Abstract
Crustose coralline algae (CCA) are key reef-building primary producers that are known to induce the metamorphosis and recruitment of many species of coral larvae. Reef biofilms (particularly microorganisms associated with CCA) are also important as settlement cues for a variety of marine invertebrates, including corals. If rising sea surface temperatures (SSTs) affect CCA and/or their associated biofilms, this may in turn affect recruitment on coral reefs. Herein, we report that the CCA Neogoniolithon fosliei, and its associated microbial communities do not tolerate SSTs of 32 °C, only 2–4 °C above the mean maximum annual SST. After 7 days at 32 °C, the CCA exhibited clear signs of stress, including bleaching, a reduction in maximum quantum yield (Fv/Fm) and a large shift in microbial community structure. This shift at 32 °C involved an increase in Bacteroidetes and a reduction in Alphaproteobacteria, including the loss of the primary strain (with high-sequence similarity to a described coral symbiont). A recovery in Fv/Fm was observed in CCA exposed to 31 °C following 7 days of recovery (at 27 °C); however, CCA exposed to 32 °C did not recover during this time as evidenced by the rapid growth of endolithic green algae. A 50% reduction in the ability of N. fosliei to induce coral larval metamorphosis at 32 °C accompanied the changes in microbiology, pigmentation and photophysiology of the CCA. This is the first experimental evidence to demonstrate how thermal stress influences microbial associations on CCA with subsequent downstream impacts on coral recruitment, which is critical for reef regeneration and recovery from climate-related mortality events.
Keywords: crustose coralline algae, biofilms, temperature, microbes, bleaching, coral settlement
Introduction
Larvae of numerous marine invertebrate species, including sponges, echinoderms, polychaetes, barnacles, molluscs, tunicates, bryozoans and cnidarians settle and metamorphose in response to bacterial biofilms (reviewed by Wieczorek and Todd, 1998). Coral larvae actively select their site of permanent attachment using chemical cues to induce metamorphosis (Morse et al., 1988, 1994, 1996; Heyward and Negri, 1999). The primary source of these chemical morphogens is various species of non-geniculate calcareous coralline algae (CCA). Bacterial biofilms occur on the surface of CCA (Galbary and Veltkamp, 1980; Garland et al., 1985; Lewis et al., 1985; Johnson et al., 1991) and also have a role in inducing larval metamorphosis for some invertebrates. A strain of Pseudoalteromonas, isolated from CCA, is able to induce significant levels of metamorphosis of coral larvae (Acropora willisea and A. millepora) (Negri et al., 2001); numerous bacterial strains from CCA induce settlement of the sea urchin Heliocidaris erythrogramma (Huggett et al., 2006) and metamorphosis of crown-of-thorns starfish larvae (Acanthaster planci) is also triggered by microorganisms associated with CCA (Johnson and Sutton, 1994).
Although it is well-established that settlement and recruitment of marine invertebrates varies with biotic and abiotic factors (hydrodynamics, light, temperature, gravity, surface texture and presence of conspecifics) (Rodriguez et al., 1993), little work has been done to test whether environmental variables affect the inductive ability of marine microbial biofilms. Biofilm age and depth can affect the inductive and inhibitory capacities of biofilms (Wieczorek and Todd, 1997; Webster et al., 2004), and localised anthropogenic impact can also shift the biofilm composition (Meyer-Reil and Koster, 2000; Lawrence et al., 2004) and potentially influence subsequent invertebrate recruitment (Webster et al., 2006). However, to date no research has addressed how variables associated with a changing climate (elevated sea-surface temperature (SST), ocean acidification and eutrophication) will affect the inductive ability of marine microbial biofilms.
The potential impacts of climate change on marine microorganisms are rarely assessed, despite the critical contribution of microbes to marine biomass, diversity and ecosystem function. One of the primary effects of climate change is a projected 1.8–4 °C increase in global air temperature by 2100, along with similar rises in SST (range of best estimates from the six emissions marker scenarios of Special Report on Emission Scenarios (SRES)) (IPCC, 2007). Elevated sea temperatures could significantly impact on marine microbes, potentially altering microbial diversity, function and community dynamics (Webster and Hill, 2007). Elevated SST may affect recruitment in a number of ways: (1) by altering the ability of microbes to produce morphogenic signalling compounds, or (2) by causing a shift in the community composition and microbial succession of the biofilm, which may adversely (or positively) affect subsequent recruitment of macro-organisms, including corals. These outcomes could have serious implications for reef-building, maintenance and recovery processes. Environmental conditions that adversely affect the distribution and abundance of microbes involved in settlement and metamorphosis of reef invertebrates could therefore have large-scale impacts on ecosystem structure and the distribution and reproductive fitness of some keystone species.
Owing to the importance of both inert (sediment-associated) and CCA-associated biofilms in the recruitment of coral larvae, this study aimed to (1) investigate the impact of elevated SST on the microbial biofilms of CCA and reef sediments and (2) assess whether the ability of CCA to induce metamorphosis in coral larvae can be affected by exposure to elevated SST.
Materials and methods
Sample collection and temperature exposure
Fragments of the crustose coralline algae Neogoniolithon fosliei (8 × 8 cm) and 1 kg of surface sediments were collected between 5–8 m at Davies Reef (19°09. 35′S; 146°52.87′E), Queensland, Australia in October 2008. The upper 2 cm of reef sediments were scraped into glass beakers using a sterile spatula. All samples were transported in flowing seawater to the indoor temperature-controlled aquarium at the Australian Institute of Marine Science, Townsville, Australia. CCA discs of identical size were prepared using a 20-mm diamond hole saw.
The experimental design incorporated four temperatures (27 °C, 29 °C, 31 °C and 32 °C; range ±0.2 °C) in three replicate 30-l flow-throughs (600 ml min−1) aquaria per temperature. Sampling times were T=0 day (all treatments at 27 °C), T=1 day, T=2 days, T=7 days and T=14 days. Temperatures were maintained for the first 7 days and then all temperatures were returned to 27 °C for the final 7 days of the experiment as a recovery period. In each tank, there were 6 × 6 wells, 12-ml cell-culture plates (Iwaki, Japan) with three wells per plate containing reef sediment at 6 mm depth and the other three wells containing the CCA discs. An individual six-well plate was randomly removed per tank at each time point (nine total replicates per temperature at each time for each CCA and sediment). Incoming seawater was filtered to 1 μm. All samples were maintained under a diel cycle of (12:12 h) at 80 μmol quanta m−2 s−1, reflecting light intensity at 8 m on the reef. Initially, all aquaria were left at 27 °C for 72 h and then temperatures were adjusted gradually (0.15 °C h−1) until reaching the final temperatures (at T=0 day).
At each time point all CCA and sediment samples were photographed and frozen in liquid nitrogen for molecular analysis. Seawater samples (1 l) from each temperature at each time point were filtered through a 0.2-μm sterivex filter (Durapore, Millipore, North Ryde, New South Wales, Australia), which were filled with 1.8 ml of lysis buffer (40 m EDTA (pH 8.0), 50 m Tris and 0.75 M sucrose) and frozen at −80 °C.
Thermal stress can result in the loss of pigments (bleaching) in CCA (Anthony et al., 2008). Damage to photosystem II (PSII) of CCA was assessed using pulse amplitude modulation fluorometry. Fluorescence measurements were taken following a 30-min dark adaptation period using a Mini-PAM fluorometer (Walz, Effeltrich, Germany) by placing a 6-mm fibre-optic probe perpendicular to the centre of the CCA discs. Initial fluorescence (F0) was determined by applying a weak pulse-modulated red-measuring light (0.15 μmol photons m−2 s−1). To quantify maximum fluorescence (Fm), a short pulse (800 ms) of saturating actinic light (>3000 μmol photons m−2 s−1, 650 nm) was applied (Jones et al., 2003). Maximum quantum yield (Fv/Fm) of photochemical energy conversion in PSII was calculated based on F0 and Fm as per Equation (1) (Genty et al., 1989).
a reduction in Fv/Fm indicates damage to the D1 protein in PSII (Osmond et al., 1999).
Coral metamorphosis assays
Larvae were raised from gametes collected from live colonies of the reef-building coral A. millepora (Ehrenberg, 1834). Corals were collected by snorkelling from a depth of 6–8 m on a fringing reef of Pelorus Island, Great Barrier Reef (GBR) (18°32 S, 146°29 E) and transported to flow-through aquaria at the Australian Institute of Marine Science. Synchronous spawning occurred on 14 November 2008, and the released gametes, in the form of buoyant egg–sperm bundles, were cross-fertilised as described in Negri and Heyward (2000). Larvae were raised in a 500-l plastic tank with flowing 1 μm-filtered seawater at 27 °C. Larval metamorphosis assays were performed in 10-cm diameter sterile plastic petri dishes maintained at 28 °C. Coral larvae (8 days old) (n=10–20) were introduced to each dish containing a single CCA disc that had been exposed to each of the treatment temperatures for 7 days and seawater was added to a final volume of 30 ml. Larvae were defined as metamorphosed when they had changed from either free-swimming or casually-attached pear-shaped forms to squat, firmly-attached disk-shaped structures with pronounced flattening of the oral–aboral axis and typically obvious septal mesenteries radiating from the central mouth region (Heyward and Negri, 1999). A time period of 24 h was chosen as the end point for scoring early-stage metamorphosis.
Denaturing gradient gel electrophoresis
Frozen CCA tissue and reef sediments (approximately 0.5 g per sample) were aseptically transferred to 1.5 ml Eppendorf tubes using sterile scalpels. Grinding buffer (0.5 ml) was added to each replicate sample (100 m Tris (pH 9.0), 100 m EDTA (pH 8.0), 1% SDS and 100 m NaCl). Tubes were immersed in liquid nitrogen and ground with plastic pestles. Samples were incubated at 65 °C for 60 min before addition of 187 μl 5 M potassium acetate. Samples were incubated on ice for 30 min and centrifuged at 8000 g for 15 min. The supernatants were transferred to fresh tubes and DNA was precipitated with 0.8 vol of isopropanol. DNA was extracted from seawater filters by addition of 200 μl lysozyme (10 mg ml−1), incubation at 37 °C for 45 min, addition of 200 μl of proteinase K (0.2 μg ml−1) and 1% SDS and incubation at 55 °C for 1 h. Lysates were recovered into fresh eppendorf tubes and DNA was extracted with a standard phenol:chloroform:isoamyl alcohol procedure and precipitated with 0.8 vol of isopropanol.
The 16S rRNA gene from each CCA disc, sediment and seawater sample was amplified by PCR with primers 1055f and 1406r (Ferris et al., 1996). PCR reactions were performed as described by Ferris et al. (1996) and products from duplicate PCR reactions were combined and 15 μl applied to 8% w/v polyacrylamide (37.5:1) gels containing a 50–70% denaturing gradient of formamide and urea. Gels were electrophoresed at 60 °C for 17 h in 1 × TAE buffer at 50 V using the Ingeny D-Code system (Amundsenweg, The Netherlands) and stained with 1 × Sybr Gold for 30 min, visualised under UV illumination and photographed. The denaturing gradient gel electrophoresis (DGGE) banding profiles using these universal 16S rRNA gene primers could not be resolved with the CCA and sediment samples because of extreme bacterial diversity producing a smear of bands on the gels. Additional DGGE-PCR was conducted for the CCA samples with primers rpoB1698f and rpoB2041r specific to the rpoB gene (Dahllöf et al., 2000), F203α with 788R to amplify the 16S rRNA gene from the Alphaproteobacteria (Gelsomino and Cacco, 2006; Huys et al., 2008) and CFB319 with 788R to amplify the 16S rRNA gene from the Cytophaga–Flavobacteria–Bacteroidetes group (Fierer et al., 2005; Huys et al., 2008). The reverse primers rpoB2041 and 788R were modified to incorporate the GC clamp as per Ferris et al. (1996). DGGE analysis was performed as described above and band analysis was performed on Fragment NT Version 1.1a (Quantity One, Bio-Rad Laboratories Pty Ltd., Gladesville, New South Wales, Australia). Individual band numbers were assigned based on the bands migration relative to control bands. Bands assigned the same number had identical migration end points. Bands that did not migrate to the same point on duplicate gels were not considered in subsequent analyses.
The 18S rRNA gene from each CCA sample was also amplified by PCR with universal eukaryotic primers as follows: NS1f, 5′-GTAGTCATATGCTTGTCTC-3′ and NS2r, 5′-GGCTGCTGGCACCAGACTTGC-3′ (White et al., 1990). The PCR and DGGE were performed as described above with the exception that the products were electrophoresed on a 35–70% denaturing gradient.
16D rRNA gene cloning and sequencing
For a more comprehensive phylogenetic comparison between the different temperatures, the 16S rRNA gene from each CCA disc sampled at 2- and 7 day was amplified by PCR with primers 63f and 1387r (Marchesi et al., 1998). The PCR products were combined for all replicate CCA discs per temperature at each time point. To assess whether biotic (CCA) and abiotic (sediment) biofilms responded differently to elevated temperature, the sediment samples at 27 °C and 32 °C were also collected at 2- and 7 day and assessed by 16S rRNA gene sequence analysis. PCR products were ligated into the TOPO TA cloning vector (Invitrogen Australia Pty Ltd., Mulgrave, Victoria, Australia). Ligations were sent to the Australian Genome Research Facility (University of Queensland, Brisbane) for transformation and sequencing of 192 clones for each CCA library and 96 clones for each sediment library. Australian Genome Research Facility transformed the ligations into Genehog electrocompetent cells (Invitrogen), grew the cells in 2 × YT+ampicillin media, performed a standard alkaline lysis plasmid preparation and sequenced the clones with ABI Bigdye v3.1 chemistry on the AB3730xl (standard 50-cm array run module; Mulgrave, Victoria, Australia). Clone sequences were submitted to the Genbank under the accession numbers HM177481 to HM178920.
Phylogenetic analysis
Clone sequence quality was checked manually using Sequencher (Genesearch, Brisbane, Queensland, Austraila). Chimeric sequences were identified using the programs CHECK_CHIMERA (Maidak et al., 1999) and Bellerophon (Huber et al., 2004). Sequences were imported into the ARB software package (http://www.arb-home.de) (Ludwig et al., 1998), automatically aligned using FastAligner and manually edited. Phylogenetic trees were calculated with almost complete 16S rRNA (1400 bp) sequences for all the close relatives of target sequences using the neighbour-joining and maximum parsimony methods in ARB. Partial sequences were subsequently imported to the tree without changing branch topology using the ARB parsimony-interactive method. The robustness of inferred tree topologies was evaluated after 1000 bootstrap resamplings of the neighbour-joining data in the Phylip program (Felsenstein, 1993).
Data analyses
Clone sequence results were visualised as phyla level pie charts to assess the phylogenetic affiliation of the shifting microbes and heat maps were constructed at a 97% sequence similarity level to reveal the extent of operational taxonomic unit (OTU) differences between the libraries.
The variability in percentage of coral metamorphosis on CCA at each temperature treatment was assessed using one-way analysis of variance (Statistica 6.0, StatSoft, Tulsa, OK, USA).
Principal components analysis was performed to analyse similarity in microbial community composition using DGGE data (Statistica 6.0, StatSoft). Matrices were constructed separately for the rpoB gene DGGE, the combined analysis of the Alphaproteobacteria and Cytophaga–Flavobacteria–Bacteroidetes DGGE gels, the eukaryotic DGGE gel and the seawater 16SrRNA DGGE gel using the presence (1) or absence (0) of a band in each sample.
Pie charts were constructed using the phylogenetic assignments produced in ARB and plotted using Sigmaplot (v7.101, SPSS).
Distance matrices were generated in ARB by using Jukes–Cantor correction and OTU assignments; rarefaction curves and diversity estimates were produced in DOTUR using a distance of 0.03 (Schloss and Handelsman, 2005). Statistical analysis for CCA and sediment libraries were performed on the genetic distance matrices using LIBSHUFF (Schloss et al., 2004) to determine whether variability in library composition between treatments was because of chance or a real biological effect. OTU heat maps were constructed using a log2 scale in MOTHUR version 1.8.1 (Schloss et al., 2009). A community Newick-formatted tree with a distance of 0.03 was produced for the CCA libraries using MOTHUR version 1.8.1 according to the Yue and Clayton theta structural diversity measure.
Results
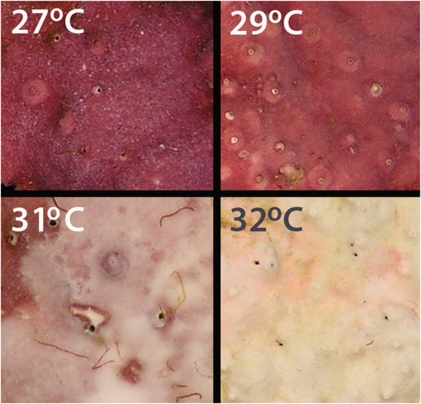
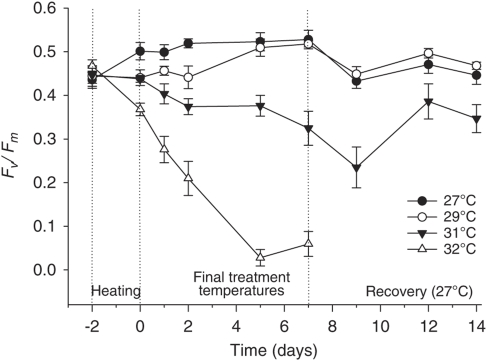
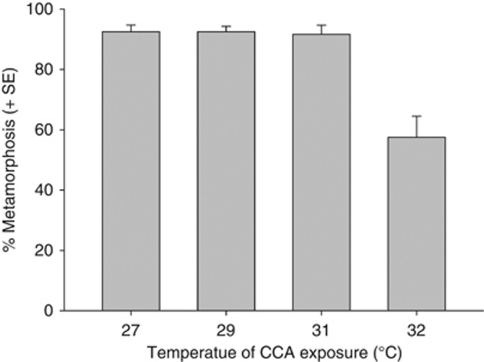
Elevated SST resulted in visible CCA bleaching (Figure 1) and a corresponding reduction in maximum quantum yield (Fv/Fm) in CCA at 31 °C and 32 °C (Figure 2). A subsequent increase in Fv/Fm was observed when CCA treated at 31 °C was returned to 27 °C after 7 days. The CCA discs treated at 32 °C for 7 days (Figure 1c) developed a green shade under the surface during recovery, indicating colonisation by endolithic green algae. The Fv/Fm values were not compared with the 32 °C discs during recovery, as these endolithic algae would have contributed to the fluorescence responses. Live CCA discs treated at 27 °C for 7 days induced settlement and metamorphosis of 93±2% of coral larvae (Figure 3). This level of metamorphosis was consistent for CCA treated at 29 °C and 31 °C, but dropped significantly (to 58±2%) for heavily bleached CCA exposed to 32 °C for 7 days (P<0.01, ANOVA F3,2.05=15.3).
Figure 1.
Photographic panel of CCA after 7 days at each of the experimental temperature treatments showing loss of pigment at 31 °C and complete bleaching at 32 °C.
Figure 2.
Maximum potential quantum yield (Fv/Fm) of CCA in each temperature treatment during the 7 days exposure and subsequent 7 days recovery period.
Figure 3.
Percent metamorphosis of coral larvae on CCA that had been treated at 27 °C, 29 °C, 31 °C and 32 °C for 7 days.
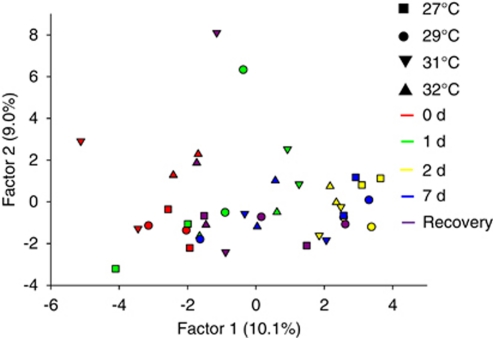
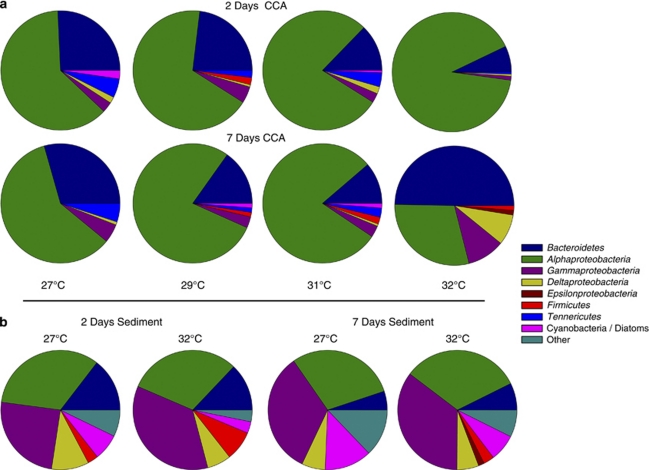
Screening of the CCA microbial communities by DGGE with universal 16S rRNA gene primers was not possible because of the high microbial diversity, which resulted in banding patterns too dense and complex for adequate resolution. Conversely, screening of the CCA microbial communities by DGGE with rpoB gene primers showed no distinct clustering because of temperature treatment (Figure 4; Supplementary Figure 1). However, more detailed community analysis using 16S rRNA clone libraries revealed a distinct shift in the CCA microbial community composition at the phyla/class level at 32 °C after 7 days (Figure 5a). Sediment biofilms, on the other hand, exhibited no obvious community differences between the two temperature extremes (Figure 5b). The 32 °C CCA libraries were dominated by Bacteroidetes with a lesser abundance of Alpha-, Gamma- and Deltaproteobacteria (Figure 5a). All other CCA libraries were dominated by Alphaproteobacteria, with a moderate abundance of Bacteroidetes and a low abundance of all other phyla. The sediment biofilms all contained high proportions of Alpha and Gammaproteobacteria and moderate abundances of Deltaproteobacteria, Bacteroidetes, Cyanobacteria, Firmicutes and ‘other' comprising the Chloroflexi, Nitrospira and Actinobacteria (Figure 5b).
Figure 4.
Principal components analysis of CCA bacterial community composition at each of the temperature treatments. DGGE banding pattern data generated with rpoB gene primers was used to construct a similarity matrix.
Figure 5.
Phyla and class-level differences in bacterial biofilm composition on CCA (a) and reef sediment (b) at each temperature treatment. The ‘other' category contains sequences from the Chloroflexi, Nitrospira and Actinobacteria phyla. Graphs were constructed using the frequency of 16s rRNA sequences belonging to each bacterial group from clone library analysis.
LIBSHUFF statistical analysis of the libraries revealed a highly significant difference between the 2- and 7-day 32 °C CCA libraries and all other temperature treatments (Table 1) and confirmed that there was no significant difference between any of the sediment biofilm treatments (P>0.01). Although the 2-day CCA library at 31 °C was significantly different to the 2-day CCA libraries from 27 °C and 29 °C, respectively; the 7-day CCA library at 31 °C was only significantly different to the 27 °C library at 7 days. The community composition identified in the 27 °C libraries for 2- and 7 day were also found to be statistically different to all other communities (Table 1).
Table 1. LIBSHUFF analysis of variability in CCA clone library community composition.
| Library comparison | Significance | Notes |
|---|---|---|
| Time 2 days | ||
| 27 °C vs ALL 2 days | P<0.002 | T 2 days 29 °C subset of T 2 days 27 °C (P>0.01) |
| 29 °C vs 31 °C | P=0.002 | |
| 32 °C vs ALL 2 days | P<0.0002 | |
| Time 7 days | ||
| 27 °C vs ALL 7 days | P<0.001 | T 7 days 29 °C subset of T 7 days 27 °C (P>0.01) |
| 32 °C vs ALL 7 days | P=0.0001 | |
| Between time | ||
| 7 days 27 °C vs ALL 2 days | P=0.0001 | |
| 7 days 29 °C vs 2 days 32 °C | P=0.0001 | |
| 7 days 31 °C vs 2 days 29 °C | P<0.001 | |
| 7 days 31 °C vs 2 days 32 °C | P=0.0001 | T 2 days 32 °C subset of T 7 days 31 °C (P>0.01) |
| 7 days 32 °C vs ALL 2 days | P=0.0001 | |
Abbreviations: ALL, all other treatments; CCA, crustose coralline algae; T, time.
Only statistically significant comparisons are shown.
**No significant differences were detected between any of the four sediment libraries.
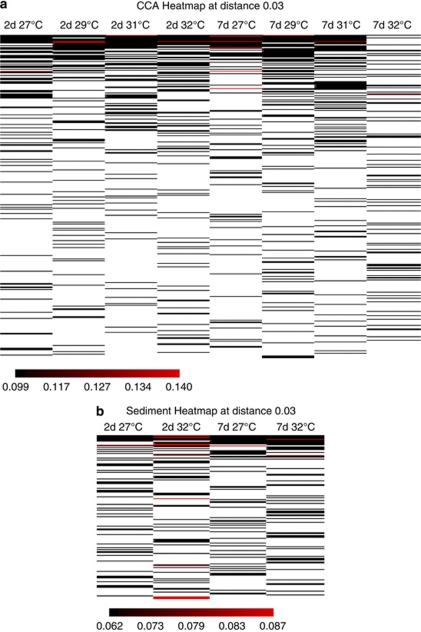
To assess the differences in community composition at a higher level of phylogenetic resolution, heat maps were constructed based on an OTU assignment at a species-level (97%) sequence-similarity threshold (Figure 6). Although the heat map of OTUs revealed some variability within most of the CCA libraries, the 7-day 32 °C treatment was a clear outlier (Figure 6a). In contrast, much less variability was detected between the sediment biofilm communities exposed to the two temperature extremes after both 2 and 7 days (Figure 6b). Subsequent analysis of the CCA biofilm community phylogenetic tree in ARB revealed three major differences between the 7-day 32 °C treatment and all other libraries. First, there was a loss of an Alphaproteobacteria (HM177926), which made up 30% of the sequences from the 2-day 32 °C library, but only 0.6% of the sequences from the 7-day 32 °C library. This strain dominated all the other CCA libraries at both time points and had closest sequence similarity (96%) to a symbiont from the coral Montastrea faveolata (accession number FJ202556, (Sunagawa et al., 2010). The second difference was the appearance of a Deltaproteobacteria (HM178559), which was absent from all other libraries but made up 8% of the sequences from the 7-day 32 °C library and had highest similarity (98%) to a Desulfovibrio from a study of healthy and diseased corals (accession number FJ425598). The third major difference was a substantial increase in the abundance of a Bacteroidetes strain (HM178650) with 99% sequence similarity to a Flavobacterium (accession number AB270586). This strain made up 39% of the total sequences from the 7-day 32 °C library, but only 0.5% and 1.6% of the 2-day 32 °C and 7-day 27 °C libraries, respectively.
Figure 6.
OTU heat maps generated with a distance of 0.03 for CCA (a) and sediment (b) libraries. The 7-day 32 °C CCA library is a distinct outlier and there is greater variability between the CCA derived libraries than the sediment-derived libraries.
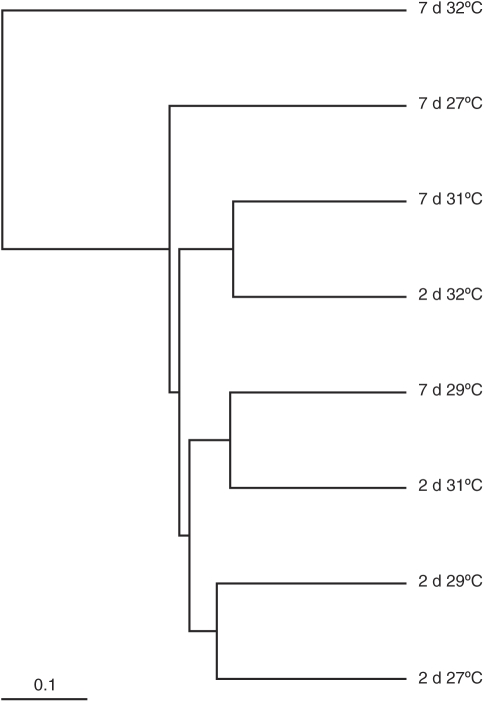
To further visualise the similarity between CCA biofilm communities at different SSTs, a community tree was constructed based on an OTU assignment at a 97% sequence-similarity threshold (Figure 7). The tree demonstrates the clear differentiation of the CCA exposed to 32 °C for 7 days. This analysis also reveals that the 7-day 31 °C and 2-day 32 °C biofilms cluster together and apart from the biofilms exposed to lower temperatures.
Figure 7.
Community tree constructed from an OTU assignment using a distance of 0.03. The cluster analysis shows similarity between each of the CCA-derived 16S rRNA gene clone libraries.
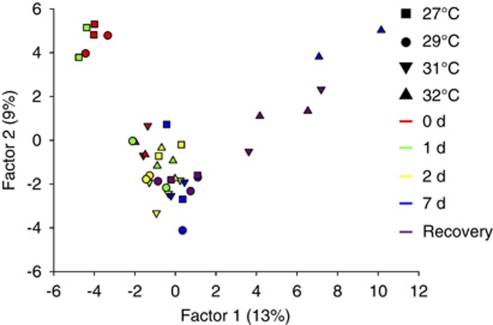
To verify the species-level shift detected in CCA clone library analysis, additional DGGE was conducted using primers specific to the Alphaproteobacteria and Bacteroidetes groups (Supplementary Figures 3 and 4). Principle components analysis of these DGGE profiles revealed clear separation of the 27 °C and 29 °C libraries from the 0- and the 1-day 27 °C libraries (Figure 8). Consistent with the clone library analysis, the profile from CCA at 32 °C was distinct from the other samples after 7 days. However, the samples from CCA at 32 °C after 2 days did not generate different banding profiles to the samples from the lower temperatures. Although the CCA at 31 °C for 7 days clustered with the 2- and 7-day samples from the lower temperatures, the 31 °C-CCA profile from the recovery period did clearly separate from the other samples (Figure 8).
Figure 8.
Principal components analysis of CCA bacterial community composition at each of the temperature treatments. DGGE banding pattern data generated with primers specific to the Alphaproteobacteria and Bacteroidetes groups.
In terms of diversity and evenness of the clone libraries, there was no apparent trend with temperature. Rarefaction analysis indicated that between 71% and 88% of the diversity was sampled in the various CCA biofilm treatments, but only 23–36% of the diversity was sampled in the sediment biofilm libraries (Table 2; Supplementary Figures 2a and b). The most diverse CCA library using both Chao1 and Ace indices was the 7-day 29 °C library followed by the 7-day 31 °C and 2-day 27 °C libraries (Table 2). The lowest-diversity CCA library was the 7-day 27 °C treatment.
Table 2. Diversity indices calculated from sequences of 16S rRNA genes using a 97% sequence similarity threshold.
| Treatment | Total clones | Unique OTUs | Diversity sampled (%) | Shannon–Weaver | Chao1 | Ace |
|---|---|---|---|---|---|---|
| CCA 27 °C 2 days | 155 | 63 | 81 | 3.69 | 98 | 111 |
| CCA 29 °C 2 days | 135 | 53 | 80 | 3.48 | 76 | 95 |
| CCA 31 °C 2 days | 144 | 53 | 79 | 3.30 | 82 | 105 |
| CCA 32 °C 2 days | 171 | 51 | 88 | 3.21 | 76 | 77 |
| CCA 27 °C 7 days | 117 | 50 | 81 | 3.54 | 66 | 82 |
| CCA 29 °C 7 days | 157 | 70 | 72 | 3.73 | 174 | 179 |
| CCA 31 °C 7 days | 155 | 63 | 71 | 3.41 | 98 | 116 |
| CCA 32 °C 7 days | 142 | 51 | 76 | 3.23 | 93 | 126 |
| Sed 27 °C 2 days | 65 | 54 | 29 | 3.90 | 234 | 296 |
| Sed 32 °C 2 days | 62 | 55 | 23 | 3.97 | 196 | 244 |
| Sed 27 °C 7 days | 69 | 55 | 36 | 3.91 | 165 | 212 |
| Sed 32 °C 7 days | 64 | 54 | 29 | 3.93 | 202 | 215 |
Abbreviations: CCA, crustose coralline algae; OTU, operational taxonomic unit, Sed, sediment.
To assess how CCA eukaryotic biofilms were affected by temperature, DGGE was performed with universal 18S rRNA gene primers. Although more bands were detected in the 7 days and recovery samples (Supplementary Figure 5), there was no trend associated with temperature treatment of the CCA samples. This was further illustrated with principle components analysis revealing no clear separation of the samples according to temperature treatment (Supplementary Figure 6). Similarly, 16S rRNA gene DGGE analysis of the seawater samples from each of the temperature treatments revealed a greater number of bands in the 7 days and recovery time points (Supplementary Figure 7), but no clear effect of temperature. Principle components analysis of this DGGE revealed clear clustering of the microbial communities according to sampling time rather than treatment temperature (Supplementary Figure 8).
Discussion
This study revealed that the CCA N. fosliei, and its associated microbial communities do not tolerate SSTs of 32 °C, only 2–4 °C above the mean maximum annual SST at Davies Reef where the CCA was collected (http://data.aims.gov.au/awsqaqc/do/start.do). After 7 days at 32 °C, the CCA exhibited clear signs of stress, including visual evidence of bleaching, a reduction in maximum quantum yield and a large shift in microbial community structure involving a move away from Alphaproteobacteria towards a dominance by Bacteroidetes. A recovery in photosynthetic capacity (Fv/Fm) was observed in CCA exposed to 31 °C following 7 days of recovery (at 27 °C); however CCA exposed to 32 °C did not appear to recover during this time as evidenced by the rapid growth of endolithic green algae. A reduction in the ability of N. fosliei to induce coral larval metamorphosis at 32 °C accompanied the changes in microbiology, pigmentation and photophysiology of the CCA. This is the first experimental evidence to demonstrate how thermal stress on coralline algae may influence coral recruitment and this may have implications for reef regeneration and recovery from climate-related mortality events.
The thermal threshold for bleaching in N. fosliei was similar to that of the GBR corals A. formosa, A. elseyi and Pocillopora damicornis (Berkelmans and Willis, 1999). The visible bleaching at 31 °C and 32 °C was preceded by drops in Fv/Fm, which declined by 53% and 80%, respectively after 7 days compared with CCA maintained at 27 °C. This reduction in Fv/Fm indicates potential chronic photoinactivation, caused by thermal and oxidative damage to (or loss of) the D1 reaction centre in PSII (Warner et al., 1999). In a recent study, the shallow CCA species Porolithon onkodes from the GBR also demonstrated partial bleaching following exposure to elevated SST (Anthony et al., 2008). N. fosliei is abundant on the GBR at depths of 5–10 m, and may be less adapted to cope with increases in SST. The loss of pigmentation in P. onkodes was accompanied by reductions in productivity (Anthony et al., 2008), possibly because of chronic photoinactivation as described here. The slow recovery in Fv/Fm of CCA exposed to 31 °C in this study is consistent with the gradual repair of PSII damaged by thermal stress; however, this could not be confirmed in CCA discs exposed to 32 °C because of the presence of endolithic green algae under the surface of the CCA.
The microbial communities associated with CCA shifted at both 31 °C and 32 °C, but the shift was much more rapid at the higher temperature. Although the Alphaproteobacteria clearly dominates the CCA microbial community at ambient temperatures, there is a distinct shift towards the Bacteroidetes in CCA exposed to 32 °C for 7 days. The primary Alphaproteobacteria strain to be lost from CCA at 32 °C had highest sequence similarity to a described coral symbiont. The role of this microbe within the CCA biofilm has not been ascertained, but there is some evidence that the loss of bacterial symbionts due to elevated SST can have significant implications on the health of the host (Webster et al., 2008a). The increase in Bacteroidetes is consistent with previous studies of marine organisms subjected to stressors such as elevated SST or disease. In the GBR sponge Rhopaloeides odorabile, the Bacteroidetes were absent from sponges maintained at temperatures up to 31 °C but dominated the microbial community in sponges exposed to 33 °C (Webster et al., 2008a). Similarly, diseased corals and sponges host a much greater abundance of Bacteroidetes than their healthy counterparts (Frias-Lopez et al., 2002; Pantos and Bythell, 2006; Webster et al., 2008b). The closest relative to the Bacteroidetes sequence that became highly abundant at 32 °C was a novel marine Flavobacteria with anti-oxidative properties (Shindo et al., 2007). The Bacteroidetes are diverse and abundant in the marine ecosystem (O'Sullivan et al., 2006) and are major utilizers of high-molecular mass dissolved organic matter (Cottrell and Kirchman, 2000). For this reason, they are often abundant in nutrient-rich waters in which biomacromolecules accumulate (Reichenbach, 1989). It is possible that heat-stressed or decaying CCA cells provided an abundant and varied source of nutrients for these bacteria, resulting in an increased abundance of Bacteroidetes in the 7-day 32 °C samples. The increased abundance of a Deltaproteobacteria in CCA exposed to 32 °C for 7 days is consistent with studies showing an increase in Delatproteobacteria in diseased corals and sponges. The closest described relative to this sequence was a Desulfovibrio spp. retrieved from an unpublished study of diseased and healthy corals. Closely related Desulfovibrio spp. have been implicated in coral black band disease (AY750147) and in black patch syndrome affecting the sponge Alpysina aerophoba (EU267167), (Webster et al., 2008b). An alternative explanation for the community differences observed in CCA at 32 °C may be due to microbes associated with the green endolithic algae that colonised the bleached CCA surface after 5 days. Future research is urgently required to assess the implications of these microbial shifts to CCA health and coral recruitment.
The bacterial biofilms on the surface of CCA exhibited a different response to elevated temperature than bacterial communities associated with reef sediments, which were largely conserved at all temperatures. In addition, the microbial community of the seawater from each of the CCA tanks showed no correlation to the treatment temperature. This indicates that either (1) the thermal stress response of the host CCA contributes to the shift in the biofilm community or (2) the composition of the biofilm on the surface of CCA is more sensitive to environmental change (either because of a more specific association, as may occur in symbiosis, or due to the initial CCA bacterial community composition containing more temperature-sensitive biofilm species). For the large number of marine invertebrates that settle and metamorphose in response to sediment biofilms, this community stability suggests that these organisms may be less affected by increasing SST than those that rely on more sensitive bacterial biofilms associated with biotic surfaces such as CCA. DGGE analysis showed that the CCA biofilm community was unable to recover its original composition within 7 days once the SST had been returned to ambient. It is possible that the prolonged temperature stress (as occurred in the 7-day experiment) caused a phase shift in the microbial community to an alternate stable state. Alternatively, the impact of elevated SST on CCA health may have adversely affected the ability of the CCA immune system to regulate microbial colonisation and growth.
This study has highlighted the importance of using multiple methods for assessing microbial community responses. DGGE facilitated the analysis of large numbers of replicates, whereas clone libraries were utilised to explore the composition of the shifting microbes. LIBSHUFF analysis of 16S rRNA clone libraries found many more differences than the DGGE analysis (for example, microbial community differences in libraries at 27 °C and 29 °C). DGGE with the rpoB gene was unable to distinguish the microbial shift that occurred at 32 °C after 7 days. The results from 16S rRNA gene clone libraries and 16S rRNA gene DGGE with Alphaproteobacteria and Bacteroidetes-specific primers were largely consistent, although the DGGE did not detect the shift at 32 °C after 2 days. This is possibly because of the fact that only the predominant microbes will be detected with this type of fingerprinting approach, whereas deep clone sequencing can assess more subtle effects or the response of the ‘rarer' microbial community members.
Interestingly, the elevated SST did not have an impact on microbial community diversity or evenness. This is in contrast to other studies using marine models in which stressors such as SST (Webster et al., 2008a) eutrophication (Uthicke and McGuire, 2007) or disease (Cooney et al., 2002; Pantos et al., 2003; Pantos and Bythell, 2006; Webster et al., 2008b) cause a shift (either an increase or decrease) in microbial diversity. A reduced microbial diversity was observed in sediments from an inshore reef with high inorganic carbon compared with a nearby offshore reef (Uthicke and McGuire, 2007); however, a 53% increase in bacterial diversity was detected in a GBR sponge at 33 °C compared with the sponge microbial community at 27 °C.
A significant reduction in coral metamorphosis occurred in response to CCA that had been exposed to 32 °C for 7 days. The reduction in inductive capacity at 32 °C may be due to a number of factors including: (1) the loss of a critical microbial species on the surface of CCA that is directly responsible for the induction of metamorphosis (Johnson and Sutton, 1994), (2) the loss of a microbial species required to enable the CCA to produce an inducer for larval metamorphosis (disruption of a shared metabolic pathway), (3) the inhibition of a biochemical process within the CCA alone to produce an indicator for larval metamorphosis, (4) the inhibition of larval metamorphosis by specific microbes (Dobretsov and Qian, 2004), or (5) an interaction (for example, pathogenicity) between the microbial community and the CCA (for instance Deltaproteobacteria and Desulfovibrio in CCA exposed to 32 °C have been identified in association with diseased corals and sponges).
N. fosliei is a key reef-building primary producer on the GBR and other coral reefs (Harrington et al., 2004). The low-thermal tolerance of this species is of concern as CCA are especially vulnerable to increases in ocean acidification (pCO2), which is projected to intensify under continued conditions of climate change (Kuffner et al., 2008). Combinations of elevated SSTs and acidification further enhance CCA bleaching, productivity and reduce calcification (Anthony et al., 2008; Martin and Gattuso, 2009). Furthermore, CCA (including N. fosliei) are critical to the recruitment of many coral species (Heyward and Negri, 1999; Harrington et al., 2004), including many Acropora species, which often dominate reef habitats of the GBR. Any reduction in the capacity of CCA to induce coral larval metamorphosis by thermal stress, as described here, would clearly add to the pressures already faced by corals under conditions of a changing climate. Although shifts in CCA-associated microbial communities correlate with reductions in coral larval metamorphosis, further work is required to determine the role of microbiology in the recruitment process.
Acknowledgments
We thank Dr Sven Uthicke for helpful discussions about data analysis and Dr Ken Wasmund for assisting with sample processing.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anthony KRN, Kline DI, Diaz-Pulido G, Dove S, Hoegh-Guldberg O. Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc Natl Acad Sci. 2008;105:17442–17446. doi: 10.1073/pnas.0804478105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelmans R, Willis BL. Seasonal and local spatial patterns in the upper thermal limits of corals on the inshore Central Great Barrier Reef. Coral Reefs. 1999;18:219–228. [Google Scholar]
- Cooney RP, Pantos O, Le Tissier MD, Barer MR, O'Donnell AG, Bythell JC. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ Microbiol. 2002;4:401–413. doi: 10.1046/j.1462-2920.2002.00308.x. [DOI] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahllöf I, Baillie H, Kjelleberg S. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl Environ Microbiol. 2000;66:3376–3380. doi: 10.1128/aem.66.8.3376-3380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov S, Qian PY. The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. J Exp Mar Biol Ecol. 2004;299:35–50. [Google Scholar]
- Felsenstein J.1993PHYLIP (Phylogenetic Inference Package)Version 3.5c.Department of Genetics, University of Washington: Seattle, WA [Google Scholar]
- Ferris MJ, Muyzer G, Ward DM. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson JA, Bilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW. Partitioning of bacterial communities between seawater and healthy, black-band diseased, and dead coral surfaces. Appl Environ Microbiol. 2002;68:2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbary D, Veltkamp CJ. Observations on Mesophyllum lichenoides (Corallinaceae, Rhodophyta) with scanning electron microscope. Phycologia. 1980;19:49–53. [Google Scholar]
- Garland CD, Cooke SL, Grant JF, McMeekin TA. Ingestion of the bacteria on and the cuticle of crustose (non-articulated) coralline algae by post-larval and juvenile abalone (Haliotis ruber leach) from Tasmanian waters. J Exp Mar Biol Ecol. 1985;91:137–149. [Google Scholar]
- Gelsomino A, Cacco G. Compositional shifts of bacterial groups in a solarised and amended soil as determined by denaturing gradient gel electrophoresis. Soil Biol Biochem. 2006;38:91–102. [Google Scholar]
- Genty B, Briantais JM, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta. 1989;990:87–92. [Google Scholar]
- Harrington LM, Fabricius K, De'ath G, Negri A. Recognition and selection of settlement substrata determine post-settlement survival in corals. Ecology. 2004;85:3428–3437. [Google Scholar]
- Heyward AJ, Negri AP. Natural inducers for coral larval metamorphosis. Coral Reefs. 1999;18:273–279. [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatic Applications Note. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Huggett MJ, Williamson JE, de Nys R, Kjelleberg S, Steinberg PD. Settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia. 2006;149:604–619. doi: 10.1007/s00442-006-0470-8. [DOI] [PubMed] [Google Scholar]
- Huys G, Vanhoutte T, Vandamme P. Applications of sequence-dependent electrophoresis fingerprinting in exploring biodiversity and population dynamics of human intestinal microbiota: What can be revealed. Interdiscipl Perspect Infect Dis. 2008;2008:26. doi: 10.1155/2008/597603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . Climate Change 2007: The Physical Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press Cambridge: UK; 2007. [Google Scholar]
- Johnson CR, Muir DG, Reysenbach A-L. Characteristic bacteria associated with the surfaces of coralline algae: a hypotheses for bacterial induction of marine invertebrate larvae. Mar Ecol Prog Ser. 1991;74:281–294. [Google Scholar]
- Johnson CR, Sutton DC. Bacteria on the surface of crustose coralline algae induce metamorphosis of the crown of thorns starfish Acanthaster planci. Mar Biol. 1994;120:305–310. [Google Scholar]
- Jones RJ, Muller J, Haynes D, Schreiber U. Effects of herbicides diuron and atrazine on corals of the Great Barrier Reef, Australia. Mar Ecol Prog Ser. 2003;251:153–167. [Google Scholar]
- Kuffner IB, Andersson AJ, Jokiel PL, Rodgers K, Mackenzie FT. Decreased abundance of crustose coralline algae due to ocean acidification. Nat Geosci. 2008;1:114–117. [Google Scholar]
- Lawrence JR, Chenier MR, Roy R, Beaumier D, Fortin N, Swerhone GD, et al. Microscale and molecular assessment of impacts of nickel, nutrients, and oxygen level on structure and function of river biofilm communities. Appl Environ Microbiol. 2004;70:4326–4339. doi: 10.1128/AEM.70.7.4326-4339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TE, Garland CD, McMeekin TA. The bacterial biota on crustose (nonarticulated) coralline algae from Tasmanian waters. Microb Ecol. 1985;11:221–230. doi: 10.1007/BF02010601. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, et al. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Parker CT, Jr, Garrity GM, Larsen N, Li B, et al. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA [published erratum appears in Appl Environ Microbiol 1998 Jun; 64: 2333] Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Gattuso JP. Response of Mediterranean coralline algae to ocean acidification and elevated temperature. Glob Change Biol. 2009;15:2089–2100. [Google Scholar]
- Meyer-Reil L-A, Koster M. Eutrophication of marine waters: effects on benthic microbial communities. Mar Pollut Bull. 2000;41:255–263. [Google Scholar]
- Morse ANC, Iwao K, Baba M, Shimoike K, Hayashibara T, Omori M. An ancient chemosensory mechanism brings new life to coral reefs. Biol Bull. 1996;191:149–154. doi: 10.2307/1542917. [DOI] [PubMed] [Google Scholar]
- Morse DE, Hooker N, Morse ANC, Jensen RA. Control of larval metamorphosis and recruitment in sympatric agariciid corals. J Exp Mar Biol Ecol. 1988;116:193–217. [Google Scholar]
- Morse DE, Morse AN, Raimondi PT, Hooker N. Morphogen-based chemical flypaper for Agariciahumilis coral larvae. Biol Bull. 1994;186:172–181. doi: 10.2307/1542051. [DOI] [PubMed] [Google Scholar]
- Negri AP, Heyward AJ. Inhibition of fertilization and larval metamorphosis of the coral Acropora millepora (Ehrenberg, 1834) by petroleum products. Mar Pollut Bull. 2000;41:420–427. [Google Scholar]
- Negri AP, Webster NS, Hill RT, Heyward AJ. Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar Ecol Prog Ser. 2001;223:121–131. [Google Scholar]
- O'Sullivan LA, Rinna J, Humphreys G, Weightman AJ, Fry JC. Culturable phylogenetic diversity of the phylum ‘Bacteroidetes' from river epilithon and coastal water and description of novel members of the family Flavobacteriaceae: Epilithonimonas tenax gen. nov., sp. nov. and Persicivirga xylanidelens gen. nov., sp. nov. Int J Syst Evol Microbiol. 2006;56:169–180. doi: 10.1099/ijs.0.63941-0. [DOI] [PubMed] [Google Scholar]
- Osmond CB, Anderson JM, Ball MC, Egerton JJG.1999Compromising efficiency: the molecular ecology of light resource utilisation in terrestrial plantsIn: Scholes C, Baker M (eds).Advances in Physiological Plant Ecology Blackwell: Oxford [Google Scholar]
- Pantos O, Cooney RP, Le Tissier MD, Barer MR, O'Donnell AG, Bythell JC. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ Microbiol. 2003;5:370–382. doi: 10.1046/j.1462-2920.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- Pantos O, Bythell JC. Bacterial community structure associated with white band disease in the elkhorn coral Acropora palmata determined using culture independent 16S rRNA techniques. Dis Aquat Org. 2006;69:79–88. doi: 10.3354/dao069079. [DOI] [PubMed] [Google Scholar]
- Reichenbach H.1989Nonphotosynthetic, nonfruiting gliding bacteria. Genus 1. Cytophaga Winogradsky 1929, 577AL, emendIn: Staley JT, Bryant MP, Pfennig N, Holt JG (eds).Bergey's Manual of Systematic Bacteriology Baltimore [Google Scholar]
- Rodriguez SR, Ojeda FP, Inestrosa C. Settlement of benthic marine invertebrates. Mar Ecol Prog Ser. 1993;97:193–207. [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Larget BR, Handelsman J. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol. 2004;70:5485–5492. doi: 10.1128/AEM.70.9.5485-5492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo K, Kikuta K, Suzuki A, Katsuta A, Kasai H, Yasumoto-Hirose M, et al. Rare carotenoids, (3R)-saproxanthin and (3R,2′S)-myxol, isolated from novel marine bacteria (Flavobacteriaceae) and their antioxidative activities. Appl Microbiol Biotech. 2007;74:1350–1357. doi: 10.1007/s00253-006-0774-y. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLoS One. 2010;5:e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uthicke S, McGuire K. Bacterial communities in Great Barrier Reef calcareous sediments: contrasting 16S rDNA libraries from nearshore and outer shelf reefs. Est Coast Shelf Sci. 2007;72:188–200. [Google Scholar]
- Warner ME, Fitt WK, Schmidt GW. Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. P Natl Acad Sci. 1999;96:8007–8012. doi: 10.1073/pnas.96.14.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Battershill CN, Negri AP. Recruitment of Antarctic marine eukaryotes onto artificial surfaces. Polar Biol. 2006;30:1–10. [Google Scholar]
- Webster NS, Cobb RE, Negri AP. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2008a;2:830–842. doi: 10.1038/ismej.2008.42. [DOI] [PubMed] [Google Scholar]
- Webster NS, Hill RTH.2007Vulnerability of marine microbes on the Great Barrier Reef to climate changeIn: Johnson JE, Marshall PA (eds).Climate Change and the Great Barrier Reef Great Barrier Reef Marine Park Authority and Australian Greenhouse Office: Australia, Townsville [Google Scholar]
- Webster NS, Smith LD, Heyward AJ, Watts JE, Webb RI, Blackall LL, et al. Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl Environ Microbiol. 2004;70:1213–1221. doi: 10.1128/AEM.70.2.1213-1221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Xavier JR, Freckelton M, Motti CA, Cobb R. Shifts in microbial and chemical patterns within the marine sponge Aplysinaaerophoba during a disease outbreak. Environ Microbiol. 2008b;10:3366–3376. doi: 10.1111/j.1462-2920.2008.01734.x. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J.1990Amplification and direct sequencing of fungal ribosomal RNA genes for phylogeneticsIn: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds).PCR Protocols: A Guide to Methods and Applications Academic Press: New York; 315–322. [Google Scholar]
- Wieczorek SK, Todd CD. Inhibition and facilitation of bryozoan and ascidian settlement by natural multi-species biofilms: effects of film age and the roles of active and passive larval attachment. Mar Biol. 1997;128:463–473. [Google Scholar]
- Wieczorek SK, Todd CD. Inhibition and facilitation of settlement of epifaunal marine invertebrate larvae by microbial biofilm cues. Biofouling. 1998;12:81–118. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.