Bacteria have long been believed to exist as separate, individual cells that take up nutrients as they are available and multiply if conditions are favorable. However, the view that these organisms are unable to undergo complex interactions is gradually being abandoned as scientists are gaining insight into the fact that bacteria can sense and respond to their environment and to each other and that they are capable of coordinated activity. One of the mechanisms that has recently received much attention is quorum sensing—bacterial cell-to-cell communication. Quorum sensing is a mechanism of gene regulation in which bacteria coordinate the expression of certain genes in response to the presence or absence of small signal molecules (Defoirdt et al., 2008). One of the signal molecules involved in bacterial quorum sensing is autoinducer 2 (AI-2). AI-2 quorum sensing has been documented in different species belonging to highly diverse taxa, both Gram-negative and Gram-positive (De Keersmaecker et al., 2006), and was reported about a decade ago in the gastrointestinal bacterium Escherichia coli (Surette and Bassler, 1998). In E. coli, AI-2 induces transcription of the lsrACDBFGE operon, which encodes proteins involved in import, phosphorylation and further degradation of the signal molecule itself (Xavier and Bassler, 2005).
AI-2 has also been reported to regulate motility and flagella synthesis in E. coli (Ren et al., 2001; Sperandio et al., 2001). Interestingly, Bansal et al. (2008) recently reported that enterohemorrhagic E. coli are attracted by the quorum sensing signal AI-2 in an agarose plug assay. Very recently, Englert et al. (2009) reported that a 0–500-μ gradient of AI-2 was almost as effective as an attractant as a 0–100-μ gradient of -aspartate, a known attractant, in a microfluidic device. Based on these findings, one might hypothesize that E. coli uses AI-2 to actively join groups of bacteria (Figure 1). Indeed, a cell will start producing flagellae when it detects a certain concentration of AI-2. The cell will subsequently move toward increasing concentrations of AI-2, which are likely to be encountered close to clusters of AI-2-producing cells. When the source of AI-2 is reached, the concentration of AI-2 will be relatively high. At high AI-2 concentrations, E. coli starts active uptake and degradation of AI-2, which results in decreased production and activity of the motility apparatus (Bansal et al., 2008). As motility is an energy-consuming process, inactivation of AI-2-driven chemotaxis could explain why these bacteria produce a signal that activates its own degradation.
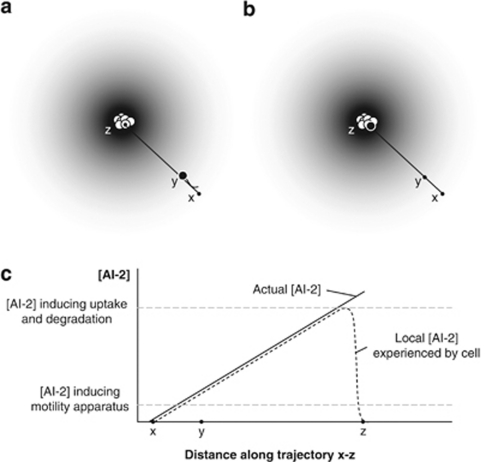
Figure 1.
Hypothesis of active group formation in E. coli based on AI-2 quorum sensing. A cluster of cells producing the signal molecule AI-2 will generate a spheric concentration gradient (indicated by the gray color). (a) AI-2 induces flagella synthesis and, therefore, an E. coli cell sensing AI-2 will start producing structures involved in motility (black cell). Because AI-2 attracts E. coli, the cell will start moving toward the source of the signal. (b) High concentrations of AI-2 induce active uptake and degradation of the signal. As a result, expression of motility structures will decrease and the cell will stop moving when it has reached a location with a high concentration of AI-2. This will occur when it has reached the cluster of AI-2-producing bacteria. (c) AI-2 concentration profile along trajectory x–z in panels a and b. The full line shows the actual AI-2 concentration profile, which increases as the black cell approximates the cell cluster. The dotted line shows the local concentration experienced by the black cell, which increases as the AI-2 concentration surrounding the cell increases. At a certain concentration, however, uptake and degradation of the signal is activated, resulting in a decrease of the local AI-2 concentration experienced by the cell and a consequent decrease in motility.
Further in-depth research is necessary to confirm this intriguing hypothesis that E. coli cells cannot only sense and respond to other bacterial cells, but also actively search to join groups of bacteria. A critical issue is the concentration of AI-2 needed to activate the subsequent processes. Indeed, in order for this mechanism to work, activation of chemotaxis towards AI-2 should start at lower AI-2 concentrations than uptake and degradation of the signal. Another issue is the extent to which degradation of AI-2 results in a decrease of local AI-2 levels to below the level needed for activation of the motility apparatus. If this mechanism works, it would be interesting to investigate over what distances it is active and under what environmental conditions it is important. In the case of E. coli, this could be the gastrointestinal tract of vertebrates and/or the external environment. Finally, as AI-2 has been reported to regulate the motility of other bacteria such as Campylobacter jejuni and Helicobacter pylori as well (Elvers and Park, 2002; Rader et al., 2007), this active grouping behavior might not be restricted to E. coli and might add yet another dimension to microbial ecology.
Acknowledgments
I, a postdoctoral fellow of FWO-Vlaanderen, thank the ‘Fonds voor Wetenschappelijk Onderzoek' for financial support. Special thanks also go to Greet Dewaele for critically reading the manuscript.
The author declares no conflict of interest.
References
- Bansal T, Jesudhasan P, Pillai S, Wood TK, Jayaraman A. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol. 2008;78:811–819. doi: 10.1007/s00253-008-1359-8. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker SCJ, Sonck K, Vanderleyden J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006;14:114–119. doi: 10.1016/j.tim.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. Quorum sensing and quorum quenching in Vibrio harveyi: lessons learned from in vivo work. ISME J. 2008;2:19–26. doi: 10.1038/ismej.2007.92. [DOI] [PubMed] [Google Scholar]
- Elvers KT, Park SF. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology. 2002;148:1475–1481. doi: 10.1099/00221287-148-5-1475. [DOI] [PubMed] [Google Scholar]
- Englert DL, Manson MD, Jayaraman A. Flow-based microfluidic device for quantifying bacterial chemotaxis in stable, competing gradients. Appl Environ Microbiol. 2009;75:4557–4564. doi: 10.1128/AEM.02952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The quorum sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol. 2007;189:6109–6117. doi: 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Sims J, Wood TK. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5 H)-furanone. Environ Microbiol. 2001;3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Giron JA, Kaper JB. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli (EHEC) O157:H7. J Bacteriol. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]