Abstract
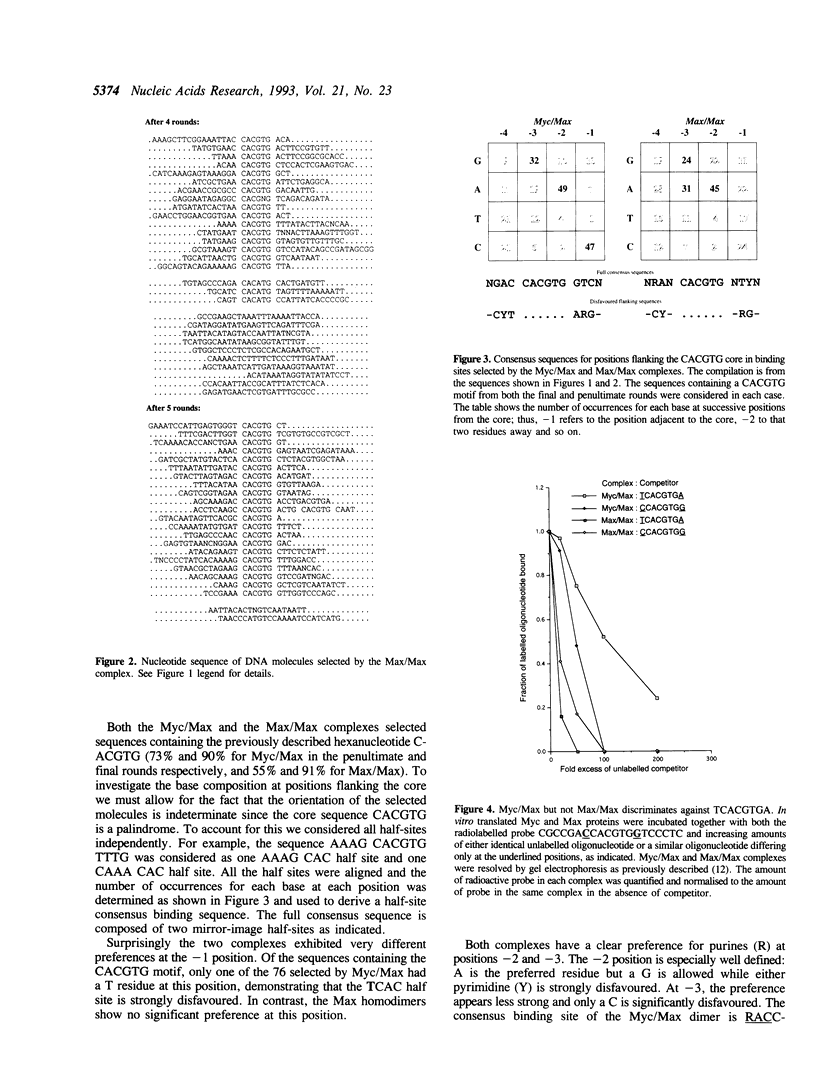
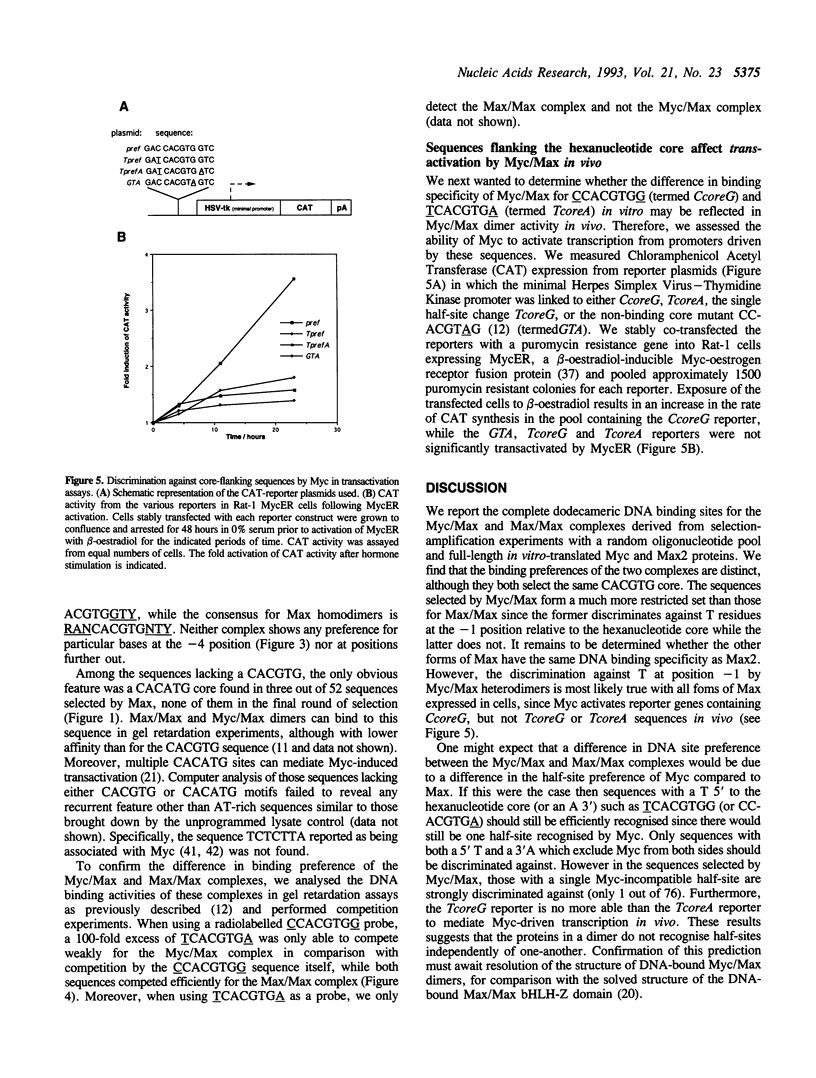
The transcription factor c-Myc and its dimerisation partner Max are members of the basic/helix-loop-helix/leucine-zipper (bHLH-Z) family and bind to the DNA core sequence CACGTG. Using a site-selection protocol, we determined the complete 12 base pair consensus binding sites of c-Myc/Max (RACCACGTGGTY) and Max/Max (RANCACGTGNTY) dimers. We find that the c-Myc/Max dimer fails to bind the core when it is flanked by a 5'T or a 3'A, while the Max/Max dimer readily binds such sequences. Furthermore we show that inappropriate flanking sequences preclude transactivation by c-Myc in vivo. In conclusion, Max/Max dimers are less discriminatory than c-Myc/Max and may regulate other genes in addition to c-Myc/Max targets.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993 Jan 29;72(2):233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Amati B., Dalton S., Brooks M. W., Littlewood T. D., Evan G. I., Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992 Oct 1;359(6394):423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- Amin C., Wagner A. J., Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993 Jan;13(1):383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew D. S., Ashmun R. A., Simmons B. C., Cleveland J. L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991 Oct;6(10):1915–1922. [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993 Jan 29;72(2):211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Benvenisty N., Leder A., Kuo A., Leder P. An embryonically expressed gene is a target for c-Myc regulation via the c-Myc-binding sequence. Genes Dev. 1992 Dec;6(12B):2513–2523. doi: 10.1101/gad.6.12b.2513. [DOI] [PubMed] [Google Scholar]
- Berberich S., Hyde-DeRuyscher N., Espenshade P., Cole M. max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene. 1992 Apr;7(4):775–779. [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Kretzner L., Eisenman R. N. Myc and Max function as a nucleoprotein complex. Curr Opin Genet Dev. 1992 Apr;2(2):227–235. doi: 10.1016/s0959-437x(05)80278-3. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Crouch D. H., Fisher F., Clark W., Jayaraman P. S., Goding C. R., Gillespie D. A. Gene-regulatory properties of Myc helix-loop-helix/leucine zipper mutants: Max-dependent DNA binding and transcriptional activation in yeast correlates with transforming capacity. Oncogene. 1993 Jul;8(7):1849–1855. [PubMed] [Google Scholar]
- Davis L. J., Halazonetis T. D. Both the helix-loop-helix and the leucine zipper motifs of c-Myc contribute to its dimerization specificity with Max. Oncogene. 1993 Jan;8(1):125–132. [PubMed] [Google Scholar]
- Eilers M., Picard D., Yamamoto K. R., Bishop J. M. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989 Jul 6;340(6228):66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992 Apr 3;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fisher F., Goding C. R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992 Nov;11(11):4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher F., Jayaraman P. S., Goding C. R. C-myc and the yeast transcription factor PHO4 share a common CACGTG-binding motif. Oncogene. 1991 Jul;6(7):1099–1104. [PubMed] [Google Scholar]
- Gu W., Cechova K., Tassi V., Dalla-Favera R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Okazaki T., Itani T., Ogata M., Sato Y., Ariga H. An initiation site of DNA replication with transcriptional enhancer activity present upstream of the c-myc gene. EMBO J. 1988 Oct;7(10):3135–3142. doi: 10.1002/j.1460-2075.1988.tb03180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992 Oct 1;359(6394):426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Littlewood T. D., Amati B., Land H., Evan G. I. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene. 1992 Sep;7(9):1783–1792. [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Morgenstern J. P., Land H. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990 Feb 25;18(4):1068–1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B., Morgenbesser S. D., DePinho R. A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992 Aug;6(8):1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- Mäkelä T. P., Koskinen P. J., Västrik I., Alitalo K. Alternative forms of Max as enhancers or suppressors of Myc-ras cotransformation. Science. 1992 Apr 17;256(5055):373–377. doi: 10.1126/science.256.5055.373. [DOI] [PubMed] [Google Scholar]
- Negishi Y., Iguchi-Ariga S. M., Ariga H. Protein complexes bearing myc-like antigenicity recognize two distinct DNA sequences. Oncogene. 1992 Mar;7(3):543–548. [PubMed] [Google Scholar]
- Papoulas O., Williams N. G., Kingston R. E. DNA binding activities of c-Myc purified from eukaryotic cells. J Biol Chem. 1992 May 25;267(15):10470–10480. [PubMed] [Google Scholar]
- Penn L. J., Laufer E. M., Land H. C-MYC: evidence for multiple regulatory functions. Semin Cancer Biol. 1990 Feb;1(1):69–80. [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B. Biphasic effect of Max on Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions. Genes Dev. 1992 Dec;6(12A):2429–2439. doi: 10.1101/gad.6.12a.2429. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Reddy C. D., Dasgupta P., Saikumar P., Dudek H., Rauscher F. J., 3rd, Reddy E. P. Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation. Oncogene. 1992 Oct;7(10):2085–2092. [PubMed] [Google Scholar]
- Reisman D., Elkind N. B., Roy B., Beamon J., Rotter V. c-Myc trans-activates the p53 promoter through a required downstream CACGTG motif. Cell Growth Differ. 1993 Feb;4(2):57–65. [PubMed] [Google Scholar]
- Seed B., Sheen J. Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988 Jul 30;67(2):271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R., Schreiber-Agus N., Morgenbesser S. D., DePinho R. A. Myc and Max: a putative transcriptional complex in search of a cellular target. Curr Opin Cell Biol. 1992 Jun;4(3):468–474. doi: 10.1016/0955-0674(92)90013-3. [DOI] [PubMed] [Google Scholar]
- Västrik I., Koskinen P. J., Alitalo R., Mäkelä T. P. Alternative mRNA forms and open reading frames of the max gene. Oncogene. 1993 Feb;8(2):503–507. [PubMed] [Google Scholar]
- Zervos A. S., Gyuris J., Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993 Jan 29;72(2):223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]



