Abstract
Recent applications of culture-independent, molecular methods have revealed unexpectedly high diversity in a variety of functional and phylogenetic groups of microorganisms in the ocean. However, none of the existing research tools are free from significant limitations, such as PCR and cloning biases, low phylogenetic resolution and others. Here, we employed novel, single-cell sequencing techniques to assess the composition of small (<10 μm diameter), heterotrophic protists from the Gulf of Maine. Single cells were isolated by flow cytometry, their genomes amplified, and 18S rRNA marker genes were amplified and sequenced. We compared the results to traditional environmental PCR cloning of sorted cells. The diversity of heterotrophic protists was significantly higher in the library of single amplified genomes (SAGs) than in environmental PCR clone libraries of the 18S rRNA gene, obtained from the same coastal sample. Libraries of SAGs, but not clones contained several recently discovered, uncultured groups, including picobiliphytes and novel marine stramenopiles. Clone, but not SAG, libraries contained several large clusters of identical and nearly identical sequences of Dinophyceae, Cercozoa and Stramenopiles. Similar results were obtained using two alternative primer sets, suggesting that PCR biases may not be the only explanation for the observed patterns. Instead, differences in the number of 18S rRNA gene copies among the various protist taxa probably had a significant role in determining the PCR clone composition. These results show that single-cell sequencing has the potential to more accurately assess protistan community composition than previously established methods. In addition, the creation of SAG libraries opens opportunities for the analysis of multiple genes or entire genomes of the uncultured protist groups.
Keywords: marine protists, diversity, single-cell genomics, picobiliphytes
Introduction
Protists, both phototrophic and heterotrophic, are important constituents of marine ecosystems. Phototrophic protists contribute significantly to biomass, primary production and respiration (Li, 1994; Maranon et al., 2001; Jardillier et al., 2005; Tarran et al., 2006). Heterotrophic protists also have an important role in the microbial loop and the marine carbon cycle through respiration, grazing on phytoplankton and heterotrophic microbes and by being a food source to zooplankton (Azam et al., 1983; Fuhrman and McManus, 1984; Yokokawa and Nagata, 2005; Jeong et al., 2008). Many marine protists function as mixotrophs (Zubkov and Tarran, 2008; de Castro et al., 2009), harbor endosymbionts (Buck and Bentham, 1998; Not et al., 2007b) or parasitize other eukaryotes (Harada et al., 2007; Brown et al., 2009), demonstrating their diverse and complex ecological roles.
Although there is a wealth of knowledge about marine protists from years of study by traditional methods, the diversity and identity of many of these organisms, such as most marine microbes, remains unknown. Labor-intensive microscopy methods, culturing or molecular techniques examining DNA sequences can be used to identify species or phylotypes. For nano- and picoplankton, very little information on the identity of these cells can be obtained by light microscopy because of the small size and typically indistinct morphologies. Recent studies using culture-independent 18S rRNA gene surveys have revealed novel groups, highlighting the need for further research in this area (Diez et al., 2001; Moon-van der Staay et al., 2001; Massana et al., 2004a; Stoeck et al., 2006; Worden, 2006; Countway et al., 2007; Shalchian-Tabrizi et al., 2007; Amaral-Zettler et al., 2009; Brown et al., 2009; Vigil et al., 2009).
Currently used molecular methods for determining protist community composition and diversity involve fluorescence in situ hybridization (Biegala et al., 2003), metagenomics (Not et al., 2009), environmental PCR-based fingerprinting (Vigil et al., 2009), sequencing of cloned PCR products (Massana et al., 2004b; Not et al., 2007a) and pyrotag sequencing (Amaral-Zettler et al., 2009; Brown et al., 2009). Quantitative real-time PCR (qPCR) has also been used to study known groups of picoeukayotes (Zhu et al., 2005). However, each of these techniques has serious limitations, such as PCR and/or cloning artifacts, biases caused by the variable copy number of target genes per cell, low phylogenetic resolution and so on.
Sequencing the DNA of individual cells offers a novel way to analyze microbial community composition and diversity (Raghunathan et al., 2005; Zhang et al., 2006; Marcy et al., 2007; Stepanauskas and Sieracki, 2007; Woyke et al., 2009). Recent advances in single-cell analyses to examine microbial function and identity are beginning to change our knowledge of microbial systems. These methods have the advantage of avoiding certain PCR and cloning biases that distort results obtained from community DNA. They also enable the linkage of phylogeny and potential metabolism at the cellular level (Stepanauskas and Sieracki, 2007). We report here the comparison of environmental PCR clone library to the single-cell sequencing approach. We compare these methods to estimate the diversity of the relatively understudied small heterotrophic marine protists. We find that single-cell sequencing reveals higher protist diversity than environmental PCR-based clone libraries, and therefore, a more robust estimate of community structure.
Methods
Sample collection and cell sorting
A 50 ml coastal water sample was collected from 1 m depth in Boothbay Harbor in the Gulf of Maine, USA (43°50′39.87′′N 69°38′27.49′′W). Sampling was conducted at high tide (0815 hours) on 25 July 2007 using a Niskin bottle. The water temperature was 18 °C and chlorophyll was 2.65 μg l−1 (96% passed a 20 μm mesh). The abundances of heterotrophic bacteria and heterotrophic and phototrophic protists were 3.1 × 106, 2200 and 30 500 per ml, respectively, all indicative of typical summer conditions there. Samples were kept in the dark at in situ temperature until processing (less than 6 h). Subsamples (3 ml) were incubated for 10 min with Lysotracker Green DND-26 (75 nmol l−1; Invitrogen, Carlsbad, CA, USA), a pH-sensitive green fluorescing probe that stains food vacuoles in protists (Rose et al., 2004). Target cells were identified and sorted using a MoFlo (Beckman-Coulter, Brea, CA, USA) flow cytometer equipped with a 488 nm laser for excitation. Before sorting, the cytometer was cleaned thoroughly with bleach. All tubes, plates and buffers were ultraviolet-treated before use, to remove any DNA contamination. A 1% NaCl solution (0.2 μm filtered and ultraviolet-treated) was used as sheath fluid. Full details of the cleaning and preparation techniques are described in Stepanauskas and Sieracki (2007).
Heterotrophic protists were identified by the presence of Lysotracker fluorescence and the absence of chlorophyll fluorescence. Side scatter was used to select only the smaller protists, approximately <10 μm in diameter. Three types of cell sorting were performed: community DNA, single-cell DNA and microscopy sorts. Community sorts were used in clone library construction. Target cells (ca. 4000) were deposited per each, triplicate 1.5 ml tube containing 20 μl Lyse-n-Go (Thermo-Fisher Scientific, Waltham, MA, USA). For single-cell sorts, individual target cells were deposited into 96-well plates, in which some wells were dedicated for positive controls (10 cells per well) and negative controls (0 cells per well). All wells on the microplates contained 5 μl of either 1 × phosphate-buffered saline (137 m NaCl, 2.7 m KCl, 10 m Na3PO4 and 2 m K3PO4 adjusted to pH 7.4) or Lyse-n-Go (Pierce). Samples were centrifuged briefly and stored at −80 °C. For microscopy sorts, 1000 target cells were collected in 1 × phosphate-buffered saline, preserved with 0.5% paraformaldehyde and then dual-stained with proflavine (5 μg ml−1, Sigma-Aldrich, St Louis, MO, USA) and DAPI (5 μg ml−1, Sigma). Samples were immediately imaged using epifluorescence microscopy to confirm the sort targets.
Whole-genome amplification
Cells deposited into Lyse-n-Go (both community and single-cell sorts) were lysed using a thermal cycle protocol provided by the manufacturer. The single cells that were deposited into phosphate-buffered saline were lysed using cold KOH (Raghunathan et al., 2005). Genomic DNA from the lysed cells was amplified using multiple displacement amplification (MDA) (Dean et al., 2002; Raghunathan et al., 2005). For single-cell lysates, MDA reagents were added directly to the microplate wells. Single-cell MDA reaction volumes were 50 μl for Lyse-n-Go lysates and 83 μl for KOH lysates. All MDA reactions contained 2U μl−1 Repliphi polymerase, 1 × reaction buffer, 0.4 m dNTPs, 2 m DTT (Epicentre, Madison, WI, USA), 1 μ SYTO-9 (Invitrogen) and 50 n random hexamer primer (IDT, Coralville, IA, USA). Samples were incubated at 30 °C for 6 h using a real time thermal cycler with fluorescence measured at 6 min intervals. The Repliphi polymerase was inactivated by incubation for 3 min at 65 °C, and the amplified DNA was stored at −80 °C until further processing. We refer to the MDA products originating from individual cells as single amplified genomes (SAGs).
Lysed cells from the triplicate community sort tubes were combined and 1 μl aliquots of the combined lysate were used as templates in eight, replicate MDA reactions with final reaction volumes of 10 μl. Products from the eight replicate MDA reactions, each containing gDNA of approximately 170 cells, were pooled before their use as a template in PCR. This community MDA replication was employed to reduce potential biases due to uneven MDA amplification (Pinard et al., 2006).
PCR and cloning
Products of MDA reactions were diluted 100-fold with ultraviolet-treated elution buffer (Qiagen, Germantown, MD, USA) and used as templates in real-time PCR targeting the 18S rRNA gene, using the primers shown in Table 1. The 22 μl PCR reactions contained 1 × pre-made mastermix (either Epicentre Failsafe Buffer L or SYBRmaster (Roche, Basel, Switzerland)), 0.3 μ each primer and 2 μl of the diluted MDA product. The templates were first denatured at 95 °C (1 min for the Failsafe buffer, 5 min for the SYBRmaster buffer), then amplified using 40 cycles of denaturing for 20 s at 94 °C, annealing for 20 s at 55 °C and extension for 60 s per 1 kbp at 72 °C. A final extension for 10 min at 72 °C and a melt curve analysis were performed on each reaction. Electrophoresis on a tris-acetate-EDTA agarose gel containing 0.02% ethidium bromide was used to isolate and verify the size of the PCR products. Amplicons of the correct size were cut out and extracted using the QIAquick DNA extraction kit (Qiagen) according to the manufacturer's instructions. Gel-purified community PCR products were cloned using TOPO TA cloning kits (Invitrogen) according to the manufacturer's instructions using PCR products from the Euk1A /516r primer set (clone names start with CS618) and the 528F/EukB primer set (clone names start with QS664 and TS698).
Table 1. PCR primers used in SAG screens and clone library construction.
| Forward primer | Reverse primer | Region [bp]a | References |
|---|---|---|---|
| Euk1A 5′-CTGGTTGATCCTGCCAG-3′ | 516r 5′-ACCAGACTTGCCCTCC-3′ | 4–563 | Amann et al., 1990; Sogin and Gunderson, 1987 |
| 528f 5′-CCGCGGTATTCCAGCTC-3′ | EukB 5′-TGATCCTTCTGCAGGTTCACCTAC-3′ | 572–1795 | Elwood et al., 1985; Medlin et al., 1988 |
| Euk1A 5′-CTGGTTGATCCTGCCAG-3′ | EukB 5′-TGATCCTTCTGCAGGTTCACCTAC-3′ | 4–1795 | Amann et al., 1990; Medlin et al., 1988 |
| 515f 5′-GTGCCAAGCAGCCGCGGTAA-3′ | 1209r 5′-GGGCATCACAGACCTG-3′ | 515–1209 | Giovannoni et al., 1988; Reysenbach et al., 1992 |
| 345f 5′-AAGGAAGGCAGCAGGCG-3′ | 499r 5′-CACCAGACTTGCCCTCYAAT-3′ | 345–499 | Zhu et al., 2005 |
Region targeted by PCR on Saccharomyces cerevisiae (accession number AY251629.1) 18S rRNA gene.
DNA sequencing and phylogenetic analyses
The 18S rRNA gene PCR products of SAGs and clones were sequenced from both ends by Agencourt Bioscience Corporation using Sanger technology. Sequences were assembled and manually curated with Sequencher 4.8 (Gene Codes, Ann Arbor, MI, USA), then checked for similarity with published sequences using BLASTn against GenBank (Altschul et al., 1990). Some SAG sequences were identified as contaminants and were removed from further analyses. Sequences were designated as contaminants if they were >99% identical to non-marine taxa. These contaminants constituted between 0 and 10% of the SAGs analyzed for each primer set and included matches to terrestrial basidiomycetes, angiosperms and insecta. Negative blanks showed no significant amplification.
Maximum likelihood techniques with 100 bootstraps were used to construct phylogenetic trees with PhyML (Dereeper et al., 2008). The phylogenetic diversity, obtained from the SAGs and the clone libraries, was compared using UniFrac significance tests (Lozupone and Knight, 2005; Lozupone et al., 2006). Simpson and Shannon diversity indices, along with rarefaction curves and abundance-based coverage estimator richness estimations were calculated using DOTUR (Chao and Lee, 1992; Chao et al., 1993; Schloss and Handelsman, 2005) with distance matrices produced by MEGA4 (Tamura et al., 2007). In the latter computations, clone libraries were randomly sub-sampled to equalize their size to SAG libraries.
Results
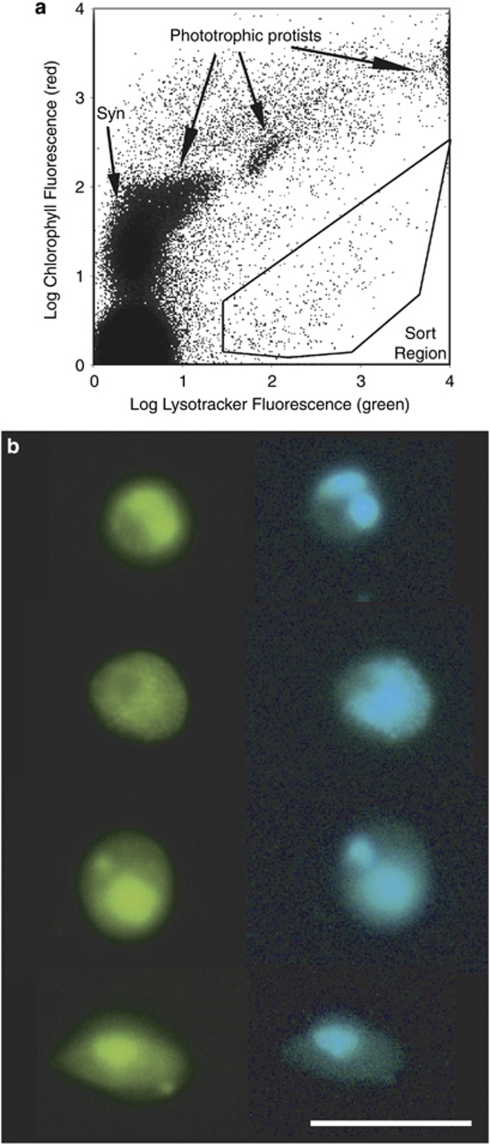
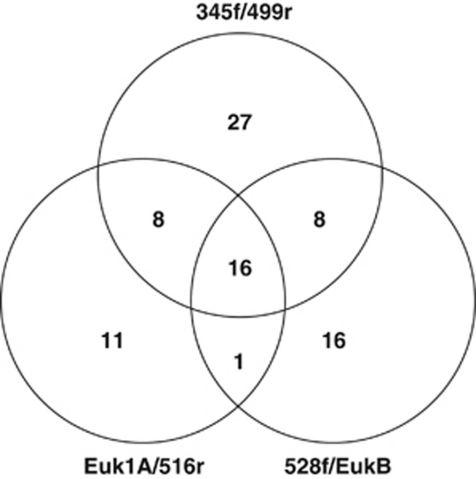
Microscopy showed that sorting of Lysotracker-stained cells successfully isolated target heterotrophic protists from the complex planktonic microbial community (Figure 1). We obtained partial 18S rRNA sequences of marine protists from 87 SAGs and 191 clones. Of the latter, 90 clones were generated using Euk1A/516r and 101 clones were generated using 528f/EukB primer sets. The number of SAGs successfully amplifying with 18S rRNA PCR primers was dependent on the choice of ‘universal' primers, as many SAGs amplified only with one or two of the tested primer pairs (Figure 2). Neither SAGs nor the community gDNA amplified with the Euk1A/EukB primer pair. The 515f/1209r primer pair was not found to be specific for eukaryote 18S rDNA and instead amplified many 16S rDNA partial gene sequences. The rate of recovery of high-quality, non-contaminant 18S rRNA gene sequences from SAGs was 15%, 17% and 25% for the individual primer sets Euk1A/516r, 528f/EukB and 345f/499r, respectively. Although the 345f/499r primer set amplified the greatest number of SAGs, the obtained amplicons were too short for robust phylogenetic analysis. The combined results of all three primer pairs produced partial 18S rRNA genes from 36% of the analyzed protist SAGs.
Figure 1.
(a) Flow cytometric sort region for heterotrophic protists. Target cells were inside the sort region, with high green fluorescence (Lysotracker-stained vacuoles) and low red fluorescence (chlorophyll). Phototrophic protists have high chlorophyll and Lysotracker fluorescence. Cells between the sort region and the phototrophs are likely heterotrophs with ingested chlorophyll or mixotrophs. Prokaryotes (for example, Synechococcus sp.) do not stain with Lysotracker. The large population in the lower left is noise and/or heterotrophic bacteria. Side scatter (not shown) was also used to limit the sort to smaller cells. (b) Epifluorescence photomicrographs of sorted heterotrophic protists. Cells were stained with proflavine (left, blue excitation) and DAPI (right, ultraviolet excitation) after sorting. Images of each cell were contrast-stretched. Scale bar is 10 μm.
Figure 2.
Comparison of the yields of SAGs for the three 18S PCR primer sets used. Number of SAGs from which marine 18S rDNA gene sequences were amplified are shown.
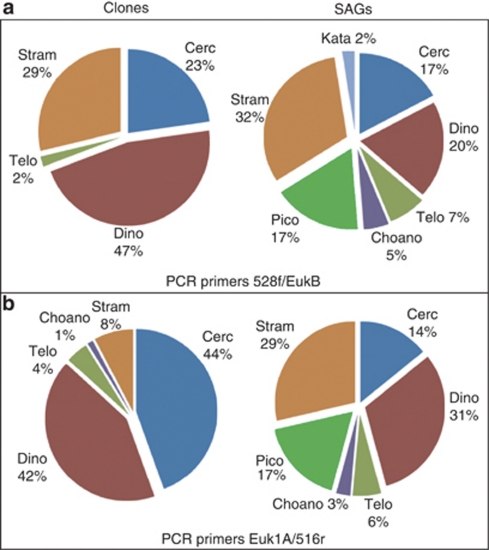
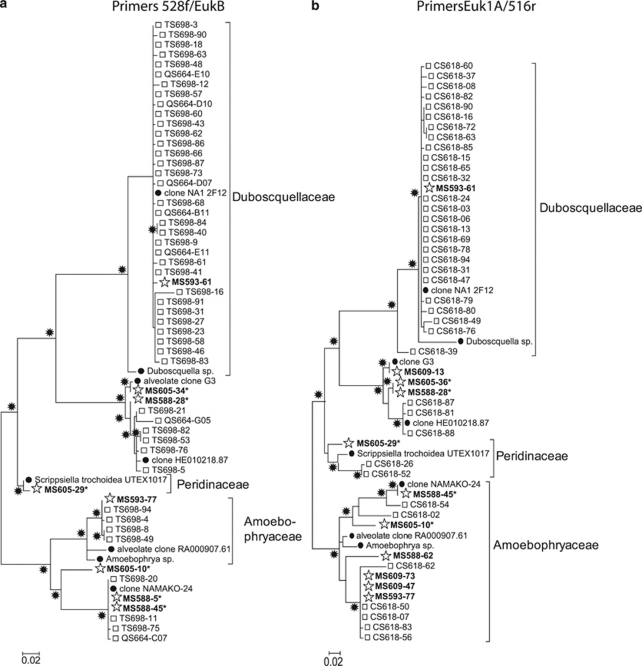
Compared with the clone libraries, SAGs represented a greater number of protist phyla (7 versus 5), even though the total number of 18S rRNA sequences obtained from SAGs was less than one third the total number of clones (Figure 3). Clone libraries were dominated by Cercozoa, Dinophyceae and Stramenopiles, with a small number of Telonemida and Choanoflagellida. In addition to those groups, the SAG library also contained six representatives of Picobiliphyta and one member of Katablepharidaceae, which were absent in clone libraries. Dinophyceae and Cercozoa constituted a consistently larger fraction of clones compared with SAGs. Both clone libraries contained large numbers (33 and 26) of identical and nearly identical sequences closely related to the dinophyte Duboscquella (Figure 4). In contrast, only one of the SAGs represented this cluster. Likewise, several other phylogenetic clusters contained significantly larger numbers of near-identical clones compared with SAGs, in particular, those among Dinophyceae and Cercozoa (Figure 4, Supplementary Figures A–C).
Figure 3.
Comparison of community composition of heterotrophic protists determined by clone libraries and SAG libraries. Results are shown from 18S rRNA gene primer pairs: (a) 528f/EukB and (b) Euk1A/516r. Phyla abbreviations are (clockwise, from 12 o'clock): Cercozoa, Dinophyceae, Telonemida, Choanoflagellida, Picobiliphyte, Stramenopile and Katablepharidaceae. In both cases SAG libraries revealed more diversity than clone libraries.
Figure 4.
Dinophyceae phylogenetic tree of 18S rDNA sequences obtained from clones (squares), SAGs (stars and bold) and their closest relatives in GenBank (closed circles) using (a) 528f/EukB and (b) Euk1A/516r primer pairs. Stars at nodes indicate bootstrap values >70%. Asterisks (*) indicate single cells lysed by KOH, all others were lysed using Lyse-N-Go.
The composition of SAGs detected with the Euk1A/516r and 528f/EukB primer sets was very similar. Moreover, there was no obvious difference in the phylogenetic composition of SAGs obtained using Lyse-N-Go or KOH lysis protocols (Figure 4, Supplementary Figures A–C). In contrast, there were distinct differences between clone libraries constructed with the two primer sets. Most notably, Stramenopiles constituted a larger fraction of clones obtained with Euk1A/516r (29% versus 8%), whereas Cercozoa were more prevalent among the clones constructed using 528f/EukB primer set (44% versus 23%).
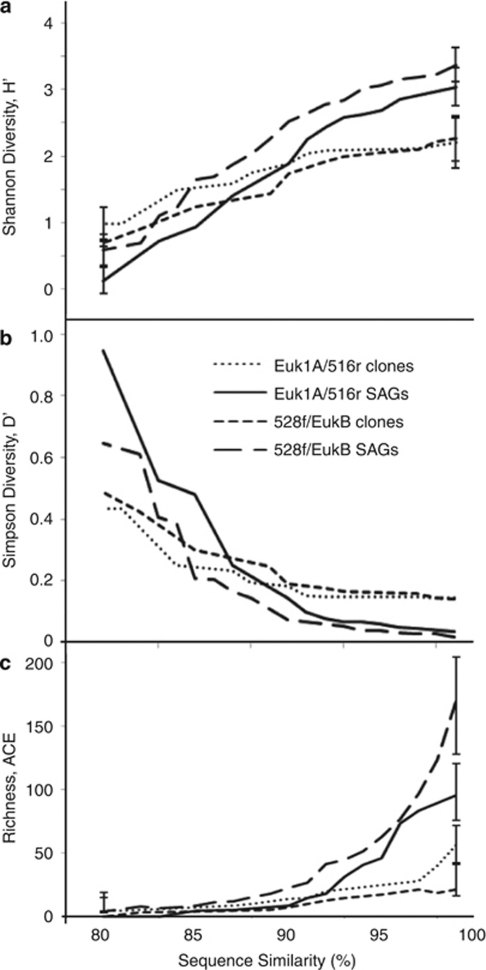
The phylogenetic diversity differed significantly between the SAG and clone libraries from both the Euk1A/516r (P=0.4) and the 528f/EukB (P<0.001) primer sets as determined by UniFrac significance tests. We found greater diversity among SAGs than clones using Shannon and Simpson diversity indices (H and D) when sequence identity thresholds delineating operational taxonomic units were set above 90% (Figures 5a and b). Likewise, the abundance-based coverage estimator indicated significantly higher species richness for SAGs compared with clones with sequence identity thresholds >92% and >80% for the Euk1A/516r and 528f/EukB primer sets, respectively (Figure 5c). In the case of all three indices, no significant differences between SAG and clone libraries were detected when using lower sequence identity thresholds for operational taxonomic unit delineation. Rarefaction analysis calculated using sequence similarity of 99% (Supplementary Figure D) indicated that neither SAG nor clone libraries were sufficiently large to represent the majority of protist taxa in the analyzed sample.
Figure 5.
Shannon (a) and Simpson (b) diversity indices and abundance-based coverage estimator (ACE) richness estimator (c) for clone and SAG libraries as a function of the sequence similarity threshold to define an operational taxonomic unit (OTU). Error bars in a and c represent 95% confidence intervals (only shown at 82% and 97% similarity, for clarity).
Discussion
The diversity and community composition of marine heterotrophic protists remains poorly understood. Protists are critical links in microbial food webs, and these trophic interactions control the biological carbon pump in the ocean. They may drive the structure of marine macrobial assemblages, and high diversity may add ecological resiliency to these key functions (Caron and Countway, 2009).
Two aspects distinguish our study from previous research: (1) we focused specifically on small (<10 μm), heterotrophic eukaryotes, rather than the entire microbial community and (2) we employed a novel, single-cell sequencing methodology and compared the results with the traditional PCR-based cloning.
At the phylum level, the composition of SAGs and clones obtained in this study corroborates previous reports from coastal waters, such as the abundance of Cercozoa, Stramenopiles and Alveolates (Romari and Vaulot, 2004). The absence of ciliates in this study confirms successful fluorescence-activated cell sorting (FACS) separation of pico- and nanoplankton from larger cells. Even though we sorted cells without detectable chlorophyll fluorescence, several of the obtained SAGs and clones are closely related to well-known phototrophs, for example, Scrippsiella sp. It is possible that some of these organisms represent phylogenetic lineages that recently lost phototrophy. Alternatively, these cells may be phototrophs (or mixotrophs) with pigment fluorescence below FACS detection limit due to photobleaching (Zvezdanovic et al., 2009) or other physiological suppression of fluorescence. The rarefaction analysis demonstrates that neither SAG nor clone libraries were sufficiently large to assess the complete diversity of the analyzed protist sample at an operationally defined ‘species' phylogenic resolution of 99% sequence similarity (Supplementary Figure D). This has been found in previous studies (Romari and Vaulot, 2004; Behnke et al., 2006; Countway et al., 2007) and highlights the high microbial diversity still to be uncovered even in waters, such as the Gulf of Maine, with a long history of traditional plankton research (Bigelow, 1924). The organisms captured by our SAG and clone libraries should, therefore, be considered as representatives of the most abundant taxa in the studied sample.
The recently discovered and yet uncultured Picobiliphytes (Not et al., 2007b) comprised 17% of the SAGs but were absent from the clone libraries (Figure 3). This raises the possibility that traditional, environmental PCR-based methods may have underrepresented this group of protists in previous studies as well. They may have fewer ribosomal gene copies, leading them to be underestimated in clone libraries relative to eukaryotes with high copy numbers. Picobiliphytes have so far been identified by fluorescence in situ hybridization in low abundance (less than 1%) from the Arctic Ocean, the Norwegian Sea and coastal European waters (Not et al., 2007b). The size of these organisms has been the subject of recent discussion, with related sequences obtained from cells in the nanoplankton size range (2–20 μm). These larger ‘biliphytes' were found in greater abundance (28% of all protist clones) in tropical eddy-influenced surface waters (Cuvelier et al., 2008). Our single-cell sequencing-based analysis indicates that biliphytes comprised a significant fraction of small protists in the studied sample from the Gulf of Maine. Optical properties of these cells suggest absence of photosynthetic pigments, which would contradict the original observations of picobiliphyte pigmentation (Not et al., 2007b). It is possible, however, that the cells had reduced chlorophyll fluorescence due to photobleaching, or some other photophysiological state of the cells. We are currently conducting further molecular studies of these SAGs to verify the intriguing possibility that some picobiliphytes are true heterotrophs.
Several SAGs and clones clustered with the recently identified Stramenopile group MAST-3 (Massana et al., 2004a, 2004b). One clone was also related to the MAST-2 group (Supplementary Figure B). Both groups, especially MAST-3, appear widespread in oceanic and coastal waters. However, not a single representative of these groups has yet been cultured. One SAG was closely related to the heterotrophic flagellate Leucocryptos marina (Clay and Kugrens, 1999), representing Katablepharidaceae (Supplementary Figure C). We obtained no clones representing this phylum. Choanoflagellida were represented by two SAGs and one clone (Supplementary Figure C). This group of exclusively heterotrophic protists was found in low abundance in surface waters of the Sargasso Sea (Countway et al., 2007), coastal waters of the English Channel (Romari and Vaulot, 2004) and the NW Mediterranean (Massana et al., 2004b). Several SAGs and clones clustered with Telonemida, another recently described group of protists with poorly understood biology (Supplementary Figure C; Shalchian-Tabrizi et al., 2006) global Ocean Sampling.
Single-cell sequencing revealed higher diversity of marine heterotrophic protists compared with environmental PCR clone libraries in the same sample from the Gulf of Maine. This is evidenced by the higher number of protist phyla, higher Shannon and abundance-based coverage estimator indices and lower Simpson index obtained from the SAG library compared with the clone libraries (Figures 3 and 5). Relative to SAGs, clone libraries were overrepresented by Dinophyceae, Cercozoa and certain groups of Stramenopiles (Figure 3, Supplementary Figure B). The same cell sorting and lysis methods were used generating SAGs and clones and the same PCR primers were used to generate clones and to screen SAGs. No other steps were involved in SAG analysis that could generate phylogenetic biases. Therefore, the discrepancy in SAG versus clone composition most likely indicates biases in clone libraries. A similar bias towards dinoflagellates in clone libraries has been reported previously in a study of an artificial microbial assemblage (Potvin and Lovejoy, 2009). As discussed above, environmental PCR clone libraries are susceptible to multiple sources of biases, such as variable target gene copy number per cell, preferential PCR amplification and variable cloning efficiency. The number of 18S rDNA gene copies in protists can vary by at least four orders of magnitude, with some correlation to genome size (Prokopowich et al., 2003) and cell size (Zhu et al., 2005). Unfortunately, information on the 18S gene copy number for most of the protists closely related to those detected in this study is not known. The likely role of variable gene copy number per cell is supported by the presence of several clusters of identical or nearly-identical sequences among clones, but not SAGs, which are insensitive to per-cell gene copy number bias. For example, the cluster of close relatives to Duboscquella contains many clones from both Euk1A/516r and 528f/EukB clone libraries, but has only one SAG (Figure 4). The consistency of the two clone libraries suggests that multiple copies of 18S rRNA genes in Duboscquella likely contributed to the cluster formation. Thus, previous findings of large fractions of clones and pyrotags being related to Duboscquella and other parasitic dinophytes (Harada et al., 2007; Brown et al., 2009) may overestimate the contribution of these groups to the composition of marine protist communities. Alternatively, differences in Duboscquella frequency in clone versus SAG libraries may have been caused by multiple parasitic cells residing inside individual, sorted protists. In the latter case, a larger number of parasites per host cell would increase parasite sequence frequency in clone, but not SAG libraries. However, this explanation is less likely, as the sorted cells may be too small to host Duboscquella–like parasites, known to infect ciliates (Harada et al., 2007; Brown et al., 2009). Some other phylogenetic groups exhibited substantial compositional differences between the two clone libraries, suggesting that PCR and/or cloning biases also had a function (Figures 3 and 4, Supplementary Figure A–C). This corroborates with the recent comparison of multiple clone libraries constructed using artificial and natural protist assemblages, demonstrating primer-specific biases in the 18S rRNA gene clone library composition (Potvin and Lovejoy, 2009). The comparative analysis of SAG and clone libraries provided here improves our understanding of microbial community composition and pinpoints specific limitations of current methodology that need to be considered in future studies.
To our knowledge, this is the first reported use of single-cell sequencing to examine protistan community composition. Although a large number of culture-independent, molecular techniques have been developed for this purpose during the last two decades, all of them are prone to significant limitations (Table 2). For example, the now traditional clone libraries and the more recent pyrotag sequencing rely on environmental PCR, which is known to cause biased representation of the various phylogenetic groups (Suzuki and Giovannoni, 1996; Potvin and Lovejoy, 2009). In addition, microbial community composition studies that are based on either environmental PCR (including qPCR) or on PCR-independent metagenomics may be significantly biased because of the highly variable per-cell copy number of phylogenetic marker genes, as discussed above. Fluorescence in situ hybridization is not susceptible to the variable gene copy number bias. However, fluorescence in situ hybridization has other serious limitations, such as bias against cells with few ribosomes and fragile cells, challenges in designing appropriate negative controls, typically low phylogenetic resolution and low throughput (the application of each probe requires a significant effort). Other limitations faced by the available methods include biases caused by the variable propensity of cells to lyse/permeabilize, the reliance of primer/probe design on limited databases, cloning biases and the need for specialized, costly equipment. Put in this context, SAG analysis offers a significant step forward in the quantitative analysis of protist community composition. It is not susceptible to the variable gene or ribosome copy number, does not rely on cloning and can easily incorporate suitable controls. The method is amenable to high throughput and automation, as demonstrated by the establishment of the Bigelow Single Cell Genomics Center (www.bigelow.org/scgc). When FACS is used for cell separation, light scatter or fluorescence can be employed to narrow the selection of target microorganisms, based on logical combinations of cell size, autofluorescence (for example, chlorophyll) and fluorescent probes (for example. Lysotracker and nucleic acid stains). A unique advantage of SAG analysis is the unlimited phylogenetic resolution, that is, multiple genes or entire genomes can be sequenced from a single cell to obtain sufficient hereditary information, independent of cultivability or community complexity. This opens unique opportunities for ecology, evolution and bioprospecting research, as demonstrated for prokaryotes in previous studies (Raghunathan et al., 2005; Zhang et al., 2006; Marcy et al., 2007; Stepanauskas and Sieracki, 2007; Woyke et al., 2009). After their generation by FACS–MDA, and after the initial screen by PCR or other methods, SAG libraries represent a long-term resource. The genomic DNA, amplified from individual cells, can be stored, re-amplified and used in an unlimited number of PCR reactions, shotgun sequencing, hybridizations, cloning-expression and other molecular biology studies in the same way as DNA extracted from pure cultures.
Table 2. Potential susceptibility of current methods to biases and other limitations for assessing protist community composition in environmental samples.
| PCR clones/454 tags | Metagenomics | qPCR | FISH | SAGs | |
|---|---|---|---|---|---|
| Sources of potential biases | |||||
| Cell permeability | + | + | + | + | + |
| Gene copy number | ++ | ++ | ++ | − | − |
| Primers and probes | ++ | − | ++ | ++ | + |
| Cloning efficiency | +/− | + | − | − | − |
| Ribosome number | − | − | − | + | − |
| Inadequate controls (false positives) | − | − | ++ | ++ | − |
| Other limitations | |||||
| Low phylogenetic resolution | +/++ | + | − | + | − |
| High cost per sample | − | + | + | ++ | + |
| Specialized equipment needed | −/+ | + | + | − | ++ |
Abbreviations: FISH, fluorescence in situ hybridization; qPCR, quantitative real-time; SAG, single amplified genome.
The susceptibility is designated as high (++), moderate (+) or absent (−).
The current implementation of SAG analysis is not entirely free from possible biases. Only 25% of the analyzed SAGs produced 18S rRNA gene sequences, and at the moment we have no proof whether these 25% are a representative subsample of the sorted cells. As PCR was used to amplify the 18S rRNA gene from SAGs, this approach is still susceptible to some PCR limitations, such as the reliance on existing databases in primer design. The similarity of SAG composition determined using either Euk1A/516r or 528f/EukB primer sets for major phyla is encouraging (Figure 3). However, we cannot exclude the possibility that some specific groups did not amplify with either of the primer sets because of sequence mismatches or inhibitory DNA secondary or tertiary structures (Potvin and Lovejoy, 2009). Incomplete cell lysis of specific taxonomic groups may be another factor leading to biases in SAG composition. However, the absence of phylogenetic differences between SAGs obtained using KOH and Lyse-N-Go protocols in this study indicates low likelihood for such biases among the studied organisms (Figure 4, Supplementary Figures A–C). Other potential reasons behind the <100% recovery rate of 18S rRNA genes from SAGs include (1) sorting of non-target cells or non-living particles, (2) unsuccessful droplet deposition by FACS and (3) sequence-unspecific failures of individual MDA and PCR reactions. None of the latter processes would bias SAG composition, as they are agnostic to the cell type. The work reported here was conducted in 96-well plates, but we now routinely sort into 384-well plates with the MoFlo sorter. We use a fluorescent bead microscope method to confirm droplet deposition by the sorter into these wells, which are about half the diameter of those in 96-well plates. This method routinely shows that the rate of unsuccessful droplet deposition into 384-well plates is well below 5%. It is unlikely that cell sorting misses account for many of the well failures. Further work is clearly needed to improve the success rate in SAG production and identification. In this context, a significant advantage of single-cell sequencing, compared with other methods, is the robust, quantitative information on what fraction of the total microbial community is represented by a SAG library.
Single-cell sequencing eliminates many biases associated with environmental PCR clone libraries and pyrotagging, such as preferential amplification of certain taxa and the over-representation of taxa with high target gene copy number. In difference to other cultivation-independent methods, a SAG library can be screened by multiple primer/probe sets or subject to whole genome shotgun sequencing, allowing for multi-locus sequence analysis and metabolic pathway reconstruction of the uncultured taxa.
Acknowledgments
We thank Ramon Massana for his advice and for participating in useful discussions relating to this project. This work was funded by NSF grants EF-0633142 and OCE-0623288, and a State of Maine, Maine Technology Institute infrastructure grant.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriology. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA Genes. PLoS ONE. 2009;4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F, Fenchel T, Field JG, Meyer-Reil LA, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Progr Ser. 1983;10:257–263. [Google Scholar]
- Behnke A, Bunge J, Barger K, Breiner HW, Alla V, Stoeck T. Microeukaryote community patterns along an O2/H2S gradient in a supersulfidic fjord (Framvaren, Norway) Appl Environ Microbiol. 2006;72:3626–3636. doi: 10.1128/AEM.72.5.3626-3636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegala IC, Not F, Vaulot D, Simon N. Quantitative assessment of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescence in situ hybridization and flow cytometry. Appl Environ Microbiol. 2003;69:5519–5529. doi: 10.1128/AEM.69.9.5519-5529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow HB.1924Plankton of the offshore waters of the Gulf of Maine Bull Bureau of Fisheries 40Part II. US Govt. Printing Office: Washington, DC [Google Scholar]
- Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM, et al. Microbial community structure in the North Pacific ocean. ISME Journal. 2009;3:1374–1386. doi: 10.1038/ismej.2009.86. [DOI] [PubMed] [Google Scholar]
- Buck KR, Bentham WN. A novel symbiosis between a cyanobacterium, Synechococcus sp., an aplastidic protist, Solenicola setigera, and a diatom, Leptocylindrus mediterraneus, in the open ocean. Mar Biol. 1998;132:349–355. [Google Scholar]
- Caron DA, Countway PD. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat Microb Ecol. 2009;57:227–238. [Google Scholar]
- Chao A, Lee SM. Estimating the number of classes via sample coverage. J Amer Statist Assoc. 1992;87:210–217. [Google Scholar]
- Chao A, Ma MC, Yang MCK. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika. 1993;80:193–201. [Google Scholar]
- Clay BL, Kugrens P. Systematics of the enigmatic kathablepharids, including EM characterization of the type species, Kathablepharis phoenikoston, and new information on K. remigera comb. nov. Protist. 1999;150:43–59. doi: 10.1016/S1434-4610(99)70008-8. [DOI] [PubMed] [Google Scholar]
- Countway PD, Gast RJ, Dennett MR, Savai P, Rose JM, Caron DA. Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream) Envron Microbiol. 2007;9:1219–1232. doi: 10.1111/j.1462-2920.2007.01243.x. [DOI] [PubMed] [Google Scholar]
- Cuvelier ML, Ortiz A, Kim E, Moehlig H, Richardson DE, Heidelberg JF, et al. Widespread distribution of a unique marine protistan lineage. Environ Microbiol. 2008;10:1621–1634. doi: 10.1111/j.1462-2920.2008.01580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al. Comprehensive human genome amplification using multiple displacement amplification. PNAS. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro F, Gaedke U, Boenigk J. Reverse evolution: driving forces behind the loss of acquired photosynthetic traits. PLoS ONE. 2009;4:e8465. doi: 10.1371/journal.pone.0008465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36 (Web Server issue:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez B, Pedros-Alio C, Massana R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwood HJ, Olsen GJ, Sogin ML. The small-subunit ribosomal RNA gene sequences from the Hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Mol Biol Evol. 1985;2:399–410. doi: 10.1093/oxfordjournals.molbev.a040362. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, McManus GB. Do bacteria-sized marine eukaryotes consume significant bacterial production. Science. 1984;224:1257–1260. doi: 10.1126/science.224.4654.1257. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, DeLong EF, Olsen GJ, Pace NR. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriology. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Ohtsuka S, Horiguchi T. Species of the parasitic genus Duboscquella are members of the enigmatic marine Alveolate Group I. Protist. 2007;158:337–347. doi: 10.1016/j.protis.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Jardillier L, Bettarel Y, Richardot M, Bardot C, Amblard C, Sime-Ngando T, et al. Effects of viruses and predators on prokaryotic community composition. Microbial Ecol. 2005;50:557–569. doi: 10.1007/s00248-005-5030-y. [DOI] [PubMed] [Google Scholar]
- Jeong HJ, Seong KA, Yoo YD, Kim TH, Kang NS, Kim S, et al. Feeding and grazing impact by small marine heterotrophic dinoflagellates on heterotrophic bacteria. J Eukary Microbiol. 2008;55:271–288. doi: 10.1111/j.1550-7408.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- Li WKW. Primary production of prochlorophytes, cyanobacteria, and Eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Ocean. 1994;39:169–175. [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, et al. Dissecting biological ‘dark matter' with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. PNAS. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranon E, Holligan PM, Barciela R, Gonzalez N, Mourino B, Pazo MJ, et al. Patterns of phytoplankton size structure and productivity in contrasting open-ocean environments. Mar Ecol Prog Ser. 2001;216:43–56. [Google Scholar]
- Massana R, Balague V, Guillou L, Pedros-Alio C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol Ecol. 2004b;50:231–243. doi: 10.1016/j.femsec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Massana R, Castresana J, Balague V, Guillou L, Romari K, Groisillier A, et al. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl Environ Microbiol. 2004a;70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryote diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- Not F, del Campo J, Balagué V, de Vargas C, Massana R. New insights into the diversity of marine picoeukaryotes. Plos ONE. 2009;4:e7143. doi: 10.1371/journal.pone.0007143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ Microbiol. 2007a;9:1233–1252. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- Not F, Valentin K, Romari K, Lovejoy C, Massana R, Tobe K, et al. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science. 2007b;315:253–255. doi: 10.1126/science.1136264. [DOI] [PubMed] [Google Scholar]
- Pinard R, de Winter A, Sarkis GJ, Gerstein MB, Tartaro KR, Plant RN, et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin M, Lovejoy C. PCR-based diversity estimates of artificial and environmental 18S rRNA gene libraries. J Eukaryot Microbiol. 2009;56:174–181. doi: 10.1111/j.1550-7408.2008.00386.x. [DOI] [PubMed] [Google Scholar]
- Prokopowich CD, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50. doi: 10.1139/g02-103. [DOI] [PubMed] [Google Scholar]
- Raghunathan A, Ferguson HR, Jr, Bornarth CJ, Song W, Driscoll M, Lasken RS. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Giver LJ, Wickham GS, Pace NR. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Ocean. 2004;49:784–798. [Google Scholar]
- Rose JM, Caron DA, Sieracki ME, Poulton N. Counting heterotrophic nanoplanktonic protists in cultures and aquatic communities by flow cytometry. Aquat Micob Ecol. 2004;34:263–277. [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Eikrem W, Klaveness D, Vaulot D, Minge MA, Le Gall F, et al. Telonemia, a new protist phylum with affinity to chromist lineages. Proc Royal Soc B: Biological Sciences. 2006;273:1833–1842. doi: 10.1098/rspb.2006.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Kauserud H, Massana R, Klaveness D, Jakobsen KS. Analysis of environmental 18S ribosomal RNA sequences reveals unknown diversity of the cosmopolitan phylum Telonemia. Protist. 2007;158:173–180. doi: 10.1016/j.protis.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Gunderson JH. Structural diversity of eukaryotic small subunit ribosomal RNAs. Ann N Y Acad Sci. 1987;503:125–139. doi: 10.1111/j.1749-6632.1987.tb40603.x. [DOI] [PubMed] [Google Scholar]
- Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. PNAS. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist. 2006;157:31–43. doi: 10.1016/j.protis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tarran GA, Heywood JL, Zubkov MV. Latitudinal changes in the standing stocks of nano- and picoeukaryotic phytoplankton in the Atlantic Ocean. Deep-Sea Res II. 2006;53:1516–1529. [Google Scholar]
- Vigil P, Countway PD, Rose J, Lonsdale DJ, Gobler CJ, Caron DA. Rapid shifts in dominant taxa among microbial eukaryotes in estuarine ecosystems. Aquat Microb Ecol. 2009;54:83–100. [Google Scholar]
- Worden AZ. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat Microb Ecol. 2006;43:165–175. [Google Scholar]
- Woyke T, Xie G, Copeland A, Gonzalez JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokawa T, Nagata T. Growth and grazing mortality rates of phylogenetic groups of bacterioplankton in coastal marine environments. Appl Environ Microbiol. 2005;71:6799–6807. doi: 10.1128/AEM.71.11.6799-6807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. Mapping of picoeukaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Zubkov MV, Tarran GA. High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature. 2008;455:224–226. doi: 10.1038/nature07236. [DOI] [PubMed] [Google Scholar]
- Zvezdanovic J, Cvetic T, Veljovic-Jovanovic S, Markovic D. Chlorophyll bleaching by UV-irradiation in vitro and in situ: absorption and fluorescence studies. Rad Phys Chem. 2009;78:25–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.