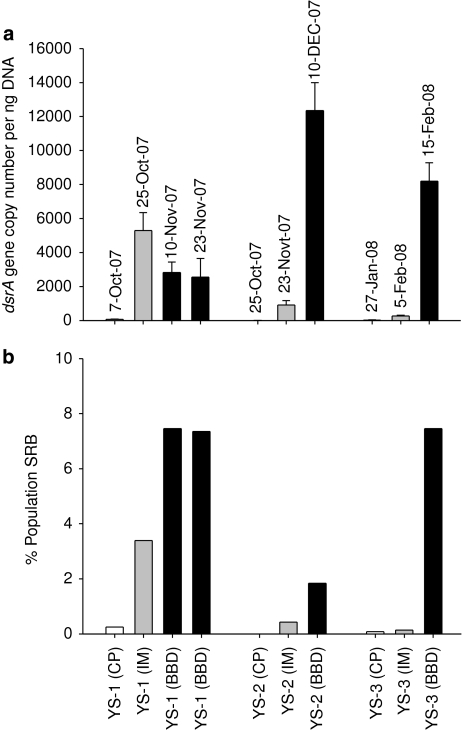
A quantitative qRT-PCR assay targeting the dsrA gene with primers DSR1-F+ and DSR-R, was adapted from Kondo et al. (2008) and LeLoup et al. (2007). The qPCR assay was modified through the use of the TaqMan chemistry (Applied Biosystems, Foster City, CA, USA). and an additional internal probe (DSRtaq (HEX)-5′-CCGATAACRCYGCCGCCGTAACCGA-3′-(TAMRA)) allowing increased specificity and discrimination in quantification of dsrA genes within samples (see Supplementary Section for detailed optimized qPCR reaction conditions). The copy number of dsrA genes detected in CP lesions was very low, ranging from only three copies detected in the YS-2 colony to 26 copies in YS-3 and 64 copies in YS-1 per ng DNA (Figure 2a). The dsrA copy number increased for colonies YS-2 and YS-3 as the lesion transitioned from CP to IM disease stages (to 9.12 × 102 and 2.58 × 102 copies per ng DNA, respectively) and substantially increased as BBD developed (to 1.24 × 104 and 8.20 × 103 copies). For the lesions on the YS-1 colony, dsrA copy numbers increased to 5.29 × 103 copies at the IM stage though was reduced to 2.81 × 103 copies at the BBD stage. When the BBD lesion on the YS-1 colony was resampled 2 weeks later (23rd Nov) the dsrA copy number within the lesion was similar (2.56 × 103 dsrA copies per ng DNA).
Figure 2.
(a) Quantitative real-time PCR (RT-PCR) targeting dsrA genes of SRB in diseased lesions of coral samples. Values represent dsrA gene copies per ng extracted DNA and are given as mean ± standard deviation of triplicate samples over a 10 × serial dilution range of template DNA (50 ng–0.05 ng). (b) Number of SRB in diseased lesions (calculated from A) relative to the total bacterial population determined by quantitative RT-PCR targeting 16S rRNA genes within lesion samples. Coral colonies YS-1, YS-2 and YS-3 were visually assessed as displaying CP, IM or BBD stages of the lesions and the sampling dates of the lesions are also provided.
The sampled BBD mat is a highly complex microbial community that in addition to bacteria may also contain fungi (Ramos-Flores, 1983) and other eukaryotic organisms, including biomass from degrading coral tissue and zooxanthellae. Normalisation of the copy number of the dsrA gene against extracted total DNA may therefore be misleading due to a large amount of co-extracted DNA from these eukaryotic cells. In addition, sampling of microbial lesions is highly variable in the amount of biomass collected and therefore accurate comparative quantification of target genes between samples is difficult. To overcome these problems and further validate the results of the dsrA quantification in these samples, an additional real-time TaqMan PCR assay was adapted from Nadkarni et al. (2002) for quantification of the bacterial abundance through targeting of the 16S rRNA gene. The assay incorporated the universal primers 331f (5′-TCCTACGGGAGGCAGCAGT-3′), 797r (5′-GGACTACCAGGGTATCTAATCCTGTT-3′) and the probe BacTaq ((6-FAM)-5′-CGTATTACCGCGGCTGCTGGCAC-3′-(TAMRA)), targeting almost all bacterial phyla except Chlorobi, Chlamydiae, Dictyoglomus and TM7 (Nadkarni et al., 2002), which were expected at low abundance from previous bacterial profiling of lesions at this study site (Sato et al., 2010).
When the dsrA gene copy number was standardized as a proportion of sulfate reducers within the total bacterial population of the lesions, SRB increased in all samples as the lesions progressed from CP to IM and then BBD (Figure 2b). For all CP lesions, the percentage of sulfate reducers was less than 1% of the total population. For colony YS-1, SRB increased to ∼3.4% of the bacterial population at the IM stage, though were still less than 1% in the other two lesion samples from colonies YS-2 and YS-3. However, all colonies displayed increased proportions of SRBs in the bacterial community in the BBD lesions, reaching >7% for colonies YS-1 and YS-3. It is interesting that the proportion of SRB in the BBD lesion of colony YS-2 was only ∼1.8% (Figure 2b) even though it possessed the highest dsrA gene copy number (Figure 2a) indicating the highly complex and variable nature of BBD lesions.
There are limitations with qPCR assays for accurate quantification of bacteria in complex samples, including multiple gene copies that may increase for actively growing populations and detection of DNA from dormant and dead cells (Nadkarni et al., 2002). In addition, DNA extraction may vary between samples, however, consistent methods were used for all lesions and thus minimizing this potential sample to sample bias. Nonetheless, all assays were carried out using time-series samples of visually identical lesions at comparable stages of disease progression, therefore, demonstrating that SRB populations are low in CP samples and increase as BBD visual signs appear on the coral colonies.
The results from this study demonstrate that within the microbial mats, the SRB populations shift in diversity and increase in abundance as the lesion transitions from CP into BBD. It is interesting that the diversity shift occurs from the CP to the IM stage and it coincides with a visual darkening of the microbial mat. These visual changes could be from an increase in cyanobacterial densities associated with the BBD lesions. In addition, the organic carbon produced during cyanobacterial photosynthesis (in the form of dissolved organic carbon) stimulates heterotrophs in the microbial mat, including the SRB. Reduced sulfur (S2−), the metabolic product of SRB in the form hydrogen sulfide (H2S), will react with metal ions such as iron that are often high in cyanobacterial-dominated microbial mats, producing metal sulfides, such as ferrous sulfide (FeS) or pyrite (FeS2) (Stal, 2000). These black or brown compounds precipitate and maybe an additional reason why the dark color is observed when visual signs of BBD are first observed. Other groups of bacteria, particularly the sulfate oxidisers, which are also present within this complex microbial mat, can oxidise the H2S to elemental sulfur or to sulfate using dissolved oxygen, metal oxides (for example, Fe oxyhydroxides and Mn oxides) or nitrate as the oxidant (Jørgensen and Nelson, 2004).
Previous microsensor studies have highlighted that microenvironments within the BBD lesions are both anaerobic and high in sulfide (Carlton and Richardson, 1995), conditions that are toxic for the underlying coral tissue (Richardson et al., 1997; Richardson, 2004). It is interesting that recent microsensor studies on both CP and BBD lesions at this study site have detected the presence of little sulfide in the CP lesions and high sulfide in the BBD lesions (unpublished results). The BBD lesions displayed higher progression rates across coral colonies when compared with CP lesions (Sato et al., 2010), likely related to the increased SRB populations resulting in higher sulfide concentrations at the tissue/lesion interface facilitating increased virulence. It is, however, unknown what drives the transition of the CP microbial community dominated by Blennothrix-related cyanobacterial species and low in SRB to a BBD microbial mat dominated by Oscillatoria-related cyanobacterial species (Sato et al., 2010) and high in SRB. Potentially, high cyanobacterial biomass may promote anoxic conditions and contribute to a shift in the SRB communities from CP to BBD, especially during dark-periods when oxygen consumption by aerobic respiration (cyanobacterial and bacterial) would occur. Alternatively a shift and increase in the SRB community may promote higher sulfide concentrations at the base of the lesion that selects for sulfide-tolerant cyanobacteria. Previous studies of BBD-derived cyanobacteria have demonstrated that they are able to perform oxygenic photosynthesis in the presence of sulfide (Richardson and Kuta, 2003) and that BBD-derived cyanobacteria are specifically adapted for survival in the sulfide-rich BBD environment (Myers and Richardson, 2009). It is warranted to test the sulfide tolerance of cultured cyanobacterial isolates derived from CP and BBD lesions to observe the role of sulfide and the SRB in driving the shift in dominating cyanobacterial types during the onset of BBD at this site on Great Barrier Reef. These two scenarios are not mutually exclusive, however, and both changes in sulfide and cyanobacterial biomass likely act synergistically along with environmental factors (for example, light and temperature Sato et al., 2009) to change the respective community populations and facilitate the onset of virulent BBD lesions.
Acknowledgments
The authors thank staff of Orpheus Island Research Station (JCU) for logistic support and field volunteers for their assistance during sampling. Lone Høj (AIMS) is also thanked for helpful comments to improve the paper.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonius A.1973New observations on coral destruction in reefs 10th Meeting of the Association of Island Marine Laboratories of the Caribbeanpp. 3.University of Puerto Rico: Association of Island Marine Laboratories of the Caribbean [Google Scholar]
- Barneah O, Ben-Dov E, Kramarsky-Winter E, Kushmaro A. Characterization of black band disease in Red Sea stony corals. Environ Microbiol. 2007;9:1995–2006. doi: 10.1111/j.1462-2920.2007.01315.x. [DOI] [PubMed] [Google Scholar]
- Ben-Dov E, Brenner A, Kushmaro A. Quantification of sulfate-reducing bacteria in industrial wastewater, by real-time polymerase chain reaction (PCR) using dsrA and apsA genes. Microb Ecol. 2007;54:439–451. doi: 10.1007/s00248-007-9233-2. [DOI] [PubMed] [Google Scholar]
- Carlton RG, Richardson LL. Oxygen and sulfide dynamics in a horizontally migrating cyanobacterial mat: Black band disease of corals. FEMS Microbiol Ecol. 1995;18:155–162. [Google Scholar]
- Cooney RP, Pantos O, Le Tissier MDA, Barer MR, O'Donnell AG, Bythell JC. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ Microbiol. 2002;4:401–413. doi: 10.1046/j.1462-2920.2002.00308.x. [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J, Klaus JS, Bonheyo GT, Fouke BW. Bacterial community associated with black band disease in corals. Appl Environ Microbiol. 2004;70:5955–5962. doi: 10.1128/AEM.70.10.5955-5962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl Environ Microbiol. 2002;68:2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen BB, Nelson DC.2004Sulfide oxidation in marine sediments: geochemistry meets microbiologyIn: Amend JP, Edwards K, Lyons TW (eds).Sulfur Biogeochemistry—Past and Presentvol. 379.Special paper Geological Society of America; 36–81. [Google Scholar]
- Kondo R, Shigematsu K, Butani J. Rapid enumeration of sulphate-reducing bacteria from aquatic environments using real-time PCR. Plankton Benthos Res. 2008;3:180–183. [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup J, Loy A, Knab NJ, Borowski C, Wagner M, Jørgensen BB. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol. 2007;9:131–142. doi: 10.1111/j.1462-2920.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- Myers JL, Richardson LL. Adaptation of cyanobacteria to the sulfide-rich microenvironment of black band disease of coral. FEMS Microbiol Ecol. 2009;67:242–251. doi: 10.1111/j.1574-6941.2008.00619.x. [DOI] [PubMed] [Google Scholar]
- Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Ramos-Flores T. Lower marine fungus associated with black line disease in star corals (Montastraea-annularis, E and S) Biol Bull. 1983;165:429–435. doi: 10.2307/1541208. [DOI] [PubMed] [Google Scholar]
- Richardson LL. Occurrence of the black band disease cyanobacterium on healthy corals of the Florida Keys. Bulletin of Marine Science. 1997;61:485–490. [Google Scholar]
- Richardson LL.2004Black band diseaseIn: Rosenberg E, Loya Y (eds.).Coral Health and Disease Springer-Verlag: Berlin, Germany; 325–336. [Google Scholar]
- Richardson LL, Kuta KG. Ecological physiology of the black band disease cyanobacterium Phormidium corallyticum. FEMS Microbiol Ecol. 2003;43:287–298. doi: 10.1016/S0168-6496(03)00025-4. [DOI] [PubMed] [Google Scholar]
- Richardson LL, Kuta KG, Schnell S, Carlton RG. Ecology of the black band disease microbial consortium. Proceedings of the eighth international coral reef symposium. 1997;1:597–600. [Google Scholar]
- Richardson LL, Miller AW, Broderick E, Kaczmarsky L, Gantar M, Stanic D, et al. Sulfide, microcystin, and the etiology of black band disease. Dis Aquat Org. 2009;87:79–90. doi: 10.3354/dao02083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Bourne DG, Willis BL. Dynamics of seasonal outbreaks of black band disease in an assemblage of Montipora species at Pelorus Island (Great Barrier Reef, Australia) Proc R Soc B: Biol Sci. 2009;276:2795–2803. doi: 10.1098/rspb.2009.0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Willis BL, Bourne DG. Successional changes in bacterial communities during the development of black band disease on the reef coral, Montipora hispida. ISME J. 2010;4:203–214. doi: 10.1038/ismej.2009.103. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stal LJ.2000Cyanobacterial mats and stromatolitesIn: Whitton BA, Potts M (eds.).The Ecology of Cyanobacteria Kluwer Academic Publishers: Dordrecht, The Netherlands; 61–120. [Google Scholar]
- Viehman S, Mills DK, Meichel GW, Richardson LL. Culture and identification of Desulfovibrio spp. from corals infected by black band disease on Dominican and Florida Keys reefs. Dis Aquat Org. 2006;69:119–127. doi: 10.3354/dao069119. [DOI] [PubMed] [Google Scholar]
- Wagner M, Roger A, Flax J, Brusseau G, Stahl D. Phylogeny of dissimilatory reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Loy A, Klein M, Lee N, Ramsing NB, Stahl DA, et al. Functional marker genes for identification of sulfate-reducing prokaryotes. Methods Enzymol. 2005;397:469–489. doi: 10.1016/S0076-6879(05)97029-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.