Abstract
Complex microbial populations are organized in relation to their environment. In the intestine, the inner lining (mucosa) is a potential focal point for such organization. The proximal murine colon contains mucosal folds that are known to be associated with morphologically distinct microbes. To identify these microbes, we used the technique of laser capture microdissection (LCM) to sample microbes associated with these folds (interfold region) and within the central lumen (digesta region). Using 16S rRNA gene tag pyrosequencing, we found that microbes in the interfold region were highly enriched for the phylum Firmicutes and, more specifically, for the families Lachnospiraceae and Ruminococcaceae. Other families such as Bacteroidaceae, Enterococcaceae and Lactobacillaceae were all enriched in the digesta region. This high-resolution system to capture and examine spatial organization of intestinal microbes should facilitate microbial analysis in other mouse models, furthering our understanding of host–microbial interactions.
Keywords: intestine, Lachnospiraceae, microbiota, mucosa associated, pyrosequencing, spatial organization
Introduction
The intestinal mucosa is a complex and dynamic system that functions as a semipermeable barrier allowing the absorption of nutrients and macromolecules required for growth and development, while protecting the blood stream from potentially invasive microorganisms (Walker et al., 1975a, 1975b; Walker 1979). These basic functions are carried out in an environment inhabited by billions of commensal microbes from the three domains of life, Bacteria, Archaea and Eukarya (Eckburg et al., 2005; Gill et al., 2006; Scanlan and Marchesi, 2008), as well as by viral particles (Zhang et al., 2006; Breitbart et al., 2008).
Advances in molecular techniques have improved our understanding of intestinal microbiota. Comprehensive 16S rRNA gene clone libraries derived from biopsies, luminal contents and feces have shown that the intestinal microbiota of mouse and humans consists of hundreds of different phylogenic species that can be classified into only four major microbial phyla: Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria, comprising 98% of the intestinal microbiota (Backhed et al., 2005; Eckburg et al., 2005; Ley et al., 2005, 2006; Gill et al., 2006; Frank et al., 2007; Rajilic-Stojanovic et al., 2007; Tap et al., 2009).
Microbes are not randomly distributed in the environment. Microbial ecology studies (that is, soil environment) have shown that microbes exhibit spatially predictable and aggregated patterns from local to regional scales (Green et al., 2004). In the intestine, one potential organizing factor is the mucosa, which forms a direct interface with the lumen. Interestingly, morphological studies of the intestinal lumen have shown that distinctive microbial populations are found near the mucosa of mice and man (Dubos et al., 1965; Savage et al., 1968, 1971; Savage, 1970; Savage and Blumershine, 1974). Later studies of 16S rRNA gene profiles showed that microbial populations associated with the intestinal mucosa are distinct from those found in fecal samples (Eckburg et al., 2005; Zoetendal et al., 2006). However, a typical limitation of these studies is that the overall structure of luminal microbes is disturbed during sample collection, thus potentially limiting the complete characterization of microbes that inhabit this location.
We used a combination of techniques that permitted the capture of intestinal microbes from specified regions of the mouse colonic lumen. First, highly penetrant organic fixatives were used to maintain the structure of the luminal contents throughout the entire sample preparation (Swidsinski et al., 2005). Second, laser capture microdissection (LCM) was used to specifically sample microbes that were either located near or distant to the mucosal surface. To best demonstrate the difference between these two populations, we focused on the ascending colon that is known to contain a morphologically distinct population of unidentified, large fusiform bacteria (Dubos et al., 1965; Savage et al., 1968, 1971; Savage, 1970; Savage and Blumershine, 1974). We found that these microbes were concentrated in areas between the mucosal folds (referred to herein as the interfold region). We also found that the corresponding region of the central lumen (referred to herein as digesta region) of the mouse ascending colon did not contain an obvious population of these fusiform bacteria. We collected microbial populations from these two regions by LCM, and characterized them by tag pyrosequencing and Sanger sequencing of 16S rRNA genes. This screen showed that Firmicutes were enriched in the interfold region, with Lachnospiraceae and Ruminococcaceae being the predominant families. This finding was confirmed by quantitative PCR (qPCR) and fingerprinting methods, showing no significant variation among mice. We propose that this technique will be useful in examining the microbial spatial organization in other intestinal locations in genetically and environmentally defined mouse strains.
Materials and methods
Mice
All animal experiments were conducted in accordance with approved protocols from the Washington University School of Medicine Animal Studies Committee. Adult (8–12 weeks old) wild-type C57/Bl6 mice were housed in microisolator cages in a specified pathogen-free barrier facility following a 12-h light cycle and fed a standard irradiated chow diet (PicoLab Rodent Chow 20, Purina Mills, St Louis, MO, USA) and water ad libitum. A detailed description of the experimental design is shown in Supplementary Figure 1.
Tissue sampled
For all analyses, entire colons (from the ascending colon to the ano–rectal junction) were removed and immediately fixed by submersion in methacarn (60% methanol, 30% chloroform and 10% acetic acid; Uneyama et al., 2002) for 15 min at 24 °C. Methacarn fixation fixative is an effective and conventional method to preserve the structure of the luminal contents and to examine the spatial organization of the intestinal lumens, including microbes and mucins (Matsuo et al., 1997; Kikuchi et al., 2005; Swidsinski et al., 2005, 2007b). The fixed colons, including their luminal contents, were cut into serial 3-mm cross-sections using a standard No. 22 surgical blade. The resulting sections were aligned and embedded in 2% agar. After routine processing and paraffin embedment, 5–10 μm sections were cut so that the resulting sections contained full cross-sections of the colon sampled along its entire length.
Laser capture microdissection
Deparaffinized 10 μm sections (xylene: one wash for 10 min and two subsequent washes of 5 min each; followed by 100% isopropanol: three washes of 5 min each) were air dried for 1 h in a desiccator. Parallel hematoxylin/eosin-stained sections were used as a guide for microdissection. Using a PixCell IIe LCM apparatus (LCM; Molecular Devices, Sunnyvale, CA, USA), two populations of microbes were sampled in the ascending colon: those in the spaces between the transverse folds (interfold) and those in the central lumen (digesta). Separate CapSure HS caps (Arcturus Engineering, Mountain View, CA, USA) were used for each region.
Microbial diversity by tag pyrosequencing analysis
Microbes from interfold and digesta regions were collected from intestinal sections (n=3 wild-type mice) by LCM. DNA was extracted from the LCM-sampled material using a QIAamp DNA Micro Kit (Qiagen, Valencia, CA, USA). Extracted DNA was used for tag-encoded pyrosequencing analysis of 16S rRNA genes using well-validated primers (27F and 338R), tags and protocols (Hamady et al., 2008). PCR amplifications were performed in quadruplicate, and products were pooled. Purified PCR products were subjected to high-throughput pyrosequencing using the GS-FLX Titanium platform (Roche, Branford, CT, USA). Analysis of pyrosequencing reads was carried out using bioinformatics tools (Library Compare and Classifier) at the Ribosomal Database Project (RDP; release 10.15) (Wang et al., 2007; Cole et al., 2009). Differences in microbial composition were considered significant if P<0.001. A detailed description of this analysis is provided in Supplementary Methods. Pyrosequencing data is accessible in the European Nucleotide Archive (Short Read Archive) under accession number ERP000288.
Microbial density by qPCR assays
To determine microbial DNA concentrations from LCM-sampled material, we developed a qPCR assay based on amplification of the RNA polymerase beta subunit (rpoB), a gene highly conserved in Bacteria (Dahllof et al., 2000; Case et al., 2007; Adekambi et al., 2008). An independent set of samples (n=6) was used to estimate densities of microbial populations. Intestinal sections were used to obtain material by LCM as described above. We used two independent sets of mice and two different DNA extraction methods as follows: (i) as described above and (ii) a modified and in-house validated DNA extraction method for paraffin wax-embedded sections (Sepp et al., 1994) (see detailed description in Supplementary Methods).
Extracted genomic DNA was used as a template for PCR amplification and quantification (ng of DNA/μl) of total Bacteria (rpoB gene) using SYBR Green PCR technology. The density of selected bacterial groups within interfold and digesta regions was determined by qPCR assays using previously validated group-specific primers. These qPCR assays targeted Bacteroidaceae–Porphyromonadaceae–Prevotellaceae, Lachnospiraceae–Ruminococcaceae, Enterococcaceae (Rinttila et al., 2004) and Lactobacillaceae (Barman et al., 2008). Primer set Lachnospiraceae–Ruminococcaceae targets 16S rRNA genes from members of Lachnospiraceae and Ruminococcaceae families (also known as Clostridium cluster XIVa or the Eubacterium rectale–Clostridium coccoides group). The theoretical 16S rRNA gene targets for this primer set are shown in Supplementary Table 1. See Supplementary Methods for a detailed description of the PCR protocols.
DNA concentrations (rpoB and 16S rRNA genes) in each LCM sample were determined using the absolute quantification method. Standard curves were constructed with fivefold dilutions of genomic DNA templates of known concentration. DNA extracted from mouse intestinal contents was used as a template for total Bacteria, whereas DNA extracted from laboratory-type strains was used for each of the selected bacterial groups. Concentrations of DNA used in the standard curves ranged from 20 ng to 1.3 pg μl−1. For each qPCR assay, standard curves were amplified at the same time as LCM samples. PCR amplifications were performed in triplicate. Bacterial group-specific qPCR signals were normalized (divided) to total Bacteria (rpoB gene) qPCR signal.
Fingerprinting analysis of Lachnospiraceae–Ruminococcaceae populations
Interindividual variations in diversity of 16S rRNA genes of families Lachnospiraceae–Ruminococcaceae were examined by PCR and terminal restriction fragment length polymorphism (Avaniss-Aghajani et al., 1994). Genomic DNA obtained from LCM samples (n=6) was used as a template for PCR amplification using Lachnospiraceae–Ruminococcaceae-specific primers targeting 16S rRNA genes (Rinttila et al., 2004) as described above, with the exception that the forward primer was 5′-end labeled with 6-carboxyfluorescein (6-FAM; Sigma-Aldrich, St Louis, MO, USA). Three independent restriction enzymes, HpyCh4IV, HhaI and MseI, were used for the digestion of Lachnospiraceae–Ruminococcaceae 16S rRNA gene amplicons. DNA fragment analysis was performed in duplicate on the ABI Prism 3730xl Analyzer (Applied Biosystems, Foster City, CA, USA) using GeneScan Liz600 marker (Applied Biosystems) as a size standard. Terminal-restriction-fragment profiles in each sample were obtained using GeneMapper software (version 3.7, Applied Biosystems). See Supplementary Methods for a detailed description of the PCR-terminal restriction fragment length polymorphism protocol.
Profiles obtained from three independent restriction analyses were concatenated to form a collective data set, and the resulting output files were used in multivariate statistical analyses using non-metric multidimensional scaling analysis and the Kulczynski similarity index (presence-absence data) (Faith et al., 1987; Noll et al., 2008). Differences in microbial composition between the two regions were analyzed by non-parametric MANOVA. All multivariate statistical analyses were performed with the PAST software package (University of Oslo, Norway) (Hammer et al., 2001; Rodriguez et al., 2006; Scupham, 2009). Pairwise comparisons of α-diversity (profile similarity between different subjects) and β-diversity (profile similarity across the interfold and digesta regions) were examined by the Kulczynski similarity index and inferential statistics.
Identification of specific Lachnospiraceae and Ruminococcaceae species was carried out in the interfold region by random cloning and Sanger sequencing
Interfold and digesta regions were sampled by LCM from an independent set of wild-type mice (n=3). DNA was extracted from the LCM-sampled material using the method for paraffin wax-embedded sections as described above. PCR amplification of near full-length 16S rRNA genes was performed as described elsewhere (Eckburg et al., 2005). See Supplementary Methods for a detailed description of the PCR and Sanger sequencing protocol. Chimera-free sequences were analyzed using the Classifier tool at RDP (Wang et al., 2007). Sequences classified into the Clostridiales order (Lachnospiraceae and Ruminococcaceae families, and unclassified Clostridiales) were retrieved from the clone library and used for phylogenetic analysis to obtain an accurate taxonomic classification. In brief, the Seqmatch search at RDP (Wang et al., 2007) was used to find and retrieve the closest matches for known-type strains. Phylogenetic analysis was performed using the maximum likelihood method (Eck and Dayhoff, 1966). See Supplementary Methods for detailed descriptions of phylogenetic analysis and statistical significance of branch order. The sequences of 16S rRNA genes identified in this work are deposited in GenBank under accession numbers: HM856189 through HM856324.
Statistical analysis
Comparisons of bacterial densities between interfold and digesta regions were performed by the Mann–Whitney–Wilcoxon test using SAS software (Statview, Version 5.0.1; SAS Institute, Cary, NC, USA). Pairwise comparisons of α-diversity and β-diversity were examined by the Kulczynski similarity index (presence-absence data) and ANOVA-Fisher's Protected Least Significant Difference test using StatView version 5.0.1. Differences were considered significant at P<0.05.
Results and discussion
Interfold and digesta regions of the mouse ascending colon contain distinct microbial populations
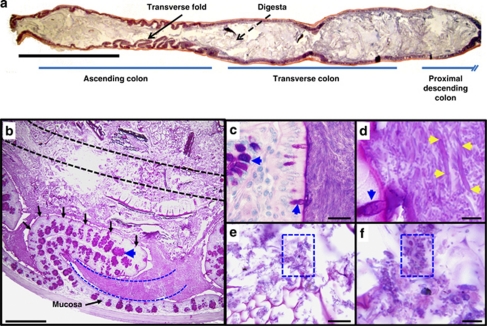
The mouse ascending colon contains transverse folds that project ≈1 mm into the lumen and are oriented in a direction perpendicular to the fecal stream (Figure 1a) (Hummel et al., 1966). This portion of the mouse colon is of additional interest as similar structural folds are also found in the human intestine (plica lunaris and plica semilunaris in the small intestine and colon, respectively). Low-power views of this region in mouse showed that the lumen contained two distinct patterns: (i) dense material between the transverse folds (interfold region) and (ii) less dense, less homogeneous material including food particles within the central lumen (digesta region) (Figure 1b). Higher-power views of these sections showed that the material within the interfold region was comprised of compact, interlacing layers of large, slightly curved fusiform-shaped bacteria (>5–10 μm) (Figures 1c and d and Supplementary Figure 2A and B). In contrast, higher-power views of the central lumen showed undigested food particles that were associated with 1–2 μm rod- and coccoid-shaped bacteria (Figures 1e and f). Microbes with similar morphology have previously been described in these regions. (Davis et al., 1973; Tannock 1987; Swidsinski et al., 2007a, 2007c). Our goal was to best characterize the interfold microbes using a high-resolution sampling method and current nucleic acid-based analytical techniques.
Figure 1.
A morphologically distinct population of predominately fusiform-shaped bacteria is located between the transverse folds of the mouse ascending colon. (a) Hematoxylin/eosin and (b–f) periodic acid-schiff -stained sections of the mouse colon. (a) The proximal portion of the colon (ascending colon) contains transverse folds that project into the lumen (denoted by arrow). The digesta is food particle-associated material in the central lumen (denoted as dashed arrow). (b) Methacarn-fixed section of a mouse ascending colon. The transverse fold (outlined in black arrows) emanates from the mucosa, and is lined by an epithelium that contains periodic acid-schiff-positive goblet cells (denoted as blue arrowhead). Interfold and digesta regions collected by LCM are denoted by blue and black dashed lines, respectively. (c, d) Higher-power views of interfold region. Interlacing fusiform-shaped microbes (denoted as yellow arrowheads) are abundant in this region. (e, f) Higher-power views of the digesta shows rod- and coccoid-shaped microbes (denoted as blue boxes). Bars=5 mm (a), 500 μm (b), 20 μm (c, e), 5 μm (d, f).
Use of LCM as a tool to characterize intact intestinal microbial populations
The sampling of region-specific microbes without a major disturbance to the luminal organization is a major challenge. Earlier studies used washes of the mucosal surface and biopsies to investigate microbes located near the mucosal surface (Eckburg et al., 2005; Franke et al., 2008; Hill et al., 2009). To capture microbes from both the interfold and digesta regions, we used LCM. We, along with others, have used LCM for analysis of host-gene expression (Wong et al., 2000; Stappenbeck et al., 2002) and to isolate microbial DNA from specified host cells (Boye et al., 2006; De Hertogh et al., 2006; Molbak et al., 2006). Although the interfold region measures on the order of 100 μm in greatest dimension, the high resolution of LCM (∼5 μm) permits easy and precise sample collection.
One of our initial concerns with isolation of bacterial DNA from methacarn-fixed material was that the number of Gram-positive bacteria can be underestimated within a complex sample because of the difficulties a Gram-positive cell wall poses to DNA extraction (Park, 2007). Therefore, we performed a preliminary mixing experiment using a Gram-positive (Enterococcus faecalis) and a Gram-negative type strain (E. coli). We placed defined ratios of these bacterial strains on slides. We then air dried, methacarn fixed and dehydrated the slides (in washes of ethanol and xylene). We then captured the bacteria and extracted their DNA. We found that we could efficiently detect both Gram-positive and Gram-negative type strains over a large linear range using qPCR assays targeting group-specific 16S rRNA genes (Supplementary Figure 3). An identical fixation and dehydration procedure was used for tissues prepared for LCM.
The interfold microbes are enriched for Lachnospiraceae and Ruminococcaceae families
To broadly screen LCM-sampled microbes from the interfold and digesta regions, we first used pyrosequencing, the current standard method to perform deep profiling of 16S rRNA genes in complex microbial samples (Hamady et al., 2008). Genomic DNA extracted from interfold and digesta samples was used for PCR amplification of 16S rRNA genes and for tag-encoded pyrosequencing analysis. A total of 42 635 and 53 748 reads were obtained from the interfold and digesta regions, respectively. After removing low-quality reads and sequences of <310 bases, we classified the remaining 29 560 sequences from the interfold region and 38 120 from the digesta region using the Library Compare tool at the RDP. The shapes of the rarefaction curves (Supplementary Figure 4) indicated that bacterial richness in the interfold and digesta regions was sufficient to ensure high coverage for both phyla and families (80% and 90% confidence intervals, respectively).
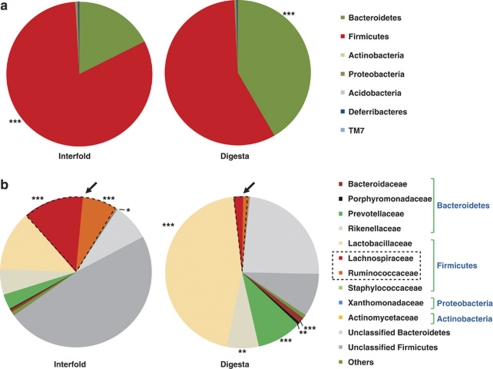
For both data sets, >96% of sequences were classified into six bacterial phyla. However, the majority of the sequences belonged to phyla Firmicutes and Bacteroidetes (94.4% versus 95.9% for interfold and digesta regions, respectively). These findings at the level of phyla are similar to those of other investigators using cecal contents, mucosal biopsies and fecal samples (Eckburg et al., 2005; Ley et al., 2005; Franke et al., 2008; Garner et al., 2009; Hill et al., 2009; Salzman et al., 2009). Interestingly, the interfold and digesta regions contained marked differences. Bacteroidetes were significantly enriched in the digesta region (40% versus 16% of reads in the interfold region), whereas Firmicutes were significantly represented in the interfold region (78% versus 56% in the digesta; Figure 2a). The classification of digesta microbes into predominately Firmicutes and Bacteroidetes concurs with previous studies of total luminal contents from the ileum, cecum and colon (Ley et al., 2005; Garner et al., 2009; Hill et al., 2009; Salzman et al., 2009).
Figure 2.
Colonic interfold microbes are enriched in Lachnospiraceae and Ruminococcaceae families. Pyrosequencing analysis was used to examine microbial diversity between interfold and digesta regions. (a) Comparisons of diversity at the phylum level. (b) Comparisons of diversity at the family level. Each chart represents the taxonomic composition. Sequences were obtained from pooled samples (n=3) of interfolds (29560 reads) and the digesta (38120 reads) region. Lachnospiraceae and Ruminococcaceae are outlined with a dotted line to highlight these families. Unclassified Bacteroidetes and Firmicutes correspond to sequences not classifiable at family level (as of March 2010). Library Compare tool at RDP estimates the probability of observing a difference in a given taxon. Differences in taxa between interfold and digesta were considered significant if P<0.001. *P<7E-03, **P<2E-11 and ***P=6E-014.
A more limited number of studies have profiled mucosa-associated microbes from samples obtained by endoscopic biopsy or dissected/washed intestines (human and mouse samples, respectively). These studies found a trend for the enrichment of Firmicutes as compared with Bacteroidetes (Eckburg et al., 2005; Frank et al., 2007; Hill et al., 2009). In our study, we found a more profound enrichment of Firmicutes.
Because of the significant enrichment of Firmicutes in the interfold region, we next examined differences at lower taxonomic levels. On the basis of the pyrosequencing analysis, the estimated microbial richness ranged between 645 and 934 Operational Taxonomic Units (OTUs) in the interfold region, and between 514 and 738 OTUs in the digesta region (95% and 97% confidence intervals, respectively). Moreover, Good's coverage, which accounts for both diversity and abundance of OTUs, revealed that 96.8% of the population (using a 97% confidence interval) was covered in the interfold region, whereas 98.0% was covered in the digesta region. These results indicated that the sequencing was of adequate depth for additional analysis. At the bacterial family level, 43% of the interfold region and 67% of the digesta region sequences were classified into 25 and 22 known families, respectively. With this subset of sequences from both regions, >98% were classified within seven bacterial families (Bacteroidaceae, Lachnospiraceae, Lactobacillaceae, Porphyromonadaceae, Prevotellaceae, Rikenellaceae and Ruminococcaceae; Figure 2b).
These differences at the level of phyla between the interfold and digesta regions were supported by specific differences in the representation of bacterial families. Strikingly, 20% of the sequences from the interfold region were classified in the Clostridium cluster XIVa group (Lachnospiraceae and Ruminococcaceae) (Collins et al., 1994; Rainey and Janssen, 1995), compared with only 3% in the digesta region. Interestingly, these two bacterial families are known to be fusiform-shaped bacteria (Cotta and Forster, 2006), which correlates with the morphological findings in the interfold region (Figure 1). Thus, on the basis of these observations, we hypothesize that these fusiform-shaped microbes inhabiting the interfold region may represent members of the Lachnospiraceae and Ruminococcaceae families. Additional studies are needed to identify and functionally characterize these microbial communities.
Similar trends of enrichment for Lachnospiraceae and Ruminococcaceae sequences in samples taken near the colonic mucosa were observed in three previous comprehensive studies of 16S rRNA gene repertoires (Eckburg et al., 2005; Franke et al., 2008; Hill et al., 2009). However, their enrichment was not as robust as our findings. In mice, comparisons of 16S rRNA genes between total luminal contents and washed proximal and distal colons showed <2-fold enrichment in Lachnospiraceae and Ruminococcaceae at the mucosal surface (Hill et al., 2009). In humans, sequences from these two bacterial families were enriched <2-fold in biopsy tissues compared with stool samples (Eckburg et al., 2005). Therefore, the use of LCM to more precisely obtain microbial samples, and the use of local reference populations (interfold versus digesta regions), may facilitate the identification of mucosa-associated microbes in a model organism.
We then validated these findings using an independent set of wild-type mice and two independent analytical techniques to confirm that the Lachnospiraceae–Ruminococcaceae families were enriched in the interfold region. As above, we used LCM to sample microbes from the interfold and digesta regions. We first used isolated bacterial DNA to perform qPCR assays of the 16S rRNA gene. We used independent sets of LCM material from interfold and digesta regions (n=6 mice) as templates for qPCR using group-specific primers. These qPCR assays targeted Bacteroidaceae–Porphyromonadaceae–Prevotellaceae, Enterococcaceae, Lachnospiraceae–Ruminococcaceae and Lactobacillaceae 16S rRNA genes. For all groups, the limit of detection was ⩽100 fg of DNA (Supplementary Figure 3).
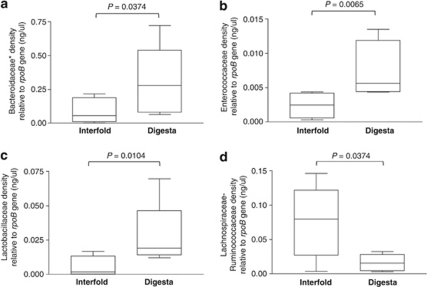
Bacterial group-specific densities were normalized within each sample using the qPCR signal of the rpoB gene, a highly conserved gene in Bacteria that encodes for the β-subunit of RNA polymerase (Dahllof et al., 2000; Case et al., 2007; Adekambi et al., 2008). These analyses showed that densities of Bacteroidaceae–Porphyromonadaceae–Prevotellaceae (P=0.0374), Enterococcaceae (P=0.0065) and Lactobacillaceae (P=0.0104) were significantly greater in the digesta region, compared with the interfold region. In contrast, the density of Lachnospiraceae–Ruminococcaceae was significantly enriched (P=0.0374) in the interfold region compared with the digesta region (Figures 3a–d). These results confirmed that the composition of microbial communities differs significantly between the interfold and digesta regions, and that Lachnospiraceae and Ruminococcaceae are predominant families inhabiting the interfold region.
Figure 3.
Colonic interfolds harbor higher density of microbes of the families Lachnospiraceae–Ruminococcaceae. (a–d) Bacterial densities of interfold and digesta regions were examined using bacterial group-specific qPCR assays. The data depict density of each bacterial group relative to total Bacteria (rpoB gene) as measured by qPCR and DNA standard curves. Bacterial group-specific qPCR signals were normalized (divided) to total Bacteria (rpoB gene) qPCR signal. The rpoB is a gene highly conserved in the Bacteria domain. The asterisk in a indicates that the group queried by this primer set also amplifies 16S rRNA genes of Bacteroidaceae, Porphyromonadaceae and Prevotellaceae. Each PCR assay was performed in triplicate. Bars in each figure represent mean values+s.e. (n=6). Comparisons were made using the Mann–Whitney–Wilcoxon test. Differences were considered significant if P<0.05. Results of qPCR assays are shown in box plots and include their five-number summaries (the smallest observation, lower quartile, median, upper quartile and largest observation).
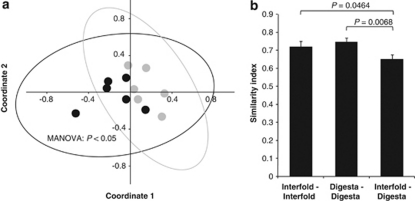
We next determined the microbial composition in the interfold and digesta regions (the same samples as were used for qPCR analysis above) using a combination of bacterial group-specific PCR and terminal restriction fragment length polymorphism techniques. Multivariate non-metric multidimensional scaling analysis ordinations derived from the Kulczynski similarity index (presence-absence data) and MANOVA statistics confirmed that the composition of Lachnospiraceae–Ruminococcaceae 16S rRNA genes from the interfold region is distinct from that observed in the digesta region (Figure 4a).
Figure 4.
Lachnospiraceae–Ruminococcaceae populations are distinct in the interfold region compared with the digesta region. Interindividual variation of Lachnospiraceae–Ruminococcaceae was examined by PCR and terminal restriction fragment length polymorphism techniques targeting Lachnospiraceae—Ruminococcaceae-specific 16S rRNA genes. (a) Differences in the structure and composition of this bacterial family between the interfold (black symbols) and digesta (gray symbols) regions were examined by multivariate analysis (n=6). Non-metric multidimensional scaling analysis ordinations derived from the Kulczynski similarity index (presence-absence data). Each symbol is representative of a single sample. Samples are plotted along the first two component axes. The ellipse corresponds to the joint 95% confidence limits. Microbial composition between the two regions was compared using non-parametric MANOVA. (b) Pairwise comparisons of α-diversity (profile similarity between different subjects, that is, interfold versus interfold) and β-diversity (profile similarity across the interfold and digesta regions) were examined by the Kulczynski similarity index (presence-absence data) and inferential statistics. ANOVA and Protected Least-Significant Difference test were used to compare differences in α-diversity and β-diversity. Differences were considered significant at P<0.05.
Using the same 16S rRNA gene profiles, we analyzed α-diversity and β-diversity to evaluate interindividual differences in the Lachnospiraceae–Ruminococcaceae composition of the interfold and digesta regions. Pairwise comparisons of the Kulczynski similarity index showed that α-diversity of these two bacterial populations between different replicate animals was more homogenous than β-diversity across the interfold and digesta regions (Figure 4b). These results further support the idea that the interfold region is inhabited by a different Lachnospiraceae–Ruminococcaceae community when compared with the digesta region, and that the structure of this bacterial family is similar in replicate animals.
Identification of specific Lachnospiraceae and Ruminococcaceae species in the interfold region by random cloning and Sanger sequencing
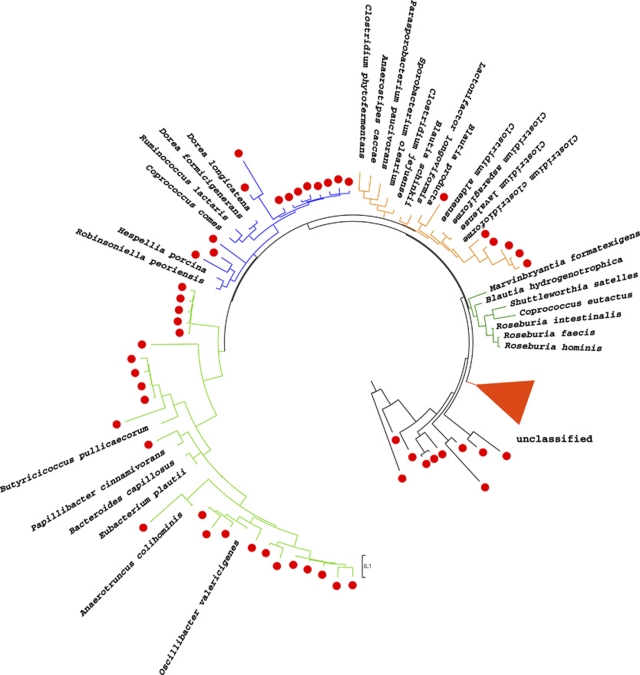
To more precisely classify Lachnospiraceae and Ruminococcaceae bacterial species (OTUs) in the interfold region, we performed Sanger sequencing using an independent set of wild-type mice (n=3). Genomic DNA sampled by LCM was used for the amplification of near full-length ‘universal'16S rRNA genes. These PCR products (≈1.5 kb) were cloned, sequenced and aligned to sequences archived in the Greengenes 16S rRNA database. A total of 873 chimera-free clones, interfold (496 clones) and digesta regions (377 clones), were compared using the Library Compare tool at RDP. From the interfold region, 136 sequences were classified into the Clostridiales order (Lachnospiraceae and Ruminococcaceae families, and unclassified Clostridiales), whereas only five sequences were similarly classified from the digesta region. The interfold sequences were retrieved from the clone library for more accurate classification using phylogenetic analyses. First, the closest matches for known-type strains were obtained using the Seqmatch search at RDP. This analysis revealed that at least 106 OTUs were enriched in the interfold region. These OTUs were classified into 17 candidate genera (≈95% confidence interval) in the family Lachnospiraceae, into 7 genera in the Family Ruminococcaceae and into 10 genera of the Clostridium cluster XIV (Collins et al., 1994) (Table 1). Topology of maximum likelihood trees confirmed that these OTUs were closely related to members of families Lachnospiraceae and Ruminococcaceae, as well as to the Clostridium cluster XIV (Figure 5). These two analyses, SeqMatch search and maximum likelihood phylogeny, revealed that most of the microbes inhabiting the interfold region represent novel OTUs (83–96% confidence interval range), and many of these microbes have not yet been cultured. Taken together, these analyses revealed that the microbes enriched in the interfold region comprise a population of largely uncharacterized microbes. Therefore, cultivation of representative microbes located in this region should be a priority.
Table 1. Operational taxonomical units of families Lachnospiraceae, Ruminococcaceae unclassified Clostridiales (Clostridium cluster XIV) enriched in the interfold region.
| Family | Number of 16S rRNA gene sequences | Similarity range | Best match (type strain)a |
|---|---|---|---|
| Lachnospiraceae | 17 | 0.91–0.83 | Robinsoniella peoriensis; PPC31; AF445285 |
| 12 | 0.90–0.87 | Clostridium jejuense; HY-35-12; AY494606 | |
| 7 | 0.93–0.90 | Coprococcus comes; ATCC 27758; EF031542 | |
| 4 | 0.90–0.87 | Roseburia intestinalis; L1-82; AJ312385 | |
| 3 | 0.93–0.90 | Dorea longicatena; III-35; AJ132842 | |
| 3 | 0.91–0.88 | Marvinbryantia formatexigens; I-52; AJ505973 | |
| 1 | 0.90 | Anaerostipes caccae; L1–92; AJ270487 | |
| 1 | 0.90 | Clostridium lavalense; CCRI-9842; EF564277 | |
| 1 | 0.88 | Clostridium phytofermentans ISDg; CP000885 | |
| 1 | 0.90 | Coprococcus eutactus; ATCC 27759; EF031543 | |
| 1 | 0.92 | Dorea formicigenerans; L34619 | |
| 1 | 0.88 | Hespellia porcina; PPC80; AF445239 | |
| 1 | 0.79 | Parasporobacterium paucivorans; SYR1; AJ272036 | |
| 1 | 0.91 | Roseburia faecis M72/1; AY305310 | |
| 1 | 0.87 | Roseburia hominis A2-183; AJ270482 | |
| 1 | 0.90 | Shuttleworthia satelles; D143K-13; AF399956 | |
| 1 | 0.88 | Sporobacterium olearium; DSM 12504; AF116854 | |
| Ruminococcaceae | 5 | 0.93–0.85 | Anaerobic bacterium ED-Mt61/PYG-s6; DQ100449 |
| 5 | 0.94–0.91 | Bacteroides capillosus; ATCC 29799; AY136666 | |
| 5 | 0.91–0.89 | Ruminococcus lactaris; ATCC 29176; L76602 | |
| 4 | 0.93–0.91 | Butyricicoccus pullicaecorum; 25-3; EU410376 | |
| 4 | 0.94–0.93 | Oscillibacter valericigenes; Sjm18–20; AB238598 | |
| 2 | 0.92–0.90 | Anaerotruncus colihominis; 14565; AJ315980 | |
| 1 | 0.92 | Papillibacter cinnamivorans; DSM12816; AF167711 | |
| Incertae Sedis XIV | 16 | 0.93–0.89 | Blautia schinkii; B; CIP 105464; DSM 10518; X94965 |
| 3 | 0.93–0.90 | Blautia hydrogenotrophica; S5a36; X95624 | |
| 2 | 0.92–0.89 | Blautia producta; ATCC 27340; L76595 | |
| Eubacteriaceae | 2 | 0.93 | Eubacterium plautii; ATCC 29863; AY724678 |
| Gracilibacteraceae | 1 | 0.89 | Gracilibacter thermotolerans; JW/YJL-S1; DQ117465 |
| Veillonellaceae | 3 | 0.96–0.90 | Veillonella parvula; ATCC 10790; AY995767 |
| 1 | 0.96 | Veillonella sp. NVG 100cf; EF108443 | |
| Unclassified | 23 | 0.93–0.84 | Clostridium aldenense; RMA 9741; DQ279736 |
| Clostridiales | 1 | 0.90 | Clostridium clostridioforme; M59089 |
| 1 | 0.91 | Clostridium sp. N6; AJ582080 |
Similarity score reported by SeqMatch at the RDP.
Closest matches for known type strains were obtained using the Seqmatch search at RDP (see Material and methods). Species, strain ID and accession number are provided.
Figure 5.
Candidate genera of families Lachnospiraceae, Ruminococcaceae and Clostridium cluster XIV in the interfold region. To obtain a more precise classification of Lachnospiraceae and Ruminococcaceae bacterial species (Operational Taxonomic Units; OTUs) in the interfold region, Sanger sequencing was performed and reads were analyzed using the Seqmatch search at RDP and phylogenetic analysis. At least 106 OTUs of families Lachnospiraceae, Ruminococcaceae and Clostridium cluster XIV were present in the interfold region (denoted by a red circle). The closest matches for known-type strains were identified and retrieved from RDP using Seqmatch search (see Table 1 for accession numbers, number of sequences in each group and similarity values). Topology of maximum likelihood trees confirmed that these OTUs were related to members of families Lachnospiraceae, Ruminococcaceae and the Clostridium cluster XIV. This tree was rooted using Aquifex pyrophilus, the closest genus to the bacterial last common ancestor. Branches of major clades are depicted by different colors. Branches of unclassified Clostridiales were collapsed (red leave, 51 sequences). The statistical significance of branch order was estimated by the generation of 1000 replications of bootstrap resampling of the originally aligned nucleotide sequences. Scale represents nucleotide substitutions per site.
Collectively, these results emphasize the need for novel experimental systems (animal models and bacterial strains) to discern basic biological principles regulating the interactions between mucosa-associated microbes and the host immune system. Interestingly, clinical evidence has linked a reduced intestinal colonization by members of Lachnospiraceae and Ruminococcaceae families to chronic intestinal disorders, such as inflammatory bowel disease (Sokol et al., 2008, 2009; Willing et al., 2009).
In summary, this study has established new insights into the spatial organization and diversity of microbes across the murine intestinal lumen. Using a high-resolution microbial capture system in conjunction with current nucleic acid analytic techniques, we showed that the microbial communities of the interfold and digesta regions were remarkably different. The significance of this finding is that the enriched families in the interfold region may be positioned to perform critical physiological/barrier functions as consortium. To understand the ecology of this consortium will be a challenge and many members of this community have not yet been cultured. We hope that this study will provide a roadmap to design strategies for culture-based studies. Finally, we propose that the studies in this model organism will provide a strategy to identify and study similar bacterial families that reside in the human intestine.
Acknowledgments
We thank Dan Frank, Ellen Li, Roderick Mackie, Wei Zhu, Peiying Hong and Emil Unanue for helpful comments on the manuscript. We thank Skip Virgin for assistance with pyrosequencing. This work was supported by R01 AI084887, Washington University Digestive Diseases Research Core Center DK52574 and the Pew foundation.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adekambi T, Shinnick TM, Raoult D, Drancourt M. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int J Syst Evol Microbiol. 2008;58:1807–1814. doi: 10.1099/ijs.0.65440-0. [DOI] [PubMed] [Google Scholar]
- Avaniss-Aghajani E, Jones K, Chapman D, Brunk C.1994A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences Biotechniques 17144–146.148–149. [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye M, Aalbaek B, Agerholm JS. Fusobacterium necrophorum determined as abortifacient in sheep by laser capture microdissection and fluorescence in situ hybridization. Mol Cell Probes. 2006;20:330–336. doi: 10.1016/j.mcp.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Case RJ, Boucher Y, Dahllof I, Holmstrom C, Doolittle WF, Kjelleberg S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 2007;73:278–288. doi: 10.1128/AEM.01177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- Cotta M, Forster R.2006The family lachnospiraceae, including the genera butyrivibrio, lachnospira and roseburia The Prokaryotes 1002–1021.Part 1, section 1.3.
- Dahllof I, Baillie H, Kjelleberg S. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl Environ Microbiol. 2000;66:3376–3380. doi: 10.1128/aem.66.8.3376-3380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CP, McAllister JS, Savage DC. Microbial colonization of the intestinal epithelium in suckling mice. Infect Immun. 1973;7:666–672. doi: 10.1128/iai.7.4.666-672.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hertogh G, Aerssens J, de Hoogt R, Peeters P, Verhasselt P, Van Eyken P, et al. Validation of 16S rDNA sequencing in microdissected bowel biopsies from Crohn′s disease patients to assess bacterial flora diversity. J Pathol. 2006;209:532–539. doi: 10.1002/path.2006. [DOI] [PubMed] [Google Scholar]
- Dubos R, Schaedler RW, Costello R, Hoet P. Indigenous, Normal, and Autochthonous Flora of the Gastrointestinal Tract. J Exp Med. 1965;122:67–76. doi: 10.1084/jem.122.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck RV, Dayhoff MO. Atlas of Protein Sequence and Structure. National Biomedical Research Foundation: Silver Spring, MD; 1966. [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP, Minchin RM, Belbin L. Compositional dissimilarity as a robust measure of ecological distance. Plant Ecol. 1987;69:57–68. [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, et al. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection. Infect Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M, et al. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. Past: paleontological statistics software package for education and data analysis. Palaeont Elec. 2001;4:e1–e9. [Google Scholar]
- Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2009;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel KP, Richardson FL, Fekete E.1966AnatomyIn: Green EL (ed).Biology of the Laboratory Mouse McGraw-Hill, Inc.: New York [Google Scholar]
- Kikuchi Y, Meng XY, Fukatsu T. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae) Appl Environ Microbiol. 2005;71:4035–4043. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. 1997;40:782–789. doi: 10.1136/gut.40.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molbak L, Klitgaard K, Jensen TK, Fossi M, Boye M. Identification of a novel, invasive, not-yet-cultivated Treponema sp. in the large intestine of pigs by PCR amplification of the 16S rRNA gene. J Clin Microbiol. 2006;44:4537–4540. doi: 10.1128/JCM.01537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M, Frenzel P, Conrad R. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol Ecol. 2008;65:125–132. doi: 10.1111/j.1574-6941.2008.00497.x. [DOI] [PubMed] [Google Scholar]
- Park D. Genomic DNA isolation from different biological materials. Methods Mol Biol. 2007;353:3–13. doi: 10.1385/1-59745-229-7:3. [DOI] [PubMed] [Google Scholar]
- Rainey FA, Janssen PH. Phylogenetic analysis by 16S ribosomal DNA sequence comparison reveals two unrelated groups of species within the genus Ruminococcus. FEMS Microbiol Lett. 1995;129:69–73. doi: 10.1016/0378-1097(95)00138-U. [DOI] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- Rinttila T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez C, Lang L, Wang A, Altendorf K, Garcia F, Lipski A. Lettuce for human consumption collected in Costa Rica contains complex communities of culturable oxytetracycline- and gentamicin-resistant bacteria. Appl Environ Microbiol. 2006;72:5870–5876. doi: 10.1128/AEM.00963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2009;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. Associations of indigenous microorganisms with gastrointestinal mucosal epithelia. Am J Clin Nutr. 1970;23:1495–1501. doi: 10.1093/ajcn/23.11.1495. [DOI] [PubMed] [Google Scholar]
- Savage DC, Blumershine RV. Surface-surface associations in microbial communities populating epithelial habitats in the murine gastrointestinal ecosystem: scanning electron microscopy. Infect Immun. 1974;10:240–250. doi: 10.1128/iai.10.1.240-250.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med. 1968;127:67–76. doi: 10.1084/jem.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, McAllister JS, Davis CP. Anaerobic bacteria on the mucosal epithelium of the murine large bowel. Infect Immun. 1971;4:492–502. doi: 10.1128/iai.4.4.492-502.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. Isme J. 2008;2:1183–1193. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- Scupham AJ. Campylobacter colonization of the Turkey intestine in the context of microbial community development. Appl Environ Microbiol. 2009;75:3564–3571. doi: 10.1128/AEM.01409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp R, Szabo I, Uda H, Sakamoto H. Rapid techniques for DNA extraction from routinely processed archival tissue for use in PCR. J Clin Pathol. 1994;47:318–323. doi: 10.1136/jcp.47.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Manchester JK, Wong MH, Gordon JI. Laser capture microdissection of mouse intestine: characterizing mRNA and protein expression, and profiling intermediary metabolism in specified cell populations. Methods Enzymol. 2002;356:167–196. doi: 10.1016/s0076-6879(02)56932-9. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Goktas O, Bessler C, Loening-Baucke V, Hale LP, Andree H, et al. Spatial organisation of microbiota in quiescent adenoiditis and tonsillitis. J Clin Pathol. 2007a;60:253–260. doi: 10.1136/jcp.2006.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007b;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Sydora BC, Doerffel Y, Loening-Baucke V, Vaneechoutte M, Lupicki M, et al. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm Bowel Dis. 2007c;13:963–970. doi: 10.1002/ibd.20163. [DOI] [PubMed] [Google Scholar]
- Tannock GW. Demonstration of mucosa-associated microbial populations in the colons of mice. Appl Environ Microbiol. 1987;53:1965–1968. doi: 10.1128/aem.53.8.1965-1968.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Uneyama C, Shibutani M, Masutomi N, Takagi H, Hirose M. Methacarn fixation for genomic DNA analysis in microdissected, paraffin-embedded tissue specimens. J Histochem Cytochem. 2002;50:1237–1245. doi: 10.1177/002215540205000911. [DOI] [PubMed] [Google Scholar]
- Walker WA, Wu M, Isselbacher KJ, Bloch KJ. Intestinal uptake of macromolecules. III. Studies on the mechanism by which immunization interferes with antigen uptake. J Immunol. 1975a;115:854–861. [PubMed] [Google Scholar]
- Walker WA, Wu M, Isselbacher KJ, Bloch KJ. Intestinal uptake of macromolecules. IV.--The effect of pancreatic duct ligation on the breakdown of antigen and antigen-antibody complexes on the intestinal surface. Gastroenterology. 1975b;69:1223–1229. [PubMed] [Google Scholar]
- Walker WA. Gastrointestinal host defence: importance of gut closure in control of macromolecular transport. Ciba Found Symp. 1979;70:201–219. doi: 10.1002/9780470720530.ch12. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn′s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- Wong MH, Saam JR, Stappenbeck TS, Rexer CH, Gordon JI. Genetic mosaic analysis based on Cre recombinase and navigated laser capture microdissection. Proc Natl Acad Sci USA. 2000;97:12601–12606. doi: 10.1073/pnas.230237997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol. 2006;59:1639–1650. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.