Abstract
Naphthenic acids (NAs) occur naturally in oil sands and enter the environment through natural and anthropogenic processes. NAs comprise toxic carboxylic acids that are difficult to degrade. Information on NA biodegradation mechanisms is limited, and there are no studies on alkyl branched aromatic alkanoic acid biodegradation, despite their contribution to NA toxicity and recalcitrance. Increased alkyl side chain branching has been proposed to explain NA recalcitrance. Using soil enrichments, we examined the biodegradation of four aromatic alkanoic acid isomers that differed in alkyl side chain branching: (4′-n-butylphenyl)-4-butanoic acid (n-BPBA, least branched); (4′-iso-butylphenyl)-4-butanoic acid (iso-BPBA); (4′-sec-butylphenyl)-4-butanoic acid (sec-BPBA) and (4′-tert-butylphenyl)-4-butanoic acid (tert-BPBA, most branched). n-BPBA was completely metabolized within 49 days. Mass spectral analysis confirmed that the more branched isomers iso-, sec- and tert-BPBA were transformed to their butylphenylethanoic acid (BPEA) counterparts at 14 days. The BPEA metabolites were generally less toxic than BPBAs as determined by Microtox assay. n-BPEA was further transformed to a diacid, showing that carboxylation of the alkyl side chain occurred. In each case, biodegradation of the carboxyl side chain proceeded through beta-oxidation, which depended on the degree of alkyl side chain branching, and a BPBA degradation pathway is proposed. Comparison of 16S rRNA gene sequences at days 0 and 49 showed an increase and high abundance at day 49 of Pseudomonas (sec-BPBA), Burkholderia (n-, iso-, tert-BPBA) and Sphingomonas (n-, sec-BPBA).
Keywords: beta-oxidation, butyl phenylbutyric acid biodegradation, naphthenic acids, oil sands, bioremediation
Introduction
Naphthenic acids (NAs) are an important group of organic pollutants comprised predominantly of saturated aliphatic and alicyclic carboxylic acids and to a lesser extent, aromatic carboxylic acids (Brient et al., 1995). NAs occur naturally in crude oil; however, of greater environmental significance is their occurrence in oil sands, in which oil extraction processes concentrate NAs in aqueous waste streams (Brient et al., 1995). NAs are among the most toxic components of both refinery effluent and oil sands wastewaters, posing significant environmental and human health risks (Dokholyan and Magomedov, 1984; Young et al., 2008).
NAs are highly toxic, recalcitrant compounds that are corrosive and may form metal naphthenate precipitates, which block pipelines and oil-processing equipment. NAs may persist in the environment for many years; for example, NA concentrations in aged wastewater have remained high at 19 mg l−1 even after several decades (Quagraine et al., 2005b). With process wastewater volume predicted to increase to >1 billion m3 by 2025 in Canada alone, this is a serious environmental issue that urgently needs addressing (Hadwin et al., 2006). Despite their persistence and toxicity, very little is known about NA biodegradation. Studies using model compounds (chemically synthesized in the laboratory) and commercial compounds (derived during petroleum fractionation) have provided an insight into the NA metabolic pathways (Rho and Evans, 1975; Taylor and Trudgill, 1978; Clemente et al., 2004; Smith et al., 2008). On the basis of these findings, biodegradation is affected by chemical structure (Whitby, 2010): generally, the more recalcitrant NAs have high molecular weights, contain multiple branched alkyl chains and methyl substituted cycloalkane rings (Smith et al., 2008; Paslawski et al., 2009; Videla et al., 2009). Although methyl groups hinder NA biodegradation (Herman et al., 1993; Smith et al., 2008), mixed bacterial populations can degrade NAs with methyl substitutions on the cycloalkane rings (Herman et al., 1993; Headley et al., 2002a, 2002b; Smith et al., 2008), demonstrating the importance of microbial consortia for complete NA biodegradation.
NA-degrading microorganisms, such as Pseudomonas putida and Pseudomonas fluorescens, have been isolated when grown on model or commercial NAs (Del Rio et al., 2006). However, commercial NAs are more readily degraded than NAs in oil sands and wastewaters, and advanced analytical techniques have shown that they are not representative of the NAs found in the environment (Scott et al., 2005; Han et al., 2008). More recent studies suggest that NAs in the environment have structures consisting of branched alkyl chains, which are less available for microbial biodegradation (Han et al. 2008; Smith et al., 2008).
NA toxicity may also inhibit microbial degradation (Brient et al., 1995; Zou et al., 1997). Although aromatic alkanoic acids make up a small proportion of NA mixtures (<10% in crude oils; Hsu et al., 2000), they are a significant component of the overall toxicity and recalcitrance of NAs found in wastewaters (Headley and McMartin, 2004). For example, Dawson et al. (1996) demonstrated the toxicity of aromatic acids, including benzoic acid and a range of phenylalkanoics, using Xenopus embryos, whereas Zhao et al. (1998) reported the toxicity of a range of benzoic acids to Vibrio fischeri, Daphnia magna and carp. More recently, Thomas et al. (2009) showed a mixture of 2′-, 3′- and 4′-isobutylphenyl-4-pentanoic acids as environmental androgen receptor antagonists. Despite this, there is insufficient information on the metabolism of aromatic alkanoic acids, and the microorganisms involved in their biodegradation are unknown. Thus, reclamation of NA-contaminated environments requires a better understanding of the microorganisms capable of degrading aromatic alkanoic acids with branched alkyl side chains.
Here, we chemically synthesized a series of aromatic alkanoic acids (Smith, 2006) to test the hypothesis that an increase in alkyl chain branching inhibits degradation, leading to a selection of microbes with different catabolic activities. Analysis of metabolites produced has allowed an aromatic alkanoic acid degradation pathway to be proposed.
Materials and methods
Aromatic carboxylic acid synthesis
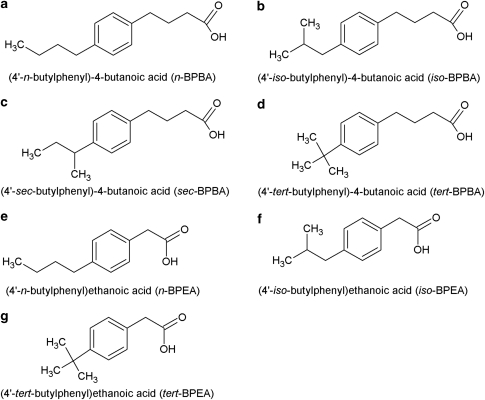
The four butylphenylbutanoic acid (BPBA) isomers used in this study were as follows: (4′-n-butylphenyl)-4-butanoic acid (n-BPBA); (4′-iso-butylphenyl)-4-butanoic acid (iso-BPBA); (4′-sec-butylphenyl)-4-butanoic acid (sec-BPBA) and (4′-tert-butylphenyl)-4-butanoic acid (tert-BPBA) (Figure 1). Three butylphenylethanoic acid (BPEA) isomers were also used in this study: (4′-n-butylphenyl)ethanoic acid (n-BPEA); (4′-iso-butylphenyl)ethanoic acid (iso-BPEA) and (4′-tert-butylphenyl)ethanoic acid (tert-BPEA) (Figure 1). All BPBAs were synthesized using a modified Haworth synthesis (Smith, 2006; Smith et al., 2008). BPEAs were synthesized using the Kindler modification of the Willgerodt reaction (March, 1985).
Figure 1.
The butylphenylbutanoic acid (BPBA) (a–d) and butylphenylethanoic acid (BPEA) (e–g) compounds used in this study.
BPBA-degrading enrichment culture
Coal-tar-contaminated soils (5–20 cm depth) from a former oil refinery in London (UK), near the Thames estuary, were enriched in sterile serum bottles (125 ml) capped with a polytetrafluoroethylene-lined crimp sealed septum and inoculated (1% w/v) in 25 ml of minimal salts medium (MSM), pH 7.0, as described by Smith et al. (2008), and containing 1% (v/v) Venezuelan heavy crude oil (Tia Juana Pesado) as the sole carbon source. Cultures were incubated with shaking (110 r.p.m.) in the dark at 20 °C for 6 months (to select for highly enriched cultures of hydrocarbon degraders) and growth monitored by turbidity. For the BPBA-degradation experiment, the coal tar enrichment culture was centrifuged (3435 × g for 15 min), cell pellets washed three times with MSM to remove trace hydrocarbons from the pre-enrichment with oil and inoculated (2% v/v) in MSM (25 ml) in sterile serum bottles (125 ml). Each BPBA isomer: n-BPBA; iso-BPBA; sec-BPBA and tert-BPBA (10 mg) dissolved in 1 ml of 0.1 NaOH was added (10 μl) individually as sole source of carbon and energy to each serum bottle (final concentration 4 mg l−1), which were capped with a polytetrafluoroethylene-lined crimp sealed septum. Killed controls (to determine whether any abiotic loss had occurred) were prepared by Tyndallization of the inocula before BPBA addition, and viability was checked by growth on nutrient agar. Abiotic controls containing the individual BPBA isomer (10 mg) dissolved in 1 ml of 0.1 NaOH and added to MSM (final concentration 4 mg l−1) were also prepared. Procedural blanks containing either the enrichment culture inoculated (2% v/v) in MSM only or un-inoculated MSM supplemented with 0.1 NaOH (10 μl) were also performed. (Due to the large number of controls performed, n- and iso-BPBA were inoculated first followed by sec-BPBA and tert-BPBA 2 weeks later). Between each inoculation there was a period of 1–2 h before samples were analyzed, due to the time required to process each set of bottles. All bottles (168 in total; that is, 36 for each BPBA including abiotic and killed controls, and 24 procedural blanks) were incubated at 110 r.p.m. in the dark at 20 °C for 49 days. Destructive sampling of triplicate bottles was carried out at 0, 14, 35 and 49 days, whereby cultures were centrifuged at 3435 × g for 15 min. Supernatants were frozen at −20 °C before ethyl acetate extraction. Cell pellets were resuspended in 1 ml sterile MSM. The cell suspension (50 μl) was used to perform total, viable, Gram-positive and Gram-negative cell counts using the ViaGram Red+ staining kit (Molecular Probes, Eugene, OR, USA) according to the manufacturer's instructions. The P-values for mean cell numbers were calculated using the Kruskal–Wallis test. The remainder of the cell suspension was frozen at −20 °C for nucleic acid extraction.
BPBA extraction and gas chromatography mass spectrometry analysis
To remove any hydrocarbon contamination, all glassware was soaked overnight in Decon90 (Decon, Hove, UK), rinsed three times with distilled water, baked until dry at 110 °C, rinsed three times with acetone (Fisher Scientific, Loughborough, UK) and autoclaved. The internal standard, 4-phenylbutanoic acid (Acros Organics, Geele, Belgium) (10 mg) dissolved in 1 ml methanol (high-performance liquid chromatography grade, Fisher Scientific) was added to each supernatant from above (final concentration 2 mg l−1). Each BPBA isomer from the supernatants was extracted by acidifying to pH <2 (using concentrated HCl) and extracted three times with 15 ml high-performance liquid chromatography grade ethyl acetate (Fisher Scientific) as described by Smith et al. (2008). Solvent extracts were pooled, dried with 5–10 g anhydrous Na2SO4 (Fisher Scientific) for >90 min and the organic acids concentrated by rotary evaporation (Büchi, Flawil, Switzerland) at 40 °C. Samples were transferred to a gas chromatography vial (Chromacol, Welwyn Garden City, UK), sparged with nitrogen and reacted with N,O-bis(trimethylsilyl)trifluoroacetamide (Supelco, Bellefonte, PA, USA) at 60 °C for 20 min to form trimethylsilyl derivatives. Derivatized samples were resuspended in 1 ml dichloromethane (high-performance liquid chromatography grade, Acros Organics) and separated by gas chromatography-mass spectrometry using an Agilent 7890 GC interfaced with an Agilent 5975C MS (Agilent, Wokingham, UK). Run conditions were a 1 μl splitless injection (injector temperature of 250 °C) onto a 30 m × 250 μm × 0.25 μm HP-5 MS column. Oven temperatures were increased from 40°C to 300 °C at 10 °C min−1 followed by a 10 min hold at 300 °C. Helium was used as the carrier gas at a flow rate of 1 ml min−1. The transfer line and source were held at 230 °C. The mass spectrophotometer was a quadrupole operated in full-scan mode (scan range 50–550 Da).
Microtox analysis
Toxicity of the four BPBA and three BPEA isomers was measured by Microtox assay (Azur Environmental, Fairfax, CA, USA). Triplicates of each isomer dissolved in 1 ml of 0.1 NaOH were added to reconstituted cells at final concentrations of 33.3, 50 and 75 mg l−1, and 0.1 NaOH (1 ml) was used as a control. Each organic acid was analyzed using the liquid-phase test protocol as described by the manufacturer.
Cloning and phylogenetic analysis
DNA was extracted from the cell resuspension (950 μl) as described by Griffiths et al. (2000) as modified by Nicol et al. (2005). Approximately 1357 bp fragments of bacterial 16S rDNA were obtained by a nested PCR using the primers pA/pH′ and pB/pG′ for the first and second amplifications, respectively (Edwards et al., 1989). PCRs (50 μl) were performed in a Gene Amp PCR system 9700 Thermocycler (Applied Biosystems, Foster City, CA, USA) as follows: 1 × buffer (Qiagen, Crawley, UK), 0.2 m dNTPs (Fermentas, St Leon-Rot, Germany), 0.4 μ each primer, 2.5 U Taq DNA polymerase (Qiagen) and approximately 25 ng of DNA. PCR cycling conditions were as follows: 95 °C for 2 min followed by 28 cycles of 94 °C for 30 s, 57 °C for 30 s and 72 °C for 1.5 min; then 72 °C for 10 min. Cloning of the PCR products was performed using the pGEM T-Easy kit according to the manufacturer's instructions (Promega, Madison, WI, USA). Plasmids were extracted using the Qiagen plasmid extraction kit (Qiagen). Clone libraries were screened by PCR amplification using primers for positions 341–534 in Escherichia coli (Muyzer et al. 1993) and Denaturing Gradient Gel Electrophoresis performed as described previously (McKew et al., 2007), except gels were silver-stained as described by Nicol et al. (2005). Representative clones from each library were selected and sequenced using the primer pG′ (Edwards et al., 1989) (Source Bioscience Ltd., Cambridge, UK). Sequences were aligned with sequences from GenBank using the RDP INFERNAL aligment tool (Michigan State University, MI, USA) (Nawrocki and Eddy, 2007). Phylogenetic analysis was performed using PHYLIP 3.4 (University of Washington, WA, USA) with Jukes-Cantor DNA distance and neighbor-joining methods (Jukes and Cantor, 1969; Saitou and Nei, 1987). Bootstrap analysis was based on 100 replicates using SEQBOOT and CONSENSE (PHYLIP 3.4). Tree construction was performed using Treeview (WIN32) version 1.5.2 (Page, 1996). Sequences were deposited into the GenBank database with the accession numbers GU816174–GU816240.
454 Pyrosequencing
Triplicate enrichments from 0 and 49 days were selected for pyrosequencing. The 16S rRNA gene was amplified with pA/pH′ primers using the primers and cycling conditions described previously (Muyzer et al. 1993), except that the forward primer had no GC-clamp and a 5′ modification with a 454 amplicon adaptor followed by a unique 10-nucleotide barcode (Parameswaran et al., 2007; Acuña Alvarez et al., 2009). PCR products were quantified with a Nanodrop ND-1000 spectrophotometer (Nanodrop, Wilmington, DE, USA) and pooled in equimolar amounts. Approximately 200 ng of pooled sample was analyzed by pyrosequencing by the NERC Biomolecular Analysis Facility. Sequences were separated using the barcodes, and sequences over 150 bp were processed using the Greengenes pyrosequencing pipeline (DeSantis et al., 2006a). Sequences were aligned against the Greengenes 16S rRNA gene database using Nearest Alignment Space Termination (DeSantis et al., 2006b). Parameters used for analysis were 150 bp minimum length. Only sequences over 90% identity were further analyzed. The taxonomic composition was obtained by comparison against the Ribosmomal Database Project using the Greengenes classification tool (DeSantis et al., 2006a). Microbial community composition patterns were evaluated using multidimensional scaling (Clarke and Ainsworth, 1993), applying the Bray-Curtis similarity indices for relative sequence abundances obtained by pyrosequencing.
Results
BPBA degradation and metabolite production
A hydrocarbon-degrading enrichment culture derived from heavy coal-tar-impacted soils provided an enriched pool of microbes putatively capable of BPBA degradation. This inoculum completely metabolized the least branched n-BPBA after 35 days (Figure 2a), whereas transformation of the more branched iso-, sec- and tert-BPBA was much slower (Figures 2b–d). There were no significant abiotic losses (P=0.117) with either the killed or the abiotic controls, and so BPBA transformation can be attributed to microbial activity.
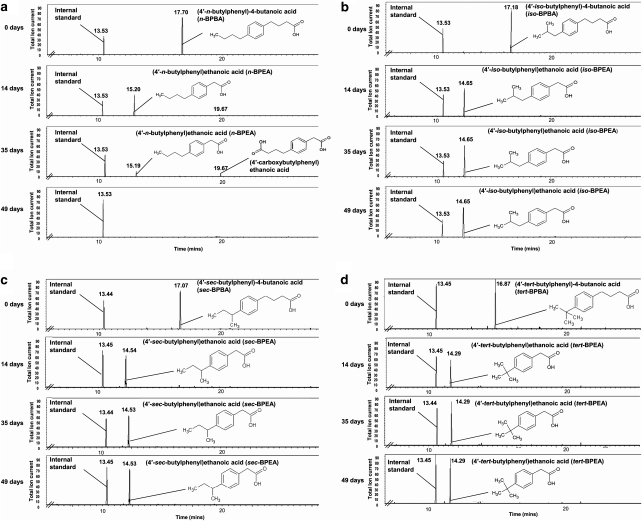
Figure 2.
Gas chromatograms showing the degradation of four BPBA isomers over a 49-day incubation period by an enrichment culture. Enrichments grown on n-BPBA (a), iso-BPBA (b), sec-BPBA (c) or tert-BPBA (d). Clearly visible is the disappearance of the n-BPBA trimethylsilyl (TMS) ester peak (RT: 17.70) and the appearance of a secondary peak at 14 days (RT: 15.20), confirmed as n-BPEA (TMS ester). This is further degraded within 49 days with the transient production of another peak (RT: 19.67), tentatively assigned as the bis-TMS ester of (4′-carboxybutylphenyl)ethanoic acid. Clearly visible is the disappearance of the iso-, sec- and tert-BPBA trimethylsilyl (TMS) ester peak and the appearance of another peak in 14 days, confirmed as iso-, sec- and tert-BPEA (TMS ester) respectively. This is not further transformed during the course of the experiment (49 days).
Degradation of each BPBA isomer resulted in a corresponding increase in the production of a major metabolite after 14 days, which had a molecular mass of 264 and was confirmed as the trimethylsilyl ester of the ethanoic acid equivalent of the butanoic acids (BPEA) (Supplementary Information Figure 1A, C–E). After 35 days, n-BPEA was degraded to a further metabolite with a retention time of 19.67 min and a molecular mass of 366 . This metabolite was identified by mass spectrometry as the bis-trimethylsilyl ester of (4′-carboxybutylphenyl)ethanoic acid, a diacid corresponding to addition of a carboxylic acid group to the alkyl side chain (Figure 2a, Supplementary Information Figure 1B). Complete degradation of n-BPEA and (4′-carboxybutylphenyl)ethanoic acid occurred by 49 days, demonstrating complete degradation of n-BPBA (Figure 2a). However, little further degradation of iso-, sec- and tert-BPEA metabolites occurred after day 14 (Figures 2b–d). Analysis of the gas chromatograms identified a small peak corresponding to sec-BPBA, which had not been degraded by day 14 (Figure 2c). The extent of BPBA transformation correlated overall to the degree of alkyl branching with the least branched n-BPBA being more readily degraded.
Toxicity of the four BPBA isomers and n-, iso- and tert-BPEA metabolites produced herein was determined by the Microtox assay (Table 1), which has been used as the ‘gold standard' for NA toxicity measurements (Frank et al., 2009). The highly branched tert-BPBA and tert-BPEA were the most toxic isomers. It is notable that the ethanoic acid metabolites were consistently (and statistically significantly) less toxic than their butanoic acid parent compounds (with iso-BPEA almost five times less toxic than iso-BPBA).
Table 1. EC50 values (determined by the Microtox assay) for n-, iso-, sec- and tert-BPBA isomers and n-, iso- and tert-BPEA isomers (ND: sec-BPEA was not determined).
| Alkyl substituents |
Alkyl phenyl alkanoic acid |
|
|---|---|---|
| EC50 BPBA (mg l−1)a | EC50 BPEA (mg l−1)a | |
| n- | 18.7±0.65 | 43.0±0.75 |
| iso- | 14.7±0.68 | 69.2±1.04 |
| sec- | 39.6±5.21 | ND |
| tert- | 9.4±0.66 | 25.9±0.71 |
Purity of all the compounds as determined by gas chromatography-mass spectrometry was between 92% and 97% (Smith, 2006).
Mean±95% confidence limits, n=3.
Community and phylogenetic analyses
Cell counts were performed during incubation on each of the BPBA isomers (Supplementary Information Table 1) and were low at day 0 for all the enrichments (between 1.35 × 102 and 8.51 × 102 cells ml−1). No significant changes occurred in cell numbers throughout the enrichment period with cultures grown on sec- (P=0.561) and tert-BPBA (P=0.288). However, for enrichment cultures grown on n-BPBA, cell numbers increased 37-fold from 5.78 × 102 cells ml−1 (at day 0) to 2.14 × 104 cells ml−1 (at day 49) (P=0.027). With enrichments grown on iso-BPBA, a twofold increase in cell numbers was observed from 8.51 × 102 cells ml−1 (at day 0) to 1.66 × 103 cells ml−1 (at day 49) (P=0.03).
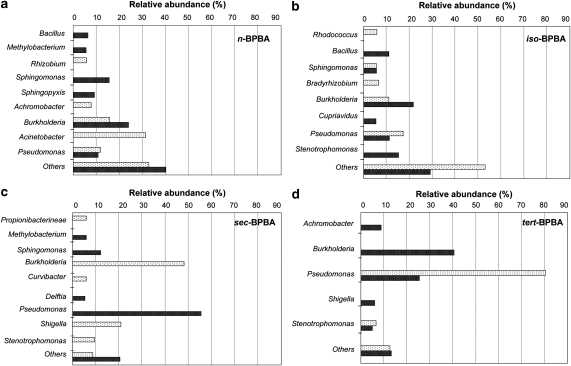
To identify the dominant microbes in the BPBA enrichments, the bacterial communities present at 0 and 49 days were characterized by 454 pyrosequencing of the 16S rRNA genes. Analysis of 15 491 sequences (over 150 bp) revealed that enrichments had a different community at day 49 compared with day 0, and microbial communities differed between enrichments grown on each BPBA isomer (Figure 3, Supplementary Information Table 2). The dominant microorganisms present in n-BPBA enrichments at day 0 were: Acinetobacter (32%), Burkholderia (16%), and Pseudomonas, Achromobater and Rhizobium (each <12% of the community). At day 0, Pseudomonas (81% of the community) dominated tert-BPBA enrichments, whereas Burkholderia (48% of the community) and Shigella (21% of the community) dominated sec-BPBA enrichments. However, at day 0, iso-BPBA enrichments were not dominated by one genus but contained several genera including Pseudomonas (17%) and Burkholderia (11%).
Figure 3.
Bacterial communities in enrichments grown on four BPBA isomers and analyzed by 454 pyrosequencing of the 16S rRNA genes. Bacterial genera comprising >5% of the total community in enrichments grown on n-BPBA (a), iso-BPBA (b), sec-BPBA (c) and tert-BPBA (d). Others: sum of abundances for genera representing <5% of the community composition. Enrichments sampled at 0 day (light bars) and 49 days (dark bars) incubation.
At day 49, after either partial or complete BPBA degradation, the community composition was different. Specifically, there was an increase in the number of Burkholderia sequences found in n-BPBA, iso-BPBA and tert-BPBA enrichments (comprising 24, 22 and 41% of the community, respectively). Conversely, in sec-BPBA enrichments the number of Burkholderia sequences decreased from 48% to 2% (Figure 3c, Supplementary Information Table 2), whereas the number of Pseudomonas sequences increased (from 1% to 56%) (Figure 3c, Supplementary Information Table 2). In contrast, after transformation of tert-BPBA to tert-BPEA there was a large decrease in Pseudomonas sequences (from 81% to 26%) (Figure 3d, Supplementary Information Table 2). The community present in iso-BPBA enrichments after 49 days was not dominated by one genus but remained as diverse as at day 0, with representatives from several genera including Burkholderia, Stenotrophomonas, Pseudomonas and Bacillus (Figure 3b, Supplementary Information Table 2). n-BPBA enrichments at day 49 were also diverse, and in addition to Burkholderia, there were Sphingomonas (16%), and Sphingopyxis, Methylobacterium and Bacillus (each comprising <10% of the community) (Figure 3a, Supplementary Information Table 2). The sec-BPBA enrichments at day 49 had several less dominant members, including Sphingomonas (12%) (Figure 3c, Supplementary Information Table 2). Microbial community composition was evaluated using multidimensional scaling using the Bray-Curtis similarity indices for relative sequence abundances obtained by pyrosequencing (Supplementary Information Figure 2). Communities become more similar during growth on n- and iso-BPBA by day 49, compared with sec- and tert-BPBA (stress factor 0.01).
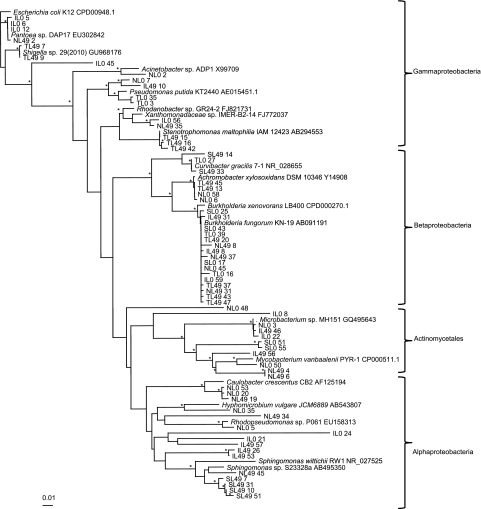
In order to obtain almost full-length 16S rRNA gene sequences for phylogenetic analysis of the microorganisms present after BPBA degradation; clone libraries were generated from 0- and 49-day enrichments (totaling 366 clones). Libraries were screened by Denaturing Gradient Gel Electrophoresis, and 67 clones (distributed across samples) were selected, sequenced (>500 bp) and used to construct a phylogenetic tree (Figure 4). Although there was no unequivocal clustering of clones, based on either incubation time or substrate, generally, there was good agreement between the sequences recovered by the clone libraries and those retrieved by 454 pyrosequencing. Clone libraries were dominated by 16S rRNA gene sequences relating to Burkholderia, with representatives from n-, iso- and tert-BPBA libraries at day 49 and day 0, and sec-BPBA at day 0. No sequences relating to Burkholderia were found in clone libraries from sec-BPBA at day 49, corroborating the decrease in Burkholderia sequences identified in sec-BPBA enrichments (from 48% at day 0 to <2% at day 49) by pyrosequencing. In addition to Burkholderia, 16S rRNA gene sequences relating to Sphingomonas spp. were also found in n- and sec-BPBA clone libraries at day 49, further supporting the data obtained by pyrosequencing (Figure 4). Although sequences recovered by clone libraries generally corroborated those identified by pyrosequencing, some differences were observed. For example, Mycobacterium species were over-represented in the clone libraries (that is, n-, iso-BPBA at days 0 and 49, and sec-BPBA at day 0) (Figure 4) compared with pyrosequencing. Conversely, Pseudomonas was under-represented in the clone libraries (that is, n-, tert-BPBA at day 0, and iso-BPBA at day 49) (Figure 4).
Figure 4.
Phylogenetic analysis of the 16S rRNA gene sequences from selected clones. Included are type strains obtained from GenBank. Sequence analysis was performed on common partial sequences (>500 bp) using Jukes–Cantor DNA distance and neighbor-joining methods. Bootstrap values represent percentages from 100 replicates of the data and percentages >80% are shown by *. The scale bars indicates 0.01 substitutions per nucleotide base.
Four clone sequences derived from iso-BPBA were related to Alphaproteobacteria. Three of these clone sequences were from the iso-BPBA enrichment at day 49 (IL49 57, IL49 26 and IL49 53; Figure 4). Clone IL49 57 clustered with clone IL0 21 (from iso-BPBA at day 0) and had 97% sequence identity to an uncultured bacterium. The closest named representative to IL49 57 was from the order Rhizobiales within the Alphaproteobacteria with 90% sequence similarity.
Discussion
We have demonstrated differential degradation of alkyl branched aromatic alkanoic acids, identified metabolic intermediates and characterized changes in the microbial communities. We obtained an enrichment culture with the ability to metabolize four BPBA isomers and found that the least branched n-BPBA was completely degraded by day 49, whereas the more branched iso-, sec- and tert-BPBA isomers were only metabolized as far as their respective ethanoic acids by 49 days. Thus, an increase in alkyl chain branching reduced the biotransformation of BPBA. Smith et al. (2008) found that the ethanoic acids derived from butylcyclohexylbutanoic acid degradation were also persistent (up to 30 days). Our results suggest that the more refractory NAs found in oil sand wastewaters may include branched alkyl phenylethanoic acids and is consistent with the findings of Smith and Rowland (2008). Thus, information on the effects of alkyl branching on biodegradation rates is important if the removal of refractory NAs in wastewaters is to be achieved.
The highly branched tert-BPBA was more toxic than n-, iso- and sec-BPBA and a general detoxification occurred during BPBA degradation. Although the ethanoic acid metabolites produced during BPBA degradation were less toxic than the parent compounds, they were nonetheless toxic (EC50 26–69 mg l−1), and unless there are microorganisms capable of oxidizing such compounds they pose an environmental risk with the potential to accumulate over time.
Previous studies have shown that NA biodegradation rates may be influenced by chemical structure and molecular weight (Herman et al., 1993; Scott et al., 2005; Biryukova et al., 2007; Smith et al., 2008). For non-branched carboxylic acids, the addition of methyl groups hindered β-oxidation (Herman et al., 1993; Alexander, 1999; Smith et al., 2008) and only mixed bacterial populations degraded recalcitrant NAs with methyl substitutions on the cycloalkane rings (Herman et al., 1993; Headley et al., 2002a, 2002b). Differences in NA degradation rates for different geometric isomers have also been observed, whereby intramolecular hydrogen bonding occurring with the cis-isomers makes them less bioavailable and difficult to metabolize (Headley et al., 2002b).
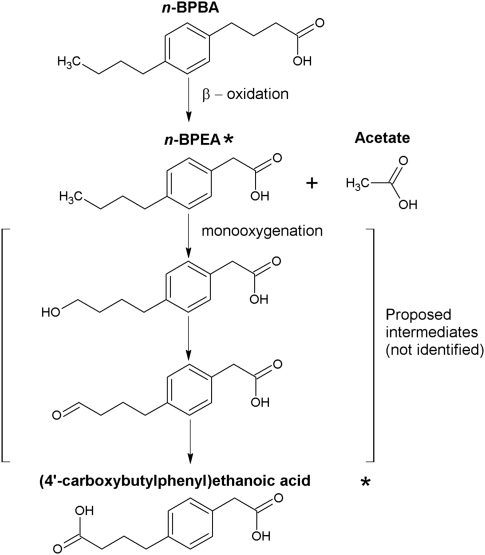
To date, there is no information on the pathways associated with aromatic alkanoic acid degradation. However, pathways have been proposed for the biodegradation of aliphatic and alicyclic carboxylic acids and include β-oxidation (Blakley, 1978; Blakley and Papish, 1982; Alexander, 1999; Quagraine et al., 2005a), combined α- and β-oxidation (Beam and Perry, 1974; Rontani and Bonin, 1992) and aromatization (Blakley, 1974; Taylor and Trudgill, 1978; Trudgill, 1984). In this work, we sought to elucidate the initial steps involved in aromatic alkanoic acid metabolism. Identification of two intermediate metabolites produced during n-BPBA transformation (that is, n-BPEA and (4′-carboxybutylphenyl)ethanoic acid) confirms that the initial degradation steps involve the removal of two carbons from the carboxyl side chain, indicative of β-oxidation. Identification of the n-alkyl diacid indicates carboxylation of the alkyl side chain had occurred. A putative BPBA degradation pathway whereby further oxidation of the alkyl side chain and cleavage of the ring is proposed (Figure 5).
Figure 5.
Proposed n-BPBA degradation pathway by an enrichment culture. n-BPEA and (4′-carboxybutylphenyl)ethanoic acid metabolites were confirmed by mass spectra analysis and indicated by an asterisk.
Several microorganisms including: Acinetobacter anitratum, Alcaligenes faecalis and Pseudomonas putida have been shown to metabolize model NAs by aerobic β-oxidation pathways (Blakley, 1974; Rho and Evans, 1975; Blakley and Papish, 1982). Alcaligenes sp. is also able to use combined α- and β-oxidation pathways to metabolize cyclohexylacetic acid (Rontani and Bonin, 1992). This ability to utilize both α- and β-oxidation pathways may be useful in the bioremediation of more recalcitrant NAs.
In our study, 16S rRNA gene sequence analysis by clone libraries and pyrosequencing identified the microbial communities present during BPBA degradation. At day 0, the differences in communities from n- and iso-BPBA enrichments compared with sec- and tert-BPBA enrichments may be attributed to either differences in the initial inoculum (owing to inoculations at different times) or differential toxic effects observed following immediate inoculation. In the case of n-BPBA enrichments where complete mineralization of n-BPBA occurred by day 49 (and resulted in a 37-fold increase in cell numbers), it may be assumed that an increase in abundance of 16S rDNA sequences by day 49 is indicative of the involvement of a microorganism in n-BPBA degradation (especially from the most abundant sequences). Here, we found a predominance of Burkholderia and Sphingomonas sequences from n-BPBA enrichments after day 49.
Previous NA-degradation studies showed that P. putida and P. fluorescens degraded cyclohexanecarboxylic acids (Blakley and Papish, 1982; Del Rio et al., 2006). We also recovered a large proportion of sequences relating to Pseudomonas (especially in sec-BPBA enrichments). Interestingly, following sec- and tert-BPBA degradation, there was an inverse relationship between the proportion of Pseudomonas and Burkholderia sequences within the community. During sec-BPBA degradation, the percentage of Pseudomonas sequences increased with a concomitant decrease in the percentage of Bukholderia sequences. Conversely, during tert-BPBA degradation, the percentage of Pseudomonas sequences decreased with a concomitant increase in Bukholderia sequences. However, in the case of iso-, sec- and tert-BPBA enrichments where complete degradation did not occur and cell numbers remained low, changes in community composition may not be conclusively attributed to growth on BPBA.
In addition to the dominance of Pseudomonas and Burkholderia at day 49, a large number of other sequences were identified (by either cloning and/or pyrosequencing) and related to Bacillus, Mycobacterium, Microbacterium, Sphingomonas and Stenotrophomonas. To date, there are no reports of any of these microorganisms being involved in NA-degradation. Achromobacter spp., which have been shown to degrade cyclohexanecarboxylic acid and carboxylated cycloalkane by others (Blakley, 1974, 1978), were in low abundance in our study. Similarly, Corynebacterium cyclohexanicum (Tokuyama and Kaneda, 1973) and Acinetobacter spp. (Rho and Evans, 1975; Herman et al., 1993) have also been shown to degrade cyclohexanecarboxylic acid and carboxylated cycloalkanes. In our study, no sequences relating to Corynebacterium spp. were recovered from any of the enrichments, suggesting that these microorganisms may have been absent or present in low abundance. Furthermore, Acinetobacter sequences were only present in n-BPBA enrichments (at day 0), suggesting that Acinetobacter spp. were either unable to degrade the branched aromatic alkanoic acids, or were susceptible to the toxicity of the BPBAs. Multidimensional scaling analysis of the pyrosequencing data on day 0 and 49 provides strong support that specific NA-degrading microorganisms were selected by growth on n- and iso-BPBA.
Although high numbers of microorganisms (106–108 cells ml−1) have been found in tailings pond wastewaters (Foght et al., 1985), cell counts obtained here at day 49 were low (<104 cells ml−1) and may be attributed to several factors, such as carbon limitation (of the parent and/or subsequent metabolites) or toxicity (of the parent compound and/or subsequent metabolites). Under starvation conditions, the carbon content per cell (Cc) is 26 fg C (Troussellier et al., 1997). Although Cc is species dependent (Troussellier et al., 1997), each of the enrichments in our study contained 3.1 mg l−1 of carbon on day 0, theoretically supporting a maximum of 1.17 × 1011 cells l−1, suggesting that initial enrichments were not carbon limited. However, it is possible that the bioavailable carbon at day 49 was insufficient to support growth. Enrichment cultures may also have been limited with respect to other inorganic substrates absent in the medium. Furthermore, it is also possible that the microorganisms were susceptible to the toxicity of either the BPBAs and/or metabolites produced during BPBA degradation. Indeed, the highly branched tert-BPBA (and tert-BPEA), which was the most toxic, had the lowest cell counts after day 49.
In conclusion, we demonstrated aromatic alkanoic acid degradation and identified the metabolites produced, thus allowing a BPBA degradation pathway to be hypothesized. During BPBA degradation it is possible that substrate availability (related to chemical structure) may select for specific microorganisms. We identified key microorganisms, such as Burkholderia, Pseudomonas and Sphingomonas spp., which may be involved in aromatic alkanoic acid transformations. Such information on aromatic NA biodegradation mechanisms and the microorganisms involved may facilitate a more rapid removal of NAs in oil sand wastewaters and obviate the accumulation of recalcitrant metabolites in the environment.
Acknowledgments
We thank Frederic Coulon for assistance with Microtox and the provision of soil samples. This work was supported by an NERC CASE studentship with Oil Plus Ltd (REF: NER/S/A/2006/14134) and the University of Essex. We thank Garth A Jones for useful comments on the paper.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acuña Alvarez L, Exton DA, Timmis KN, Suggett DJ, McGenity TJ. Characterization of marine isoprene-degrading communities. Environ Microbiol. 2009;11:3280–3291. doi: 10.1111/j.1462-2920.2009.02069.x. [DOI] [PubMed] [Google Scholar]
- Alexander M. Biodegradation and Bioremediation. Academic Press: London; 1999. [Google Scholar]
- Beam HW, Perry JJ. Microbial degradation and assimilation of n-alkyl-substituted cycloparaffins. J Bacteriol. 1974;118:394–399. doi: 10.1128/jb.118.2.394-399.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukova OV, Fedorak PM, Quideau SA. Biodegradation of naphthenic acids by rhizosphere microorganisms. Chemosphere. 2007;67:2058–2064. doi: 10.1016/j.chemosphere.2006.11.063. [DOI] [PubMed] [Google Scholar]
- Blakley ER. The microbial degradation of cyclohexane carboxylic acid:a pathway involving aromatization to form p-hydroxybenzoic acid. Can J Microbiol. 1974;20:1297–1306. [Google Scholar]
- Blakley ER. The microbial degradation of cyclohexanecarboxylic acid by a beta-oxidation pathway with simultaneous induction to the utilization of benzoate. Can J Microbiol. 1978;24:847–855. doi: 10.1139/m78-141. [DOI] [PubMed] [Google Scholar]
- Blakley E, Papish B. The metabolism of cyclohexanecarboxylic acid and 3-cyclohexenecarboxylic acid by Pseudomonas putida. Can J Microbiol. 1982;28:1324–1329. doi: 10.1139/m82-198. [DOI] [PubMed] [Google Scholar]
- Brient J, Wessner P, Doly M.1995Naphthenic AcidsIn: Kroschwitz, J (ed).Encyclopedia of Chemical Technology John Wiley and Sons: New York; 1017–1029. [Google Scholar]
- Clarke KR, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser. 1993;92:205–219. [Google Scholar]
- Clemente JS, MacKinnon MD, Fedorak PM. Aerobic biodegradation of two commercial naphthenic acids preparations. Environ Sci Technol. 2004;38:1009–1016. doi: 10.1021/es030543j. [DOI] [PubMed] [Google Scholar]
- Dawson DA, Schultz TW, Hunter RS. Developmental toxicity of carboxylic acids to Xenopus embryos: a quantitative structure-activity relationship and computer-automated structure evaluation. Teratogen, Carcinogen Mutagen. 1996;16:109–124. doi: 10.1002/(SICI)1520-6866(1996)16:2<109::AID-TCM5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Del Rio L, Hadwin A, Pinto L, MacKinnon M, Moore M. Degradation of naphthenic acids by sediment micro-organisms. J Appl Microbiol. 2006;101:1049–1061. doi: 10.1111/j.1365-2672.2006.03005.x. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006a;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucl Acids Res. 2006b;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokholyan VK, Magomedov AK. Effects of sodium naphthenate on survival and some physiological-biochemical parameers of some fishes. J Ichthyol. 1984;23:125–132. [Google Scholar]
- Edwards U, Rogall T, Bloecker H, Emde M, Boettger E. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7851. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foght JM, Fedorak PM, Westlake DWS, Boerger HJ. Microbial content and metabolic activities in the Syncrude tailings pond. AOSTRA J Res. 1985;1:139–146. [Google Scholar]
- Frank RA, Fischer K, Kavanagh R, Burnison BK, Arsenault G, Headley JV. Effect of carboxylic acid content on the acute toxicity of oil sands naphthenic acids. Environ Sci Technol. 2009;43:266–271. doi: 10.1021/es8021057. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microb. 2000;66:5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwin A, Del Rio L, Pinto L, Painter M, Routledge R, Moore M. Microbial communities in wetlands of the Athabasca oil sands: genetic and metabolic characterisation. FEMS Microbiol Ecol. 2006;55:68–78. doi: 10.1111/j.1574-6941.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- Han X, Scott AC, Fedorak PM, Bataineh M, Martin JW. Influence of molecular structure on the biodegradability of naphthenic acids. Environ Sci Technol. 2008;42:1290–1295. doi: 10.1021/es702220c. [DOI] [PubMed] [Google Scholar]
- Headley JV, McMartin DW. A Review of the occurrence and fate of naphthenic acids in aquatic environments. J Environ Sci Health. 2004;39:1989–2010. doi: 10.1081/ese-120039370. [DOI] [PubMed] [Google Scholar]
- Headley JV, Peru KM, McMartin DW, Winkler M. Determination of dissolved naphthenic acids in natural waters by using negative-ion electrospray mass spectrometry. J AOAC Int. 2002a;85:182–187. [PubMed] [Google Scholar]
- Headley J, Tanapat S, Putz G, Peru K. Biodegradation kinetics of geometric isomers of model naphthenic acids in Athabasca River water. Can Water Resour J. 2002b;27:25–42. [Google Scholar]
- Herman D, Fedorak P, Costerton J. Biodegradation of cycloalkane carboxylic acids in oil sand tailings. Can J Microbiol. 1993;39:576–580. doi: 10.1139/m93-083. [DOI] [PubMed] [Google Scholar]
- Hsu CS, Dechert GJ, Robbins WK, Fukuda EK. Naphthenic acids in crude oils characterized by mass spectrometry. Energy Fuels. 2000;14:217–223. [Google Scholar]
- Jukes TH, Cantor CR. Academic Press: New York; 1969. Mammalian protein metabolism. [Google Scholar]
- March J.1985Advanced organic chemistry Reactions, Mechanisms and Structure3rd Edn,Wiley-Interscience: New York; 1346 [Google Scholar]
- McKew BA, Coulon F, Osborn AM, Timmis KN, McGenity TJ. Determining the identity and roles of oil-metabolizing marine bacteria from the Thames estuary UK. Environ Microbiol. 2007;9:165–176. doi: 10.1111/j.1462-2920.2006.01125.x. [DOI] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. Query-dependent banding (QDB) for faster RNA similarity searches. PLoS Comput Biol. 2007;3:e56. doi: 10.1371/journal.pcbi.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol GW, Tscherko D, Embley TM, Prosser JI. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol. 2005;7:337–347. doi: 10.1111/j.1462-2920.2005.00698.x. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paslawski JC, Headley JV, Hill GA, Nemati M. Biodegradation kinetics of trans-4-methyl-1-cyclohexane carboxylic acid. Biodegradation. 2009;20:125–133. doi: 10.1007/s10532-008-9206-2. [DOI] [PubMed] [Google Scholar]
- Quagraine E, Headley J, Peterson H. Is biodegradation of bitumen a source of recalcitrant naphthenic acid mixtures in oil sands tailing pond waters. J Environ Sci Heal A. 2005a;40:671–684. doi: 10.1081/ese-200046637. [DOI] [PubMed] [Google Scholar]
- Quagraine E, Peterson H, Headley J. In situ bioremediation of naphthenic acids contaminated tailing pond waters in the Athabasca oil sands region- demonstrated field studies and plausible options: a review. J Environ Sci Health A. 2005b;40:685–722. doi: 10.1081/ese-200046649. [DOI] [PubMed] [Google Scholar]
- Rho E, Evans W. The aerobic metabolism of cyclohexanecarboxylic acid by Acinetobacter anitratum. Biochem J. 1975;148:11–15. doi: 10.1042/bj1480011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontani JF, Bonin P. Utilization of n-alkyl-substituted cyclohexanes by a marine Alcaligenes. Chemosphere. 1992;24:1441–1446. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scott AC, MacKinnon MD, Fedorak PM. Naphthenic acids in Athabasca oil sands tailings waters are less biodegradable than commercial naphthenic acids. Environ Sci Technol. 2005;39:8388–8394. doi: 10.1021/es051003k. [DOI] [PubMed] [Google Scholar]
- Smith BE. Naphthenic acids: synthesis characterisation and factors influencing environmental fate [PhD Thesis]. Plymouth (UK) The University of Plymouth. 2006. 2006.
- Smith BE, Lewis CA, Belt ST, Whitby C, Rowland SJ. Effects of alkyl chain branching on the biotransformation of naphthenic acids. Environ Sci Technol. 2008;42:9323–9328. doi: 10.1021/es801922p. [DOI] [PubMed] [Google Scholar]
- Smith BE, Rowland SJ. A derivatisation and liquid chromatography/electrospray ionisation multistage mass spectrometry method for the characterisation of naphthenic acids. Rapid Comm in Mass Spect. 2008;22:3909–3927. doi: 10.1002/rcm.3806. [DOI] [PubMed] [Google Scholar]
- Taylor DG, Trudgill PW. Metabolism of cyclohexane carboxylic acid by Alcaligenes strain W1. J Bacteriol. 1978;134:401–411. doi: 10.1128/jb.134.2.401-411.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KV, Langford K, Petyersen K, Smith AJ, Tollefsen KE. Effect-directed identification of naphthenic acids as important in vitro xeno-estrogens and anti-androgens in North Sea offshore produced water discharges. Environ Sci Technol. 2009;43:8066–8071. doi: 10.1021/es9014212. [DOI] [PubMed] [Google Scholar]
- Tokuyama T, Kaneda T. Corynebacterium cyclohexanicum n. sp.: a cyclohexanecarboxylic acid utilizing bacterium. Can J Microbiol. 1973;19:937–942. doi: 10.1139/m73-150. [DOI] [PubMed] [Google Scholar]
- Troussellier M, Bouvy M, Courties C, Dupuy C. Variation of carbon content among bacterial species under starvation condition. Aquat Microb Ecol. 1997;13:113–119. [Google Scholar]
- Trudgill PW. Microbial Degradation of Organic Compounds. Marcel Dekker: New York; 1984. Microbial degradation of the alicyclic ring; pp. 131–180. [Google Scholar]
- Videla PP, Farwell AJ, Butler BJ, Dixon DG. Examining the microbial degradation of naphthenic acids using stable isotope analysis of carbon and nitrogen. Water Air Soil Poll. 2009;197:107–119. [Google Scholar]
- Whitby C. Microbial naphthenic acid degradation. Adv Appl Microbiol. 2010;70:93–125. doi: 10.1016/S0065-2164(10)70003-4. [DOI] [PubMed] [Google Scholar]
- Young RF, Wismer WV, Fedorak PM. Estimating naphthenic acids concentrations in laboratory-exposed fish and in fish from the wild. Chemosphere. 2008;73:498–505. doi: 10.1016/j.chemosphere.2008.06.040. [DOI] [PubMed] [Google Scholar]
- Zhao YH, Ji GD, Cronin MTD, Dearden JC. QSAR study of the toxicity of benzoic acids to Vibrio fischeri, Daphnia magna and carp. Sci Total Environ. 1998;216:205–215. doi: 10.1016/s0048-9697(98)00157-0. [DOI] [PubMed] [Google Scholar]
- Zou L, Han B, Yan H, Kasperski KL, Xu Y, Hepler LG. Enthalpy of adsorption and isotherms for adsorption of naphthenic acid onto clays. J Colloid Interf Sci. 1997;190:472–475. doi: 10.1006/jcis.1997.4898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.