Abstract
Biodiversity is claimed to be essential for ecosystem functioning, but is threatened by anthropogenic disturbances. Prokaryotes have been assumed to be functionally redundant and virtually inextinguishable. However, recent work indicates that microbes may well be sensitive to environmental disturbance. Focusing on methane-oxidizing bacteria as model organisms, we simulated disturbance-induced mortality by mixing native with sterilized paddy soil in two ratios, 1:4 and 1:40, representing moderate and severe die-offs. Disturbed microcosms were compared with an untreated control. Recovery of activity and populations was followed over 4 months by methane uptake measurements, pmoA-qPCR, pmoA-based terminal restriction fragment length polymorphism and a pmoA-based diagnostic microarray. Diversity and evenness of methanotrophs decreased in disturbed microcosms, but functioning was not compromised. We consistently observed distinctive temporal shifts between type I and type II methanotrophs, and a rapid population growth leading to even higher cell numbers comparing disturbed microcosms with the control. Overcompensating mortality suggested that population size in the control was limited by competition with other bacteria. Overall, methanotrophs showed a remarkable ability to compensate for die-offs.
Keywords: biodiversity, stability, methane oxidation, pmoA, disturbance, population dynamics
Introduction
Earth's biodiversity is declining, forcing mankind to consider how ecosystems' stability and services will change in future (Mccann, 2000; Loreau et al., 2001). The biodiversity-stability discussion combines two multi-faceted concepts comprising species number and evenness, and resistance and resilience, respectively (Ives and Carpenter, 2007). Prokaryotic microbes can catalyse all biogeochemical cycles and have done so before the advent of eukaryotes. They have been assumed to be functionally redundant and virtually inextinguishable. However, recent work indicates that microbes may well be sensitive to environmental disturbance (Allison and Martiny, 2008). Experiments with constructed communities have demonstrated that not only the number of species but also their evenness may be important for the functional stability of a microbial system (Wittebolle et al., 2009). Overall, soil microbial diversity is indeed very high (Torsvik et al., 2002; Spain et al., 2009). Therefore, functional guilds with a well-defined substrate usage are prime candidates not only to study potentially endangered microbial functions but also to contribute to the general discussion about biodiversity and ecosystem functioning. These prime candidates include nitrifiers (Le Roux et al., 2006; Wertz et al., 2007) and methanotrophs (Horz et al., 2005).
Methanotrophs are able to use methane both for catabolism and for anabolism. They have a well-known physiology (Trotsenko and Murrell, 2008) and a large, yet tractable phylogenetic diversity (McDonald et al., 2008; Lüke et al., 2010). Methane is an important greenhouse gas contributing substantially to radiative forcing (Intergovernmental Panel on Climate Change, 2007). Methanotrophs serve as a biological filter, significantly mitigating methane emissions from wetlands, rice fields and landfills, and are the only biological sink to atmospheric methane (Reeburgh et al., 1993; Conrad, 2009). Traditionally, methanotrophs have been divided into two subgroups: type I and type II. Type I can be further divided into type Ia (for example, Methylobacter, Methylomicrobium, Methylomonas and Methylosarcina) and type Ib (Methylococcus and Methylocaldum) (Bodrossy et al., 2003). The key enzyme of all methanotrophs is the methane monooxygenase (MMO), existing either as a particulate membrane-bound form (pMMO) or as a soluble form (sMMO) (Trotsenko and Murrell, 2008). Although all known methanotrophs except Methylocella palustris (Dedysh et al., 2000) possess pMMO, sMMO is restricted to only some species. The pmoA gene encodes a subunit of pMMO and has been shown to be highly conserved, often being used as a marker gene in cultivation-independent studies (McDonald et al., 2008; Lüke et al., 2010).
Here we investigated the response of methanotrophs to a model disturbance causing a partial die-off of all microbial populations. We addressed the following questions: (i) How do methanotrophic community structure, diversity and evenness change upon disturbance? and (ii) How does the disturbance affect methane uptake rates? We incubated microcosms with untreated and sterilized paddy soil in different ratios over 4 months, and followed methane uptake rates with time. Temporal succession of methanotrophs was monitored by pmoA-based terminal restriction fragment length polymorphism (T-RFLP), and different subgroups of methanotrophs were enumerated by qPCR. For higher phylogenetic resolution, a pmoA-based diagnostic microarray was used to identify methanotrophs down to the genus level (Bodrossy et al., 2003).
Materials and methods
Soil and soil microcosms
Soil was sampled in a rice field at the CRA Agricultural Research Council, Rice Research Unit (Vercelli, Italy) in 2006. Soil parameters and agricultural practice have been described previously (Krüger et al., 2001). Soil was air-dried and part of it was sterilized by γ-irradiation (25 kGy; 60Co) (McNamara et al., 2003; Murase et al., 2006). The sterility of γ-irradiated soil was checked by following CO2 release from water-saturated soil incubated in serum bottles. CO2 release was minute (3%) as compared with non-sterile controls. Untreated and sterilized soil was homogenized and mixed in ratios of 1:4 and 1:40, representing a moderate and severe disturbance, respectively. Microcosms containing untreated soil were used as controls. Dry soil (20 g) was filled in sterile Petri dishes and saturated with autoclaved distilled water (0.45 ml per g dry soil). These microcosms were incubated for up to 4 months in gas-tight jars in an atmosphere of 10 vol.% CH4 in air at 25 °C in the dark. The atmosphere was replenished every third day. Microcosms representing the same treatment were incubated together in the same jar. Microcosms were sacrificed at intervals and soil was homogenized before being stored in aliquots at −20 °C for not more than 10 days.
DNA extraction from soil samples
Total DNA was extracted in triplicates from soil using the Q-Biogene soil extraction kit (MP, Heidelberg, Germany). The extraction was performed according to the manufacturer's instructions with the following modification: DNA was subjected to additional washing steps with 5.5 M guanidine thiocyanate to remove humic acids. DNA extracts were stored at −20 °C for further analysis.
Pore water analysis
Total NH4+ concentration in water-logged soil was measured fluorometrically (Murase et al., 2006). Anion concentrations (SO42−, NO2−, NO3−) were determined by ion chromatography (Bak et al., 1991) after centrifuging an aliquot of wet soil at 14 000 × g for 10 min followed by filtration of the supernatant (0.2 μm; Whatman, Dassel, Germany). Nitrate was only detected in minute concentrations (<0.06 m) and disappeared rapidly; nitrite was below detection limit (<0.005 m).
Oxygen profiles and methane fluxes
O2 was measured with a microsensor (Unisense, Aarhus, Denmark; (Revsbech, 1989)). To measure methane uptake rates, microcosms were removed from the jars and incubated individually in flux chambers (volume 172 ml; 3–5% CH4 in air). Rates were determined from linear regression by following the decrease of methane with time (5–6 h). Methane was measured by gas chromatography with a flame ionization detector (SRI-9300A; SRI Instruments, Torrance, CA, USA).
pmoA-based qPCR assays
All qPCR assays were performed as described previously (Kolb et al., 2003). We focused on TYPEII, MBAC and MCOC assays targeting the dominant groups of methanotrophs in Vercelli paddy soils. In a pilot experiment, MCAP and FOREST assays targeting Methylocapsa and sequences retrieved from forest soils (USC-α), respectively, failed to detect methanotrophs in Vercelli paddy soil; they were not applied further. qPCR assays were performed with an iCycler IQ Thermocycler (Bio-Rad, München, Germany). Each assay was performed in triplicate with primers, primers concentration and PCR profiles as described (Kolb et al., 2003). In addition, total 16S rRNA (EUBAC assay) was enumerated according to Stubner (2002). Details and minor modifications are summarized in Table 1. DNA from clones was used as standard for calibrating the assays. qPCR was carried out in 25-μl volumes using ABgene 96-well PCR plates (VWR International GmbH, Darmstadt, Germany). The plates were sealed with iCycler IQ optical tape (Bio-Rad). PCR master mix containing 12.5 μl iQ SYBR Green Supermix (Bio-Rad) and 0.4 μl of forward and reverse primers each was added to a 5.0-μl DNA template. The DNA template was diluted to obtain optimal target copy numbers (Kolb et al., 2003). Bovine serum albumin (0.8 g; Roche, Mannheim, Germany) was added to the EUBAC assay to relieve inhibition of amplification (Kreader, 1996). DNase- and RNase-free water was added to a final volume of 25 μl.
Table 1. Primer combinations for qPCR assays used in this study.
| Primer set | Forward/reverse primer (n) | PCR thermal profilea | Data acquisition | qPCR assay | Reference |
|---|---|---|---|---|---|
| A189f | 800 | 94 °C, 25 s | 82 °C, 10 s | MBAC | Kolb et al. (2003) |
| Mb601r | 800 | 54.5 °C, 20 s; 72 °C, 45 s | |||
| A189f | 1000 | 94 °C, 25 s | 82 °C, 10 s | MCOC | Kolb et al. (2003) |
| Mc468r | 333 | 64.0 °C, 20 s; 72 °C, 45 s | |||
| II223f | 800 | 94 °C, 25 s | 83 °C, 10 s | TYPEII | Kolb et al. (2003) |
| II646r | 800 | 69.5 °C, 20 s; 72 °C, 45 s | |||
| 519f | 330 | 94 °C, 25 s | 72 °C, 10 s | EUBAC | Stubner (2002) |
| 907r | 330 | 50 °C, 20 s;72 °C, 45 s |
Thermal profile showing temperature and time for denaturation, annealing and elongation.
pmoA-based T-RFLP analysis
The detailed T-RFLP protocol had been described before (Lüke et al., 2010). In short, the pmoA gene was amplified using the FAM-labelled forward primer A189f and the reverse primer A682r (Holmes et al., 1995). PCR amplicons were digested using the restriction endonuclease MspI and T-RFs were separated using the ABIPrism 310 (Applied Biosystems, Darmstadt, Germany). The GeneScan 3.71 software (Applied Biosystems) was used to determine the length of fluorescently labelled T-RFs by comparison with the internal standard (MapMarker 1000; Bioventures, Murfreesboro, TN, USA). Peaks derived from primer dimers were excluded from analysis after cross-checking with a negative control. T-RFLP analysis for each time point was performed in duplicates.
Diagnostic microarray
Diagnostic microarray analysis was performed according to the procedure developed by Bodrossy et al. (2003) with some modifications. The A189f/T7-A682r primer combination was used to prepare the target for the microarray probes. Five-fold-diluted Cy3 was added to the in vitro transcription reaction and hybridization was performed overnight in a hybridization oven (ThermoLifeSciences, Egelsbach, Germany) with the shaking platform set at maximum speed.
Statistics
Statistical analyses were done in R ver. 2.10.1 (R Development Core Team, 2009) using packages vegan ver. 1.18-2 for community ecology analysis (Oksanen et al., 2010) and ggplot2 ver. 0.8.5 for statistical graphics (Wickham, 2009). T-RFLP profiles were standardized before analysis (Dunbar et al., 2001). Probe signals were scaled to maximum signal intensity as revealed from previous validations with clone DNA (Bodrossy et al., 2003; Stralis-Pavese et al., 2004) and limited to 1, if higher. To calculate Shannon–Weaver's diversity H′=∑pi log pi, all probes were summed up and the fraction p per probe i was calculated.
Results
Methane uptake rate and environmental parameters
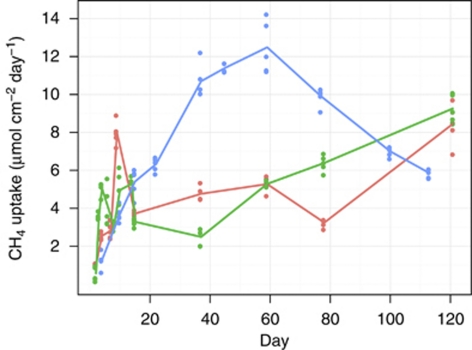
Moderate disturbance showed no adverse effect on methane uptake rates. However, methane uptake rates in severely disturbed microcosms first dropped below the control level, but increased afterwards for 2 months, attaining a maximum twice as high as in the other microcosms (Figure 1). Compared with the control, methane uptake rates in the severely disturbed microcosms were significantly lower at the beginning (day 4), but became significantly higher already at day 14 and remained so till day 77 (two-sided KS test, P⩽0.01). The moderate disturbance resulted in a somehow intermediate response, but still with significantly lower (days 14–77, P⩽0.01) methane uptake rates than in the severely disturbed microcosms. After 4 months incubation, methane uptake rates for control and moderately disturbed microcosms were virtually identical, whereas microcosms exposed to severe disturbance exhibited a slightly lower rate.
Figure 1.
Effects of disturbance on methane uptake rate (mean and individual measurements from four microcosms). Red, green and blue denote control, moderate and severe disturbances, respectively.
Oxygen penetration depth decreased rapidly during the first 10 (disturbance) to 18 days (control), being later restricted to the uppermost 0.4 mm (Supplementary Figure S1). The initial rapid change in O2 penetration depth in disturbed microcosms reflected the faster increase in methanotroph cell numbers as confirmed by qPCR (see below). Pore water sulphate was high at 1.9–2.3 m in the beginning, but virtually disappeared at day 15, while total soil ammonium showed a cyclic pattern (Supplementary Figure S2).
Population dynamics and community structure
The coverage of the qPCR assays was checked in silico using a database with 579 clone sequences retrieved from Vercelli (Lüke et al., 2010). The assays MBAC, MCOC and TYPEII covered 93%, 91% and 83% out of 164, 136 and 279 clones, respectively. The pmoA2 genotypes retrieved from Vercelli (Yimga et al., 2003) are not covered by any of the assays. Hence, the assays provide a fairly complete estimate of the target groups.
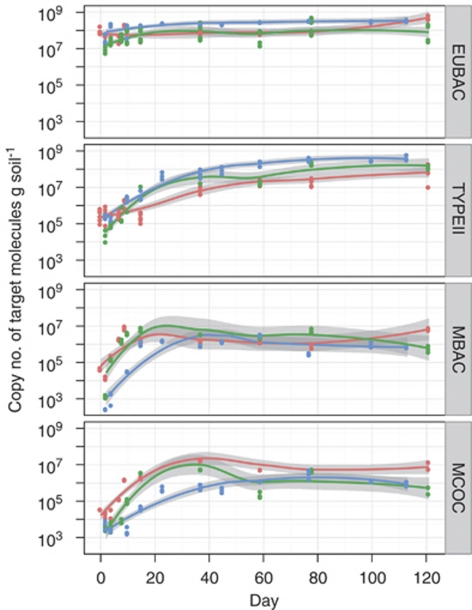
We observed a rapid initial growth of type II methanotrophs in severely disturbed microcosms, whereas growth of type I (MBAC and MCOC) was retarded (Figure 2). On the long term, however, all communities became dominated by type II. Eventually, methanotrophs reached even higher copy numbers in disturbed microcosms, nearly equalling the copy numbers of the eubacterial 16S rRNA gene.
Figure 2.
qPCR analysis of EUBAC, TYPEII, MBAC and MCOC assays. Assays were performed in triplicates at each time point. Red, green and blue denote control, moderate and severe disturbances, respectively. Local regression lines are surrounded by 95% confidence intervals (grey).
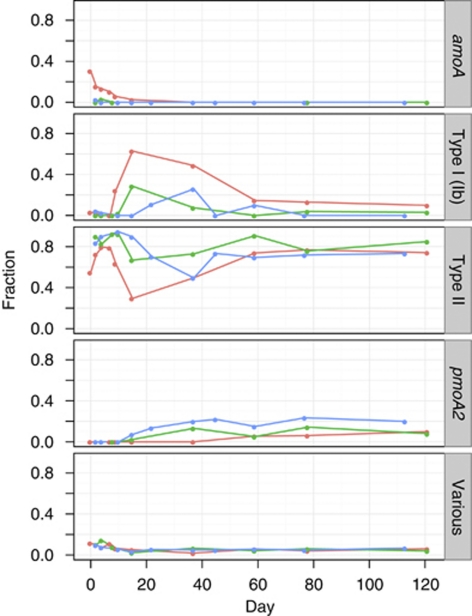
More details of community shifts became evident from pmoA T-RFLP (Figure 3). The assignment of T-RFs was based on a previous study on the same soil considering 500 clones (Lüke et al., 2010). Three dominant T-RFs were indicative for pmoA (type Ib: 79 bp; type II: 244 bp; pmoA2: 278 bp). pmoA2 encodes for a second isoenzyme in type II methanotrophs (Dunfield et al., 2002). The fraction of pmoA2 increased notably in severely disturbed microcosm. Two fragments could be affiliated with amoA (46 and 114 bp). Additional minor T-RFs were summed up and did not show any significant temporal shifts, accounting for <5% after 10 days of incubation. The T-RFLP analysis showed consistent shifts between type I and type II methanotrophs over all treatments, suggesting a succession from type I to type II methanotrophs. After 2 months, the population had stabilized and the ratio of type I to type II methanotrophs was nearly the same as at the beginning of the experiment. However, type I methanotrophs were apparently less important in the disturbed microcosms.
Figure 3.
pmoA-based T-RFLP analysis of control, moderately and severely disturbed microcosms as denoted by red, green and blue, respectively (mean of two replicates from one microcosm). Dominant T-RFs were 79, 244 and 278 bp long and indicative for type Ib, type II and pmoA2, respectively. T-RFs 46 and 114 were pooled; they are indicative of amoA. Minor T-RFs were summed up and designated as ‘Various'.
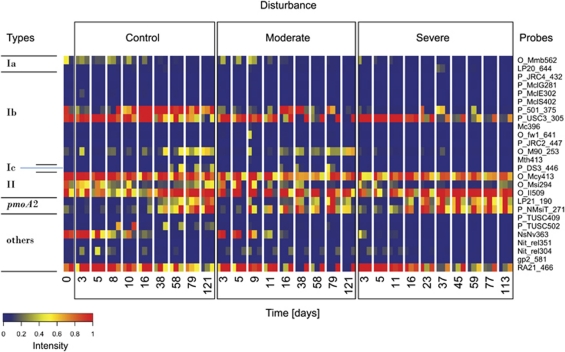
For a higher phylogenetic resolution, the pmoA-targeting microarray (Bodrossy et al., 2003) was used (Figure 4; Supplementary Figure S3). The microarray revealed high signals for probes specific for Methylocystis (type II; probe Mcys413) with no obvious temporal changes. However, Methylosinus (type II; probe Msi294) was only detected in small amounts, being nearly absent in the severely disturbed microcosm (Figure 4). Methylosarcina-like type Ia methanotrophs (probe Mmb562) and ammonia oxidizers (probe NsNv363) were only detected within the first days of incubation and seemed to have only minor roles in the disturbed microcosms. In contrast, the signal intensity of type Ib methanotrophs belonging to the rice paddy cluster 1 (probe M90_253) (Lüke et al., 2010) and to Methylocaldum-related rice paddy clusters (probe 501_375) increased with time, but was undetectable or very low in severely disturbed microcosms. Like for the T-RFLP analysis, signals for probes targeting the pmoA2 (LP21_190; NmsiT271) increased with time, becoming even more important in the disturbed microcosms.
Figure 4.
Microarray analysis of methanotroph communities in control, moderately and severely disturbed microcosms. Microarray results were normalized against reference values determined for each probe individually (Bodrossy et al., 2003), and relative fractions were calculated. Values of 1.0 and 0 indicate maximum signal intensity and no hybridization to the microarray probes, respectively. Only specific probes representing species or well-defined environmental clusters within type Ia, type Ib, type II and pmoA2 are shown. ‘Others' represent probes targeting sequences that group phylogenetically between pmoA and amoA. Owing to a lack of cultivated representatives in this intermediate group, it remains uncertain whether the corresponding proteins catalyse methane or ammonium oxidation.
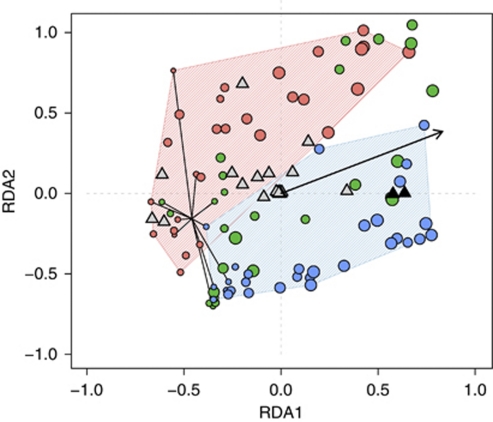
On the basis of the microarray data, the effect of disturbance was tested. We applied redundancy analysis with treatment (control, moderate and severe disturbance) as constraints and found a significant effect (ANOVA, 999 permutations, P⩽0.001) explaining 12% of the total variance. On adding time as the second constraint, treatment and time explained 12.3 and 20.7% of the variance, respectively (ANOVA, 999 permutations, P⩽0.001). Considering the interaction between treatment and time improved the ordination only marginally. Community patterns from controls and severely disturbed microcosm were well separated, whereas moderately disturbed microcosms overlapped with both (Figure 5). As suggested by Figure 4 and Supplementary Figure S3, probes indicative for pmoA2 contributed to the separation of severely disturbed microcosm (Figure 5).
Figure 5.
Microarray results as a biplot of a redundancy analysis with treatment (red, green and blue denote control, moderate and severe disturbances, respectively) and time as constraints. The black arrow indicates the time gradient. Circles refer to microcosms. The area covered by control and severely disturbed microcosms is hatched with the corresponding colours. Circle size corresponds to increasing time. Samples taken at days 0 and 3 mark the beginning of the experiment and are connected by lines starting from the mean. Triangles represent probes. Probes indicative for pmoA2 (LP21_190 and P_NMsiT_271) are marked in black.
Diversity
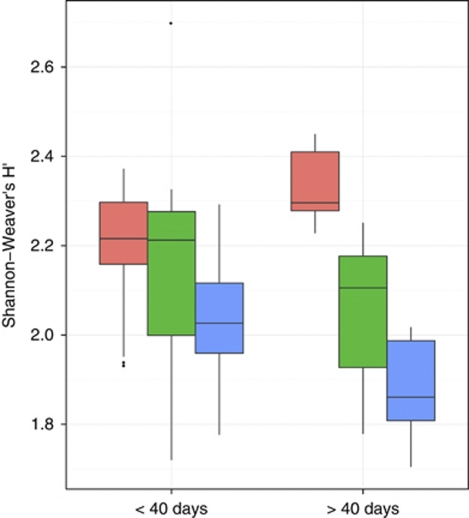
Diversity H′ has two components, number of taxa (here approximated by the number of positive probes) and evenness. One way to express evenness is by the equation V′=H′(ln s)−1 (s=number of species) (Pielou, 1977). The time course of V′ and H′ was nearly identical (data not shown) and reflected the growth dynamics of different groups. Diversity in control and upon disturbance showed drastic differences in particular after day 40, with low values in disturbed microcosms (Figure 6).
Figure 6.
Box plot showing Shanon–Weaver's diversity H′ based on pmoA microarray signals (refer to text). Red, green and blue denote control, moderate and severe disturbances, respectively.
Discussion
Here we show how a microbial guild recovers rapidly compensating for mortality. Growth of specific groups caused reduced diversity and evenness, but loss of functioning could even be over-compensated for some time. At 3 weeks after starting the experiment, methane uptake by the severely disturbed communities became even higher than in the control; however, in the long term uptake rates became similar again (Figure 1).
The abiotic environment
Among potential abiotic controls, ammonium is a top candidate acting either as an inhibitor or as a stimulator for methanotrophs and methane oxidation (Bosse et al., 1993; Bodelier et al., 2000b; Bodelier and Laanbroek, 2004). The decrease of ammonium in the first 10–15 days was probably due to assimilation, although the subsequent increase could have been caused by mineralization enhanced by grazing (Supplementary Figure S2). The role of protists as predators of soil bacteria and methanotrophs in particular has been shown before (Murase et al., 2006; Murase and Frenzel, 2007, 2008). qPCR, however, showed no net decrease of eubacteria or methanotrophs, leaving the reason for these changes still open.
Population dynamics
The rapid increase of pmoA target numbers revealed by qPCR inferred successful re-colonization of the sterile soil and ruled out that extracellular DNA (Pietramellara et al., 2009) had any major effect on population analyses. Type II methanotrophs have often been regarded as largely inactive, represented by resting stages, but target numbers increasing by two to three orders of magnitude (Figure 2) show clearly that upon disturbance they may take their chance. However, this does not imply a proportional contribution to methane oxidation. Cell-specific methane uptake rates calculated from Figures 1 and 2 decreased from 1–5 to 0.002–0.02 pmol CH4 h−1 cell−1 from days 4 to 120, respectively, suggesting that a large fraction of cells became inactive with time re-filling the microbial seed bank.
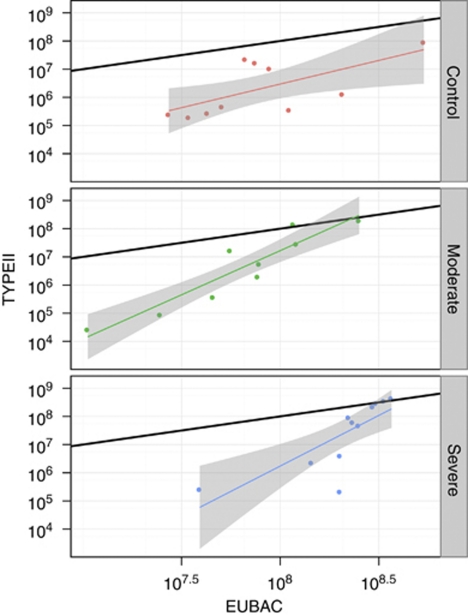
To assess competition between methanotrophs and other bacteria, a qPCR assay targeting the eubacterial 16S rRNA gene was performed. It is suggested that methanotrophs contain two pmoA copies (Semrau et al., 1995). Average 16S rRNA copy numbers, however, have been shown to be high in bacteria thriving in recently flooded rice paddies, while they decrease during succession (Shrestha et al., 2007), reflecting changing life cycle strategies (Klappenbach et al., 2000). Therefore, assuming on average two copies per cell may overestimate the cell numbers of eubacteria during early successions. Nevertheless, the ratio between the numerically dominating type II methanotrophs (pmoA copy number) and total eubacteria (16S rRNA gene copy number) shows how successfully methanotrophs over-compensate for mortality. They made up about 1–10% of the total number of bacteria in controls, but reached apparently 100% upon disturbance (Figure 7). Hence, methanotrophs are not only limited by their main substrates methane and oxygen, but also by other nutrients and potentially even by space. Aerobic heterotrophs recovering from sub-lethal radiation doses grew to population levels significantly higher than in the untreated control (McNamara et al., 2007). Only few substrates were used upon disturbance, but the same substrate spectrum as in the control could be metabolized after recovery (McNamara et al., 2007). In summary, disturbed microbial communities show a remarkable ability to compensate for losses and to re-gain functionality.
Figure 7.
Ratio between the numerically dominating type II methanotrophs (pmoA copy number) and total eubacteria (16S rRNA gene copy). Assuming two copies each per cell, the black lines are drawn assuming that methanotrophs make up 1% of all bacteria. Red, green and blue denotes control, moderate and severe disturbances, respectively. Linear regressions are surrounded by 95% confidence intervals (grey). For details see text.
Community structure
We consistently observed temporal shifts from type I to type II methanotrophs by T-RFLP and qPCR analyses. T-RFLP is a rapid and reproducible method for monitoring community structure, but it lacks the high phylogenetic resolution provided by the pmoA microarray. A redundancy analysis of the microarray signals not only verified the temporal changes, but also separated the control and severely disturbed communities showing a significant treatment effect (Figure 5). The moderately disturbed microcosms took on an intermediate position. However, both T-RFLP and microarray lack exact quantitative qualities. Confirming the trends already suggested by T-RFLP and microarray, qPCR revealed the population dynamics of the main phylogenetic groups, but could not resolve details like the increase in pmoA2 (Figure 3 and Supplementary Figure S3). In summary, the results were consistent in spite of the inherent limitations of the different methods, and lead to the same interpretation.
Both T-RFLP and microarray analyses detected a relative increase in pmoA2. The pmoA2 encodes a subunit of a pMMO isoenzyme (pMMO2) (Dunfield et al., 2002). It was observed to be common among type II methanotrophs and was found to be functionally active, using mRNA as proxy for activity (Dunfield et al., 2002; Yimga et al., 2003). The pMMO2 was constitutively expressed and oxidized methane at low mixing ratios, suggesting that pmoA2 may have an important cellular function (Baani and Liesack, 2008). The initial rapid increase of pmoA2 upon severe disturbance (T-RFLP and microarray: Figures 3 and 4) in parallel to an overall increase in pmoA target numbers (Figure 2) indicates that this was indeed caused by a quantitative increase of pmoA2. It is yet unknown whether all pMMO2 share the high affinity to methane that per se would not have provided a selective advantage in this experiment. However, other traits of the organisms carrying this gene may have been responsible for their success.
Diversity and evenness
Any disturbance from temperature shifts (Wertz et al., 2007) to fertilization (Bodelier et al., 2000a) may affect microbial diversity and performance. The most striking effect of a disturbance, however, is mortality. Although different disturbances may affect different taxa or guilds in a specific way, our experiment used radiation killing non-selectively. We focused on the time after and simulated different mortalities by mixing sterile with untreated soil. What we call here a severe disturbance corresponds to killing 97.5% of all microbes. However, it is worth to compare this ratio with the absolute number of methanotrophs. With 5 × 104 to 5 × 105 methanotrophs per g untreated soil (Figure 2), we still have to expect 2.5 × 104 to 2.5 × 105 methanotrophs per microcosm. By design, this kind of disturbance could not have affected initial evenness, and it can be assumed to have not affected species richness either, except the yet unrecovered ‘rare methanotrophic biosphere' (Lüke et al., 2010). Indeed, the number of positive probes in the microarray analysis was largely similar, varying between 21 and 26. However, intensity changed, as did diversity and evenness, which were calculated from relative probe intensities. Disturbance and low evenness had no detrimental effect on methane uptake rates, which even could increase above the level in the control (Figures 1 and 6).
Concluding remarks
In conclusion, methanotrophs showed a remarkable ability compensating and even over-compensating for die-offs. However, recovery needs time: the effects of frequent disturbances hitting the community while evenness is low may accumulate, shifting it to a different state. Calculated apparent cell-specific methane uptake rates decreased over time, suggesting that a fraction of the methanotrophic population became inactive and part of the microbial seed bank. The factors controlling this transition are still unknown, but are most probably not the main substrates oxygen and methane alone. We anticipate that future experiments will unravel how population dynamics of active microbes translate into the structure of a microbial seed bank. With methanotrophs, the seed bank may contain much more cells than are currently active (Eller et al., 2005), thus preserving and accumulating the effects of repeated disturbance making historical contingencies an important component of the debate on biodiversity and stability.
The evenness of a microbial community has been found to be important for their functioning under environmental stress (Wittebolle et al., 2009). Here we show that upon disturbance different growth rates may lead to an uneven community that, nevertheless, functions better than the control. Considering that only part of this community was active, linking diversity or evenness to functional resilience should consider not only population structure but also controls of activity.
Paddy soil methanotrophs are even able to overgrow other microorganisms after severe disturbance, provided methane is available. In contrast to our experiment, methanotrophs in upland soils live at extremely low methane concentrations and have been found to be highly vulnerable (Bodelier and Laanbroek, 2004; Priemé et al., 1997). This may reflect particular physiological adaptations, indicating that apparent functional redundancy can be constrained by other dimensions of an organism's niche. However, not only the qualitative aspect but also the energy flow through a microbial population and its effect on quantitative population dynamics deserve increased attention in biodiversity-ecosystem-functioning studies.
Acknowledgments
We thank Leventhe Bodrossy (Seibersdorf/Austria) for introducing us to the pmoA microarray and for discussion on microarray analysis. Alexandra Hahn and Bellinda Schneider provided excellent technical assistance. This study was financially supported by the International Max Planck Research School for Environmental, Cellular and Molecular Microbiology by a grant to AH, and by the Deutsche Forschungsgemeinschaft by a grant to PF. This is a contribution of METHECO to the ESF-coordinated EUROCORES-program EuroDIVERSITY.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baani M, Liesack W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp strain SC2. Proc Natl Acad Sci USA. 2008;105:10203–10208. doi: 10.1073/pnas.0702643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak F, Scheff G, Jansen KH. A rapid and sensitive ion chromatographic technique for the determination of sulfate and sulfate reduction rates in freshwater lake sediments. FEMS Microbiol Ecol. 1991;85:23–30. [Google Scholar]
- Bodelier PLE, Hahn AP, Arth I, Frenzel P. Effects of ammonium-based fertilisation on microbial processes involved in methane emission from soils planted with rice. Biogeochemistry. 2000a;51:225–257. [Google Scholar]
- Bodelier PLE, Laanbroek HJ. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol Ecol. 2004;47:265–277. doi: 10.1016/S0168-6496(03)00304-0. [DOI] [PubMed] [Google Scholar]
- Bodelier PLE, Roslev P, Henckel T, Frenzel P. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature. 2000b;403:421–424. doi: 10.1038/35000193. [DOI] [PubMed] [Google Scholar]
- Bodrossy L, Stralis-Pavese N, Murrell JC, Radajewski S, Weilharter A, Sessitsch A. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ Microbiol. 2003;5:566–582. doi: 10.1046/j.1462-2920.2003.00450.x. [DOI] [PubMed] [Google Scholar]
- Bosse U, Frenzel P, Conrad R. Inhibition of methane oxidation by ammonium in the surface layer of a littoral sediment. FEMS Microbiol Ecol. 1993;13:123–134. [Google Scholar]
- Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep. 2009;1:285–292. doi: 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, et al. Methylocella palustrisgen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol. 2000;50:955–969. doi: 10.1099/00207713-50-3-955. [DOI] [PubMed] [Google Scholar]
- Dunbar J, Ticknor LO, Kuske CR. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol. 2001;67:190–197. doi: 10.1128/AEM.67.1.190-197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunfield PF, Yimga TM, Dedysh SN, Berger U, Liesack W, Heyer J. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol Ecol. 2002;41:17–26. doi: 10.1111/j.1574-6941.2002.tb00962.x. [DOI] [PubMed] [Google Scholar]
- Eller G, Krüger M, Frenzel P. Comparing field and microcosm experiments: a case study on methano- and methylotrophic bacteria in paddy soil. FEMS Microbiol Ecol. 2005;51:279–291. doi: 10.1016/j.femsec.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Owens NJP, Murrell JC. Detection of novel marine methanotrophs using phylogenetic and functional gene probes after methane enrichment. Microbiology (UK) 1995;141:1947–1955. doi: 10.1099/13500872-141-8-1947. [DOI] [PubMed] [Google Scholar]
- Horz HP, Rich V, Avrahami S, Bohannan BJM. Methane-oxidizing bacteria in a California upland grassland soil: diversity and response to simulated global change. Appl Environ Microbiol. 2005;71:2642–2652. doi: 10.1128/AEM.71.5.2642-2652.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intergovernmental Panel on Climate Change . Climate Change 2007: The Physical Science Basis. Summary for Policymakers. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Summary for Policymakers Formally Approved at the 10th Session of Working Group I of the IPCC. IPCC Secretariat: Geneva; 2007. [Google Scholar]
- Ives AR, Carpenter SR. Stability and diversity of ecosystems. Science. 2007;317:58–62. doi: 10.1126/science.1133258. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb S, Knief C, Stubner S, Conrad R. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl Environ Microbiol. 2003;69:2423–2429. doi: 10.1128/AEM.69.5.2423-2429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M, Frenzel P, Conrad R. Microbial processes influencing methane emission from rice fields. Global Change Biol. 2001;7:49–63. [Google Scholar]
- Le Roux X, Philippot L, Degrange V, Poly F, Wertz S. Biodiversity-functioning relations in microorganisms. Biofutur. 2006;268:50–53. [Google Scholar]
- Loreau M, Naeem S, Inchausti P, Bengtsson J, Grime JP, Hector A, et al. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science. 2001;294:804–808. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- Lüke C, Krause S, Cavigioli S, Greppi D, Lupotto E, Frenzel P. Biogeography of wetland rice methanotrophs. Environ Microbiol. 2010;12:862–872. doi: 10.1111/j.1462-2920.2009.02131.x. [DOI] [PubMed] [Google Scholar]
- Mccann KS. The diversity-stability debate. Nature. 2000;405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- McDonald IR, Bodrossy L, Chen Y, Murrell JC. Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol. 2008;74:1305–1315. doi: 10.1128/AEM.02233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara NP, Black HIJ, Beresford NA, Parekh NR. Effects of acute gamma irradiation on chemical, physical and biological properties of soils. Appl Soil Ecol. 2003;24:117–132. [Google Scholar]
- McNamara NP, Griffiths RI, Tabouret A, Beresford NA, Bailey MJ, Whiteley AS. The sensitivity of a forest soil microbial community to acute gamma-irradiation. Appl Soil Ecol. 2007;37:1–9. [Google Scholar]
- Murase J, Frenzel P. A methane-driven microbial food web in a wetland rice soil. Environ Microbiol. 2007;9:3025–3034. doi: 10.1111/j.1462-2920.2007.01414.x. [DOI] [PubMed] [Google Scholar]
- Murase J, Noll M, Frenzel P. The impact of protists on activity and structure of the bacterial community in a rice field soil. Appl Environ Microbiol. 2006;72:5436–5444. doi: 10.1128/AEM.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase J, Frenzel P. Selective grazing of methanotrophs by protozoa in a rice field soil. FEMS Microbiol Ecol. 2008;65:408–414. doi: 10.1111/j.1574-6941.2008.00511.x. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RG, Simpson GL, et al. 2010Vegan: Community ecology package1.18-12. . http://R-Forge.R-project.org/projects/vegan/ .
- Pielou EC. Mathematical Ecology. John Wiley & Sons: New York; 1977. [Google Scholar]
- Pietramellara G, Ascher J, Borgogni F, Ceccherini MT, Guerri G, Nannipieri P. Extracellular DNA in soil and sediment: fate and ecological relevance. Biol Fertil Soils. 2009;45:219–235. [Google Scholar]
- Priemé A, Christensen S, Dobbie KE, Smith KA. Slow increase in rate of methane oxidation in soils with time following land use change from arable agriculture to woodland. Atmos Environ. 1997;29:1269–1273. [Google Scholar]
- R Development Core Team 2009R: A Language and Environment for Statistical Computing2.10.1. R Foundation for Statistical Computing. ISBN 3-900051-07-0.http://www.R-project.org .
- Reeburgh WS, Whalen SC, Alperin MJ.1993The role of methylotrophy in the global methane budgetIn: Murrell JC, Kelly DP (eds).Microbial Growth on C-1 Compounds Intercept: Andover; 1–14. [Google Scholar]
- Revsbech NP. An oxygen microsensor with a guard cathode. Limnol Oceanogr. 1989;34:474–478. [Google Scholar]
- Semrau JD, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, et al. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha PM, Noll M, Liesack W. Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ Microbiol. 2007;9:2464–2474. doi: 10.1111/j.1462-2920.2007.01364.x. [DOI] [PubMed] [Google Scholar]
- Spain AM, Krumholz LR, Elshahed MS. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009;3:992–1000. doi: 10.1038/ismej.2009.43. [DOI] [PubMed] [Google Scholar]
- Stralis-Pavese N, Sessitsch A, Weilharter A, Reichenauer T, Riesing J, Csontos J, et al. Optimization of diagnostic microarray for application in analysing landfill methanotroph communities under different plant covers. Environ Microbiol. 2004;6:347–363. doi: 10.1111/j.1462-2920.2004.00582.x. [DOI] [PubMed] [Google Scholar]
- Stubner S. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreenTM detection. J Microbiol Methods. 2002;50:155–164. doi: 10.1016/s0167-7012(02)00024-6. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- Trotsenko YA, Murrell JC. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol. 2008;63:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- Wertz S, Degrange V, Prosser JI, Poly F, Commeaux C, Guillaumaud N, et al. Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ Microbiol. 2007;9:2211–2219. doi: 10.1111/j.1462-2920.2007.01335.x. [DOI] [PubMed] [Google Scholar]
- Wickham H. Ggplot2. Elegant Graphics for Data Analysis. Springer: Dordrecht; 2009. [Google Scholar]
- Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458:623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- Yimga TM, Dunfield PF, Ricke P, Heyer J, Liesack W. Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl Environ Microbiol. 2003;69:5593–5602. doi: 10.1128/AEM.69.9.5593-5602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.