Abstract
In all organisms, oxidation threatens the integrity of the genome. DNA-mediated charge transport (CT) may play an important role in the generation and repair of this oxidative damage. In studies involving long-range CT from intercalating Ru and Rh complexes to 5′-GG-3′ sites, we have examined the efficiency of CT as a function of distance, temperature, and the electronic coupling of metal oxidants bound to the base stack. Most striking is the shallow distance dependence and the sensitivity of DNA CT to how the metal complexes are stacked in the helix. Experiments with cyclopropylamine-modified bases have revealed that charge occupation occurs at all sites along the bridge. Using Ir complexes, we have seen that the process of DNA-mediated reduction is very similar to that of DNA-mediated oxidation. Studies involving metalloproteins have, furthermore, shown that their redox activity is DNA-dependent and can be DNA-mediated. Long range DNA-mediated CT can facilitate the oxidation of DNA-bound base excision repair proteins to initiate a redox-active search for DNA lesions. DNA CT can also activate the transcription factor SoxR, triggering a cellular response to oxidative stress. Indeed, these studies show that within the cell, redox-active proteins may utilize the same chemistry as that of synthetic metal complexes in vitro, and these proteins may harness DNA-mediated CT to reduce damage to the genome and regulate cellular processes.
1. INTRODUCTION
Oxidative DNA damage has been implicated in a host of adverse medical conditions including aging, heart disease, and various forms of cancer [1,2]. Reactive oxygen species (ROS) such as singlet oxygen (1O2), superoxide anions (O2•−), hydrogen peroxide (H2O2), and hydroxyl radical (•OH) are a constant threat. Exogenous sources of ROS, such as cigarette smoke, air pollution, and ultraviolet radiation, have been linked to the formation of DNA strand breaks and lesions, which can lead to mutagenesis and carcinogenesis [3,4]. The danger of endogenous sources of ROS is also considerable: ROS are byproducts of oxidative respiration in mitochondria; they are produced by macrophages during immune response; and they are generated during P450 metabolism [5,6]. In order to develop diagnostics and therapeutics for the prevention of medical conditions associated with DNA damage, it is necessary to understand the chemical mechanisms which result in oxidative DNA lesions, as well as the biological pathways that exist to prevent and repair them.
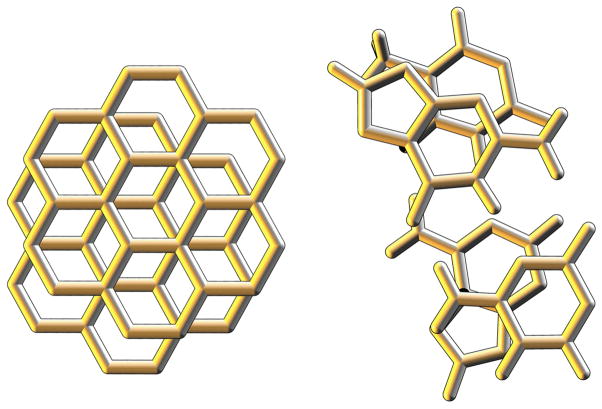
Before discussing factors that affect the oxidation of DNA, it is prudent to review the chemical characteristics of the macromolecule itself. DNA consists of long polymeric strands of nucleic acid bases, specifically the planar, aromatic heterocycles adenine (A), guanine (G), thymine (T), and cytosine (C). Pairs of strands are held together by specific hydrogen bonds formed between the nucleobases: A pairs with T, and G pairs with C. Within the strands, the bases are joined by anionic deoxyribophosphate units, which, upon formation of the duplex, wrap the stack of nucleobases in a negatively charged double helix. Consecutive base pairs are stacked closely together, allowing the aromatic π system of one to interact with that of its neighbors. In this way, the stacked aromatic bases resemble stacked sheets of graphite, as illustrated in Figure 1. In much the same way that electricity can be conducted perpendicular to stacked graphite sheets [7], DNA can mediate the transmission of charge along its length.
Figure 1.
The structure and geometry of stacked graphene sheets (left) is similar to that of stacked DNA base pairs (right).
In our laboratory, we have utilized the rich redox chemistry of transition metal complexes in conjunction with the ability of DNA to mediate charge transport (CT) reactions to generate and study oxidative damage in DNA. In this review, we will discuss the properties of metal complexes that make them ideal probes for initiating and monitoring DNA CT events. We will then discuss the ability of DNA to mediate charge transfer reactions between a charge donor and a charge acceptor, as well as the ability of DNA bases themselves to participate in DNA-mediated redox chemistry. Several biological implications of DNA CT are also described, including the accumulation of oxidative damage at sites of high guanine content, and how DNA may mediate cellular signaling and transcriptional regulation. Finally, it should be noted that this review is based on experiments conducted in our laboratory and is intended to be illustrative, not comprehensive, of the utility of metal complexes in studying DNA-mediated CT.
2. METAL COMPLEXES AS PROBES OF DNA CT
Metal complexes are powerful probes of DNA CT. By varying their redox properties and DNA-binding features, we have been able to characterize the important parameters that govern both DNA-mediated oxidation and reduction.
2.1. Advantages of Metal Complexes in Studies of DNA-Mediated CT
Any study of DNA CT must involve some means of injecting charge onto the DNA bridge and some means of reporting the CT event. Although there is a wide array of molecular probes that can carry out these tasks, the most effective ones share many chemical and physical characteristics. First, in order to utilize the electronic system of the π-stacked bases as a conduit for charge, the probe should interact strongly with the DNA base stack. Such an interaction can be difficult to achieve, considering the geometry of DNA. In general, the only access a diffusing molecule has to the base stack is either at the ends of the DNA strand or within the relatively narrow major and minor grooves which run lengthwise along the sides of the DNA molecule. Probes which are too large, or which are strongly negatively charged and therefore are repelled by the phosphate backbone of the DNA, do not easily interface with the DNA π-stack. Second, depending on the function of the probe, it must provide a straightforward means of either initiating or reporting on DNA CT, or both. Often, the photophysical or electrochemical properties of a molecule are utilized for these purposes. Some probes may also report CT events through chemical pathways such as degradation. Third, the probe should not degrade or interact chemically with the DNA or other components of the sample unless this is by design. Not only must the probe be stable enough to persist in solution, but the excited state of the molecule must also be stable if photochemical means are used to initiate or report CT, and the various redox states of the molecule must be able to withstand the charge transfer process. Finally, the ideal probe would be synthetically versatile and easy to build or modify in order to control sensitively the parameters of the experiment. Metallointercalators, transition metal complexes which bind DNA primarily by intercalation, are one class of molecules that fulfill all of these requirements.
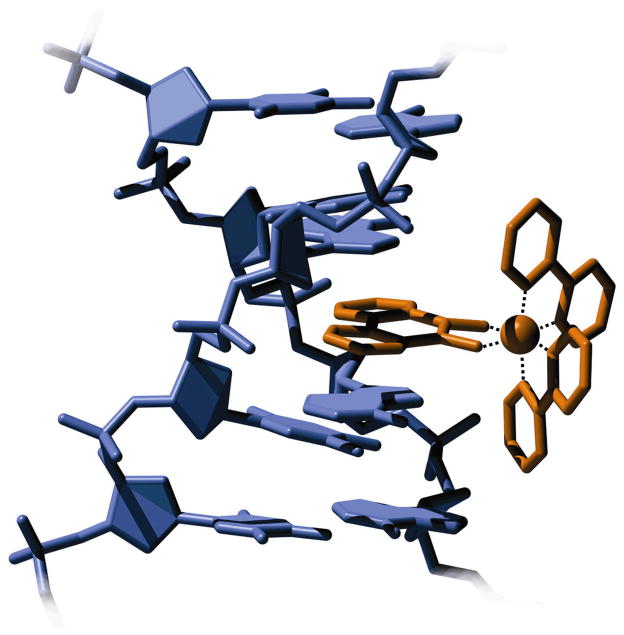
Intercalation, first reported by Lerman in 1961, is a binding mode in which the ligand, usually a planar, aromatic moiety, slips between two adjacent bases in the DNA base stack [8]. Structural changes in the DNA associated with intercalation include a slight unwinding of the helix at the intercalation site, an extension in length equal to the height of one base pair, and an increase in DNA stability, as indicated by a higher duplex melting temperature. The overall structure of the DNA is unperturbed: no bending or kinking of the helix is observed [9], and the C2’-endo sugar pucker found at non-intercalation sites is retained [10]. The effect of intercalation on the structure of DNA is shown in Figure 2. Intercalators, often large, heterocyclic structures, physically and electronically resemble the DNA bases themselves, so intimate associations may form between the DNA base stack and the binding ligand. In a sense, the intercalating molecule acts as an additional base, enabling strong π interactions with the electronic structures of the flanking bases [10]. Many molecules are known to bind DNA through this mode, including the organic intercalating drugs 9-aminoacridine, ethidium, and daunomycin [9], among others. Many metal complexes, also, may bind DNA through intercalation if they bear one or more planar, aromatic ligands such as phen (1,10-phenanthroline), phi (9,10-phenanthrenequinone diimine) or dppz (dipyrido[3,2-a:2′,3′-c]phenazine). Interestingly, by incorporating a ligand that is slightly wider than DNA, it is possible to selectively target binding at thermodynamically destabilized mismatch sites through the insertion binding mode, where the bulky ligand enters the helix from the minor groove, pushing the mismatched base pair out into the major groove [11–13]. Because insertion involves substitution in the base stack of the inserting ligand for the mismatched base pair, insertion, like intercalation, should facilitate strong electronic interactions between the inserting metal complex and the DNA base stack. Although other DNA binding modes such as electrostatic binding and groove binding have been observed [14,15], these do not offer the strong electronic coupling to the base stack that is characteristic of intercalation and insertion.
Figure 2.
Intercalative binding to DNA results in an increase of the rise at the site of binding, as well as a slight unwinding of the helix. Shown is a model of Rh(phi)(bpy)23+ (orange) bound to DNA (blue), adapted from the crystal structure of a similar construct [11].
Not only do metallointercalators bind strongly to DNA, but they also possess rich and well understood photochemistry and photophysics, which makes them advantageous for use as probes for DNA interactions, as injectors of charge onto the DNA bridge, and as reporters of DNA CT events. Particularly interesting and effective examples are the dppz complexes of ruthenium, which display the “light switch effect” [16]. Ru(bpy)2dppz2+ and Ru(phen)2dppz2+ are not luminescent in aqueous solution due to deactivation of the luminescent state via hydrogen bonding of the dppz ligand by water. However, in solutions containing duplex DNA, the complexes intercalate, the dppz ligand is protected from solution, and luminescence is restored [16–20]. Although most metal complexes do not display this remarkable discrimination, many do luminesce. In addition, many complexes absorb strongly in the visible region due to their intense metal-to-ligand charge transfer (MLCT) transitions. These properties allow for manipulation and monitoring of the electronic and redox states of the metal complexes spectroscopically. The MLCT transitions may also be exploited to initiate CT processes, since many metal complexes become strong oxidizing or reducing agents upon optical excitation. In circumstances under which the excited state of a metal complex is not a strong enough oxidizing or reducing agent to carry out the desired chemistry, it may be necessary to utilize the “flash-quench technique” [21]. This method involves the use of a diffusing molecule which is competent to oxidize or reduce the metal complex from its excited state, thus creating a strong ground state oxidant or reductant.
Other useful characteristics of metallointercalators are that they are coordinatively saturated, substitutionally inert, and rigid. This makes them extremely stable in solution, and the complexes do not coordinate with DNA. Metallointercalators are also modular. Unlike organic intercalators, the properties of metal complexes can be altered subtly and systematically by altering the structures of the constituent ligands. In addition, the three-dimensional structure of metallointercalators enables them to interact with DNA in a stereospecific, and sometimes sequence-specific, manner, while organic intercalators, which are often planar, cannot. For example, many studies have shown that Δ complexes tend to bind more tightly to right-handed B-DNA, while Λ complexes have been useful in probing left-handed Z-DNA [22–30]. This result has mainly to do with the steric agreement between the intercalated metal complex and the DNA: the ancillary ligands of Δ complexes tend to lie along the major groove of the DNA helix, whereas those of Λ complexes collide.
The versatility of metallointercalators also facilitates the sensitive tuning of their electronic and electrochemical properties. Complexes have been synthesized that absorb and emit across the visible spectrum and that sample a wide variety of redox potentials. The addition or elimination of a single functional group on either the intercalating ligand or the ancillary ligands can serve to alter the photophysical, electrochemical, or DNA binding properties of the complex. For example, addition of a carboxylic acid or benzyl group to the end of dppz, or introduction of an additional heterocyclic nitrogen, eliminates the light-switch effect and alters the absorption and emission maxima and luminescence lifetime of the complex [17].
The modularity of metal complexes also makes it possible to extend their functionality by modifying their ancillary ligands. For example, it is possible to create a covalent linkage between a metal complex and DNA through the use of a long alkyl chain [31,32]. Such linkages serve to ensure a binding ratio of unity between the metal complex and the DNA while precisely defining the binding site of the metal, without disrupting the mode of binding or solvent interactions. Alkyl chains have also been used to append organic fluorophores to metallointercalators in an effort to develop luminescent reporters of mismatches [33]. Additionally, modification of a ruthenium complex with octaarginine allows for the facile uptake of complexes into the nuclei of cancer cells [34,35]. Functionalization of the ancillary ligands may also lead to sequence-selective recognition and cleavage by metallointercalators via hydrogen bonding or van der Waals interactions with modified ethylenediamine ligands [36–40], peptide sequences [41–43], or modified phen ligands [44–47], or nuclease activity [48].
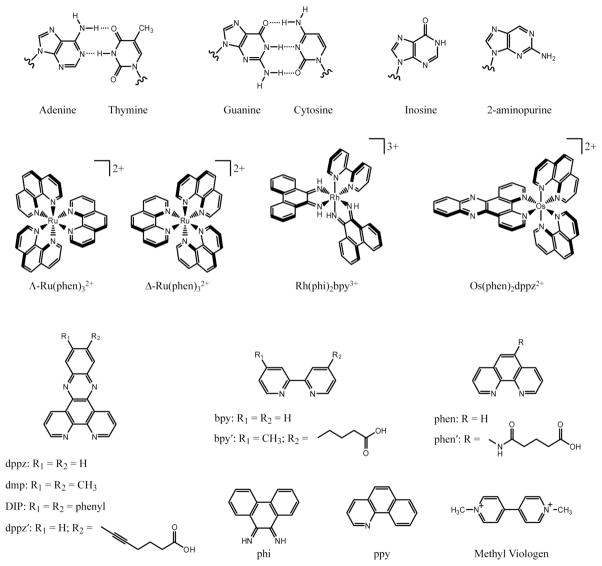
Although many classes of molecules may serve as effective intercalators for the study of DNA-mediated CT, metallointercalators provide several advantages. The array of metal complexes described in this review is shown in Scheme 1. In addition to their inherent stability in solution, they display strong coupling to the DNA base stack. Unlike organic intercalators, the photophysical, electrochemical, and DNA-binding properties of metallointercalators may be tuned in an efficient and systematic manner to modify their properties in a sensitive and subtle way. Finally, the modularity of metal complexes allows for external functionalities to be applied, expanding the utility of these probes.
Scheme 1.
Structures of DNA bases and representative metal complexes used in DNA-CT experiments.
2.2. Metal Complexes as Charge Donors and Acceptors in DNA CT
The first experiment which suggested the possibility of charge transport through the DNA base stack was an investigation of photoinduced electron transfer from Ru(phen)32+ to either Co(bpy)33+, Co(phen)33+, or Co(DIP)33+ (DIP = 4,7-diphenyl-1,10-phenanthroline) [30]. It was found that the Stern-Volmer quenching slopes scaled with the DNA binding affinity of the quencher, and that Δ-Ru(phen)32+ was quenched more efficiently than Λ-Ru(phen)32+. Further, the estimated electron transfer rate was two orders of magnitude faster than the rate observed in the absence of DNA. Although the increase in rate was primarily ascribed to the reduced dimensionality of diffusion at the DNA surface, it was suggested that electron transfer through the π-framework of DNA may have played a part.
Evidence for DNA mediation of charge transfer mounted in a study involving electron transfer from excited Ru(phen)32+* to either Co(phen)33+, Rh(phen)33+, Cr(phen)33+, or Co(bpy)33+ [49]. These complexes are known to bind intercalatively in the major groove as well as electrostatically in the minor groove. Upon addition of DNA, luminescence quenching rates for each of these pairs increased. Interestingly, in 90% glycerol solutions at 253 K, where diffusion of all species is restricted, quenching rates were lower than in buffered aqueous solutions at ambient temperature, but they were still higher than the observed quenching rates in the absence of DNA. This result suggested that for these phen complexes, DNA-mediated electron transfer is a major quenching pathway. Nonetheless, with the use of freely diffusing charge donors and acceptors, it was difficult to discern the nature of DNA mediation due to rapid equilibration between binding modes and uncertainty in the distance between donor-acceptor pairs. Further experiments were necessary to establish DNA-mediated CT as an appreciable quenching mechanism.
Due to the larger hydrophobic surface area and further extension from the metal center, the incorporation of dppz allows for stronger DNA binding by intercalation than is allowed by phen. The use of Ru(phen)2dppz2+ in electron transfer experiments rather than Ru(phen)33+ made it possible to probe ET events in which the donor was primarily bound by intercalation. Further, because non-intercalated Ru(phen)2dppz2+* is quenched by water on an ultrafast timescale, any luminescence observed must originate from the intercalated species. Steady-state and time-resolved emission quenching of Ru(phen)2dppz2+* by either the strongly intercalating Rh(phi)2phen3+ or the groove binding Ru(NH3)63+ were examined [50]. In experiments involving the intercalated quencher, no change in emission rate was observed with increasing amounts of quencher; however, the initial luminescence intensity decreased. This result meant that quenching between the two intercalated species was occurring at rates faster than the instrument could detect. When Ru(NH3)63+ was used instead as quencher, increasing its concentration yielded an increase in the rate of luminescence decay but did not alter the initial luminescence yield. These results, in addition to comparisons with results of steady-state emission quenching experiments, showed that quenching by Ru(NH3)63+ is a dynamic process, while quenching by the intercalated Rh(phi)2phen3+ is a static process.
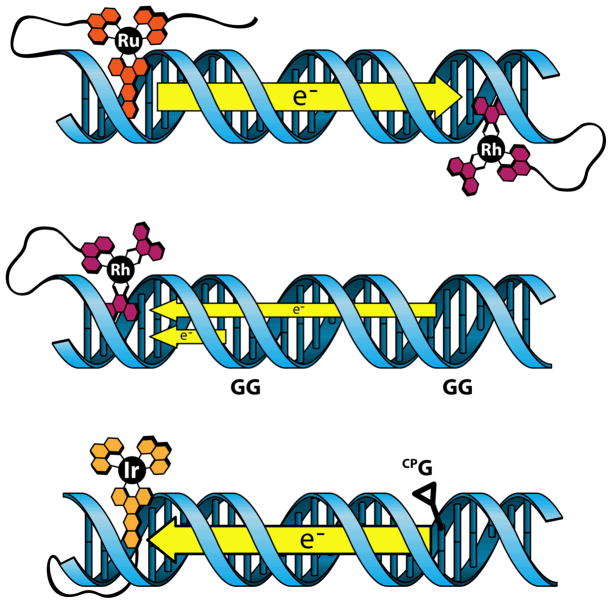
Further mechanistic insight was gained by covalently tethering Ru(phen’)2dppz2+ as an electron donor and Rh(phi)2(phen’)3+ as an acceptor (phen’ = 5-amido-glutaric acid-1,10-phenanthroline) to complementary strands of a DNA oligomer [51], as shown in Figure 3 (top). The covalent tether was long enough to allow for intercalation of the complexes, but short enough to prevent direct contact between them. By covalently attaching the donor and acceptor to opposite ends of the DNA duplex, the possibility for quenching through a diffusive mechanism was abolished, and the donor-acceptor distance was well defined. Excitation of assemblies in which the Ru-tethered strand was hybridized to its unmetallated complement resulted in strong luminescence. Addition of the covalently-tethered Rh complex to the complementary strand, however, resulted in complete quenching. Appropriate controls ensured that the quenching was taking place intraduplex, and the imposed separation between the donor and acceptor precluded quenching by diffusion. These results meant that quenching of the Ru(phen)2dppz2+ luminescence was occurring from over 35 Å away.
Figure 3.
Metal complex-DNA conjugates used to study DNA-mediated CT. Top: covalent tethering of Ru(phen’)2(dppz)2+ and Rh(phi)2(phen’)3+ to complementary DNA strands enables the study of DNA-mediated CT over large distances. Middle: bound Rh(phi)2(bpy’)3+ is competent to oxidize 5′-GG-3′ sites from a distance. Bottom: cyclopropylamine traps enable the fast capture of a charge as it travels along the DNA bridge.
That the mechanism of quenching was in fact electron transfer and not energy transfer was irrefutably established by experiments involving donors other than Ru(phen)2dppz2+. In one study, the transient absorption of systems containing varying amounts of non-covalent Ru(dmp)2dppz2+ (dmp = 4,7-dimetheyl-1,10-phenanthroline) and Rh(phi)2bpy3+ with DNA were investigated and compared with the transient spectrum obtained upon oxidative Ru(dmp)2dppz2+* quenching by Ru(NH3)63+ [52]. With increasing amounts of Rh, the luminescence decay lifetimes did not change, but the initial luminescence yield did, again signifying that the quenching in this system involved a static mechanism. The transient spectrum obtained by using the Rh complex as the quencher matched that obtained using Ru(NH3)63+ as the quencher, positively identifying the transient intermediate in the Rh experiment as the oxidation product, Ru(dmp)2dppz3+, and the mechanism of luminescence quenching as electron transfer. In another study, Os(phen)2dppz2+, rather than Ru(phen)2dppz2+, was used as the electron donor [53]. The Os complex emits at a higher wavelength, and its emission lifetime (< 10 ns) is several orders of magnitude shorter than that of Ru(phen)2dppz2+ [54]. Despite these photophysical differences, Os(phen)2dppz2+ behaves similarly: it is also a light switch, it binds DNA primarily through intercalation, and quenching by Rh(phi)2bpy3+ in the presence of DNA takes place through a static mechanism. Interestingly, the dependence of quenching yield on Rh(phi)2bpy3+ concentration is the same between Os(phen)2dppz2+ and Ru(phen)2dppz2+, meaning that the quenching mechanism is the same, despite photophysical and electronic differences. Also, transient spectra obtained upon photoexcitation of Os(phen)2dppz2+ in the presence of Rh(phi)2bpy3+ and DNA match spectra obtained through oxidative quenching of DNA-bound Os(phen)2dppz2+* by Ru(NH3)63+ and through direct ground state oxidation of Os(phen)2dppz2+ by Ce(NO3)62−. The agreement between these three spectra indicates that the oxidized Os species is being formed in each case. In addition, because the emission band of Os(phen)2dppz2+ does not overlap with the absorption band of the Rh complex, energy transfer is not available. These results together mean that both Os(phen)2dppz2+* and Ru(phen)2dppz2+* are quenched almost exclusively by Rh(phi)2bpy3+ through DNA-mediated electron transfer.
Incidentally, Ru(phen)2dppz2+ was not the complex used to identify the intermediate involved in DNA-mediated electron transfer because no long-lived transient that could be ascribed to Ru(III) was ever observed spectroscopically in mixed-sequence DNA. As was speculated and later confirmed, this was because the Ru(III) intermediate was a strong enough oxidant to oxidize the guanine bases within the DNA strand and was depleted as soon as it formed. This property was later utilized to great effect to gain a better understanding of the DNA CT process by oxidizing the bases of DNA directly.
2.3. Long-Range Oxidation of DNA
2.3.1. Characteristics of bases and base analogues
For metallointercalators of sufficiently high redox potential, the DNA bases themselves may serve as partners in charge transfer reactions. The redox potentials of the base nucleosides increase in the order: G (1.29 V) < A (1.42 V) < T (1.6 V) < C (1.7 V) [55]. Therefore, a metal complex such as Ru(phen)2dppz3+ [E0(3+/2+) = 1.63 vs. NHE] or Rh(phi)2bpy3+* [E0(3+*/2+) 2.0 vs. NHE] [50] should be competent to oxidize the bases upon photoexcitation. Interestingly, within the DNA base stack, the propensity for electron transfer to occur from a particular base is influenced by electronic interactions with its neighbors. For example, ab initio molecular orbital calculations have predicted that the electron donating ability of guanine should increase as: 5′-GT-3′, 5′-GC-3′ ≪ 5′-GA-3′ < 5′-GG-3′ < 5′-GGG-3′ [56]. Further, the HOMO of the 5′-GG-3′ doublet is calculated to lie primarily on the 5′-G, indicating that the 5′-G site should be preferentially oxidized at guanine doublets, as has been observed experimentally. The relative ease with which guanine, guanine doublets, and guanine triplets are oxidized leads to biological implications: given a random sequence of bases, regions of high guanine content are the most likely places to find large amounts of oxidative damage.
The use of non-natural base analogues further extends the ability to exploit the intimate interactions between bases in the study of DNA CT. Many base analogues only slightly perturb the geometry and energetic structure of the base stack and interact in a natural way with the other bases, becoming part of the base stack and sometimes forming hydrogen bonds with natural bases. Base analogues provide advantageous functions for the study of DNA CT. For example, 2-aminopurine is fluorescent and pairs with thymine [57]; and inosine, which shares a strong resemblance with guanine, nevertheless has a significantly higher oxidation potential (1.5 V vs. NHE) [57]. Bases that are modified by a cyclopropylamino group in the major groove serve as sensitive indicators of charge occupation. The properties of each of these classes of molecules, natural bases, non-natural base analogues, and cyclopropylamine-modified bases, can be exploited for the study of DNA CT.
2.3.2. Oxidation of Guanine by a Metallointercalator
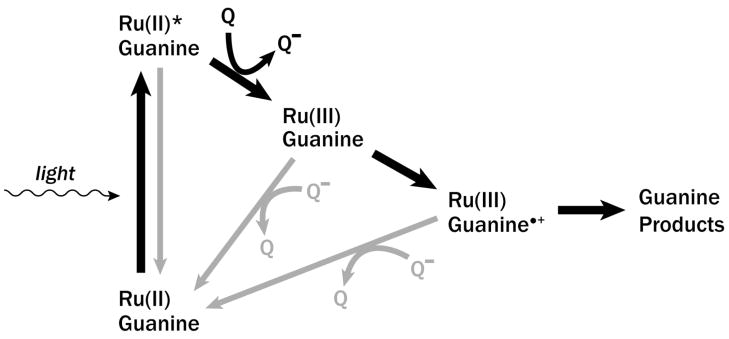
Direct proof of guanine oxidation by a ruthenium intercalator was obtained in a study involving Ru(phen)2dppz2+, DNA, and a variety of oxidative luminescence quenchers [58]. The quenchers used in the study, Ru(NH3)63+, methyl viologen (MV2+), and Co(NH3)5Cl2+, associate with DNA through groove binding and quench Ru(phen)2dppz2+* dynamically on the nanosecond timescale [50]. The study was an application of the flash-quench technique [21], shown in Scheme 2: following photoexcitation of the intercalated complex, oxidative quenching by a diffusible molecule creates the strong ground-state oxidant Ru(phen)2dppz3+ in situ, which then proceeds to oxidize guanine. The process may be interrupted by any of several processes, including depopulation of the Ru(phen)2dppz2+* excited state through luminescence, reduction of the Ru(III) oxidized species by back electron transfer (BET) from the reduced quencher, or guanine cation radical neutralization by the reduced quencher. The guanine radical thus created may then go on to react with O2 or H2O, forming permanent oxidation products.
Scheme 2.
The flash-quench technique. Following photoexcitation, Ru(II)* is oxidized by a diffusing quencher to form the powerful ground state oxidant Ru(III), which can then proceed to oxidize guanine within the base stack. Several back electron transfer pathways lower the efficiency of formation of guanine damage products.
In transient absorption experiments, the microsecond decay of a long-lived transient indicated formation of the oxidized ruthenium species in the presence of poly(dA-dT). In poly(dG-dC), no long-lived intermediate attributable to Ru(III) was observed; instead, a new transient species appeared on the timescale of Ru(II)* emission decay. This new transient was assigned to the neutral guanine radical, and its spectrum matched that previously observed by pulse radiolysis [59].
The yield of oxidized guanine product formation was then studied by gel electrophoresis. Ru(phen)2dppz2+ was irradiated at 436 nm in the presence of 18 base pair DNA duplexes containing guanine doublets or triplets and a quencher. Following radiolabeling and treatment with aqueous piperidine, which cleaves DNA at sites of guanine damage, the cleaved strands were separated by polyacrylamide gel electrophoresis and imaged by phosphorimagery. Damage occurred primarily at the 5′-G in duplexes containing 5′-GG-3′doublets, although small amounts of damage also occurred at single G sites, while strands incorporating both a 5′-GG-3′ and a 5′-GGG-3′ triplet exhibited damage mainly at the 5′-G of the triplet. Damage products were analyzed by enzymatic digestion followed by HPLC. Comparison with an authentic sample identified the major product as 8-oxo-2′-deoxyguanosine, the primary oxidative base lesion found within the cell [60].
2.3.3. GG Oxidation Occurs Over Long Distances
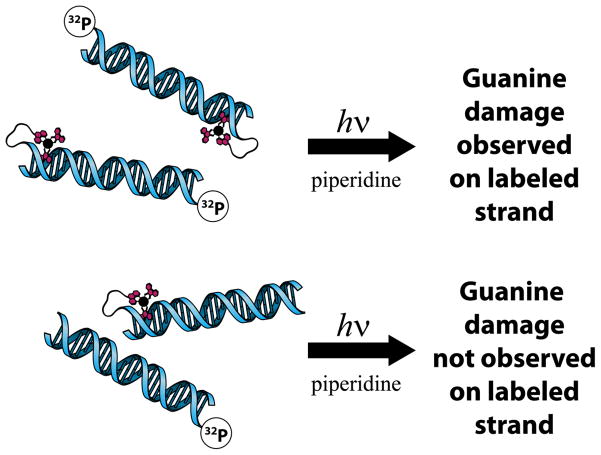
Studies of guanine oxidation were also carried out in systems containing metal-DNA conjugates. In one notable experiment, Rh(phi)2(bpy’)3+ [bpy’ = 4-methyl-4′-(butyric acid)-2,2′-bipyridine] was tethered to the end of a DNA 15-mer containing two 5′-GG-3′ doublets: one 17 Å away from the Rh binding site (proximal), and one 34 Å away from the binding site (distal) [61]. Such a construct is shown in Figure 3 (center). Rhodium complexes such as these serve as potent photooxidants when irradiated by 365 nm light, but promote direct strand cleavage at the site of intercalation when irradiated at 313 nm. When the conjugates were irradiated with 313 nm light, damage was only observed at the expected Rh binding site, three bases in from the end of the duplex. Upon excitation of the tethered complex with 365 nm light, guanine oxidation was observed primarily at the 5′-G of both 5′-GG-3′ doublets. While the irradiation experiment at 313 nm supported an intraduplex reaction, confirmation that the reaction was intraduplex was obtained in a mixed labeling experiment (Figure 4). Rhodium-DNA conjugates that were not radioactively tagged were mixed with DNA oligomers of the same sequence that were labeled but did not contain tethered Rh. Irradiation at 360 nm and subsequent piperidine treatment showed no damage to the DNA. Thus, in the Rh-tethered and labeled samples, oxidative damage was seen at distances of 17 Å and 34 Å from the bound Rh. This long-range damage was mediated by DNA CT.
Figure 4.
DNA-mediated oxidation is an intraduplex process. Top: guanine damage is observed by PAGE following irradiation and piperidine treatment of photooxidant-DNA conjugates that contain a 32P label. Bottom: no guanine damage is observed following the irradiation and piperidine treatment of mixtures which contain unlabeled photooxidant-DNA conjugates and labeled DNA that has no photooxidant bound.
Interestingly, very little difference was observed in the damage yields between distal and proximal GG sites in the experiments, meaning that radical delocalization and equilibration occurs more quickly than radical trapping and formation of permanent oxidation products, and suggesting that the distance dependence of DNA CT is quite low. In addition, guanine oxidation yields in conjugates containing the Δ isomer were higher than in those containing the Λ isomer, indicating that the efficiency of guanine damage is dependent on the interaction of the photooxidant with the base stack. Incorporation of a 5′-GGG-3′ far from the binding site led to oxidation primarily of the 5′-G of the triplet, 37 Å away from the intercalated Rh complex. Similar damage patterns were observed with the use of Ru(phen)(bpy’)(Me2dppz)2+ (Me2dppz = 9,10-dimethyl- dipyrido[3,2-a:2′,3′-c]phenazine) and the flash-quench reaction [62]. Interestingly, when only guanine singlets (no 5′-GG-3′ doublets) are incorporated into the base sequence, equal damage is observed at each guanine site, again suggesting that in the absence of a unique low energy site, charge migration and equilibration to sites of low oxidation potential occur at a faster rate than hole trapping.
Because oxidation yields at 5′-GG-3′ sites showed little variation with charge transfer distance over 11 base pairs, it was necessary to extend the length of the DNA to gain a better understanding of the distance dependence. To this end, a series of 28 base-pair duplexes were prepared with tethered Rh(phi)2(bpy’)3+ [63]. Each duplex in the series contained two 5′-GG-3′ sites that were separated from one another by increments of two base pairs, so that the distance between 5′-GG-3′ sites spanned a range from 41 to 75 Å. Upon irradiation, damage occurred at both sites, but the distal site consistently showed more damage than the proximal site. The ratio of damage at the distal site to that at the proximal site decreased only slightly and fairly linearly over the distances measured. Because the 5′-GG-3′ sites were separated by increments of only two base pairs (6.8 Å, or one-fifth of a turn in the helix), any helical phasing effects on the relative damage yields could be ruled out. In order to test the effects of CT over even greater distances, 63 base-pair DNA duplexes containing six well-separated 5′-GG-3′ sites along their length and a tethered photooxidant [either Ru(phen)(bpy’)dppz2+ or Rh(phi)2(bpy’)3+] were constructed by ligating smaller strands together. Irradiation of the ruthenated duplex by 436 nm light in the presence of MV2+ resulted in damage at the 5′ guanine of each doublet with a small diminution in oxidation with distance, showing that facile DNA-mediated oxidation can occur over 197 Å. The same experiment, carried out using the Rh-tethered duplex, yielded similar results. In these longer duplexes, damage yields decreased somewhat at longer distances, and this effect was more severe for ruthenium than for rhodium. The differences in damage yield at long distances were attributed to the ability of the flash-quench system to promote BET, differences in the extent of electronic coupling between the donor and the base stack in the two systems, and differences in the redox potentials of the donors. Interestingly, the damage yield ratio between distal and proximal sites increased dramatically with temperature, suggesting that higher temperatures facilitate charge equilibration along the length of the duplex.
In the 28 base-pair duplexes, replacement of a G·C base pair by a T·A base pair in the base sequence intervening between the two guanine doublets decreased the ratio of distal to proximal guanine damage by 38% [63]. This effect was more rigorously examined in subsequent work. Duplexes were constructed in which two guanine doublets were separated by increasing lengths of A- and T-containing sequences [64]. Photoexcitation of a tethered Rh(phi)2(bpy’)3+ complex resulted in large differences in the ratio of distal to proximal oxidative damage. Sequences that showed the lowest ratio contained 5′-TATA-3′ sequences intervening between the guanine doublets, while those showing the highest ratio contained only adenine. Interestingly, when the number of thymine bases intervening between guanine doublets was increased from two to ten by increments of two, damage ratios were 0.9, 1.2, 2.2, and 0.4, respectively. These results illustrate that factors such as DNA conformation, energetics, and base dynamics, in addition to distance, affect the efficiency of CT.
Mismatches intervening between two guanine doublets also affect the distal to proximal damage ratio, although in a manner that is not intuitive. When each of the sixteen possible combinations of matched and mismatched base pairs were incorporated between two guanine doublets, the highest distal/proximal damage ratio was observed for the C·G matched pair (2.05), while the A·T matched pair showed the third lowest ratio (0.23), after the T·C (0.15) and T·T (0.19) mismatches [65]. The observed differences in damage ratios did not correlate with the duplex stability, the thermodynamic stability of the mismatches, or the redox potential of the mismatched base. While there was a reasonable correlation with the free energies of helix destabilization of the mismatches, the best qualitative agreement was with base pair lifetimes based on imino proton exchange rates between mismatched bases, as measured by 1H NMR.
From these studies, it is apparent that many factors affect the yield of oxidative damage in DNA. Although shorter strands show little dependence on distance, damage yields are lower at longer distances in longer strands. Changes in the sequence intervening between two guanine doublets have a strong effect on the relative damage observed at the two sites, indicating that small changes in local conformation may disrupt the base stack locally, and that dynamic destacking at mismatch sites is sufficient to decrease severely the amount of damage further down the strand. The observed temperature dependence in long strands is also an indication of the major role that dynamic motions in DNA play in facilitating CT, since higher temperatures allow the DNA to sample more conformational states within the lifetime of the radical. Finally, differences in damage yields depending on the oxidant used indicate that the ability of the oxidant to couple electronically to the base stack and the propensity for BET strongly affect the efficiency of long-range DNA CT. These experiments involving metal complexes, as well as experiments involving organic oxidants such as ethidium, anthraquinone, or thionine [57,66–71] and base analogues such as 2-aminopurine [72–74] have shown that long-range DNA oxidation is a general phenomenon.
2.4. Fast Charge Trapping to Monitor Charge Occupancy on the DNA Bridge
Traditionally, models for DNA CT (see [75] for a recent review) have fallen into two basic categories: superexchange, in which the charge moves from the donor to the acceptor in a single coherent step, tunneling through an intermediating bridge; and localized hopping, in which the charge moves from base to base along the bridge, briefly occupying each site. As experimental results continued to disagree with predictions made from these models, the models were refined. For example, in hole transport, simple hopping models predict hopping to occur between guanine sites, since they are lowest in energy. The observed charge occupation on bridging adenine led to the development of thermally-assisted hopping models that resolve this inconsistency. Similarly, the influences of other bases and the solvation environment were included in even more complex polaron hopping models.
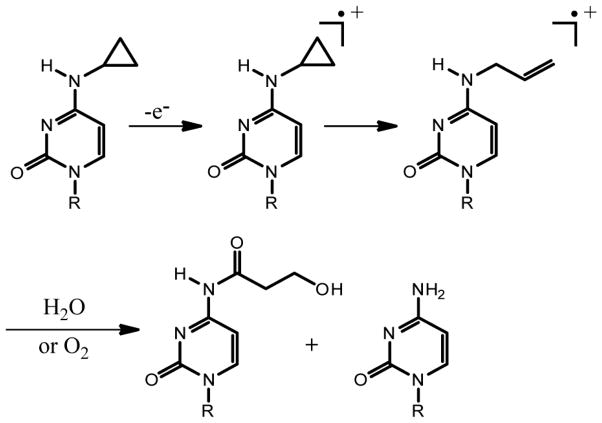
The guanine base, however, is a poor radical trap. The lifetime of a neutral guanine radical in DNA is greater than one millisecond [58], and on that timescale, the electron can migrate extensively and equilibrate in the DNA duplex. In order to gain mechanistic insight into the process of DNA-mediated CT, cyclopropylamine-modified bases, which report on short-lived charge occupancy at specific sites in DNA, were incorporated into various sequence contexts. As illustrated in Scheme 3, these modified bases, N2-cyclopropylguanine (CPG) [76], N6-cyclopropylcytosine (CPC) [77], and N6-cyclopropyladenine (CPA) [78], contain cyclopropyl groups which undergo a rapid ring-opening reaction upon oxidation. The rates of ring-opening are on the order of 1011 s−1, as suggested by comparison with similar molecules [79,80], making this reaction competitive with BET in most contexts. Further, the oxidation potentials, base pairing characteristics, and stacking properties of cyclopropyl-substituted bases are expected to be similar to those of the unmodified bases [76,77,81].
Scheme 3.
The CPC ring-opening mechanism.
Our first studies of DNA CT to CPG involved the use of photoexcited 2-aminopurine (Ap*) as the oxidant [82]. This analogue base-pairs with thymine and is well stacked in the DNA duplex. In addition, the CT process can be followed by monitoring quenching of Ap* fluorescence by guanine. In duplexes containing CPG, increasing temperatures caused an increase in the yield of ring-opened product until the melting temperature of the duplex was reached, at which point the duplex stacking was lost and almost no product was formed. The same experiment, using free Ap* rather than Ap incorporated into the base stack, showed no temperature dependence, indicating that temperature only affects the CT process, not the trapping process. This increase in ring-opening yield with increasing temperature suggests that DNA CT is a dynamic process which is facilitated by the motion of the bases. In order to study the distance dependence of CPG ring-opening yield, several strands were synthesized in which adenine bridges of increasing length were incorporated between Ap and CPG. Surprisingly, the quenching data showed a reproducible nonmonotonic periodicity in the distance dependence. In addition, little damage was observed for sequences in which the Ap and CPG were neighbors, or were separated by one intervening base pair [81]. These observations suggested that charge delocalization among small, transient, well-stacked groups of bases facilitate charge transfer, and that at short distances, BET is kinetically favored over ring-opening. To accommodate these observations, a new model for DNA CT was proposed which involves conformationally-gated hopping between well-stacked delocalized domains of charge.
This model was verified in further studies involving CPC oxidized by Rh(phi)2(bpy’)3+. When CPC was incorporated into strands 4–7 bp away from tethered Rh(phi)2(bpy’)3+, efficient ring-opening was observed upon photoexcitation, signifying that there must be some hole occupancy on cytosine during DNA CT, despite its high oxidation potential [77]. Interestingly, when CPG was incorporated at the site neighboring CPC, damage yields between the two traps were comparable, but when the distance between the CPC and the CPG traps was increased, the decomposition yield of the distal CPG decreased by a factor of two [83]. By examining CPC damage yields in various sequence contexts, the effects of neighboring bases were investigated further. In these studies, CPC decomposition depended not only on the sequence of bases intervening between the photooxidant and the hole trap, but also on the sequence distal to the hole trap. These results suggest that dynamic hole distribution on the DNA bridge is not just a function of the energies of the individual bases, and that some charge delocalization among the orbitals of neighboring bases must occur. Interestingly, while non-covalent Rh(phi)2bpy3+ is competent to oxidize both traps, non-covalent Ru(phen)(dppz)(bpy’)2+ in the presence of Ru(NH3)63+ does not show appreciable oxidation of CPC. This difference is consistent with the redox potentials of the two metal complexes.
The distance dependence of DNA CT was further studied by analyzing the decomposition yields of CPA and CPG within A tracts. Interestingly, when CPA was incorporated serially at each position along a 14 base-pair A tract, very little change in decomposition was observed with distance following irradiation of a tethered Rh(phi)2(bpy’)3+ photooxidant [84]. When CPG was incorporated at each position, however, the distance-dependent periodicity previously observed in 2-aminopurine studies was reproduced with the same apparent period, regardless of whether a Rh(phi)2(bpy’)3+, anthraquinone, or Ap photooxidant was used. [85]. Although this periodicity was similar to that observed earlier using an Ap* fluorescence quenching assay, the plots of damage yield versus distance obtained from the fluorescence quenching assay and the CPG assay were slightly different. These differences were explained recently [86]: due to the nature of the assay, fluorescence quenching informs on the yield of single-step CT, while the ring-opening assay informs on total CT; therefore, any difference between the two is the yield of multistep CT. At a distance of 8–9 bp, the yields obtained by Ap* fluorescence quenching and CPG ring-opening are equal, signifying that at this distance (27–30 Å), coherent transport takes place.
The ability of cyclopropyl traps to report on charge occupancy at various positions on the DNA bridge has allowed us to determine the relative influence of the various factors affecting the efficiency of DNA CT. Consistently and within a range of experiments, the ring-opening yield of the traps was observed to vary with distance, temperature, sequence context, and the redox potential of the donor. These observations support a model for DNA CT that consists of conformationally gated hopping of delocalized charge.
2.5. Comparing Long-Range DNA-Mediated Hole and Electron Transport with a Single Probe
Although the body of literature concerning DNA-mediated hole transport (HT) is quite extensive, complementary studies of DNA-mediated electron transport (ET) are relatively sparse. Our laboratory has extensively studied DNA-mediated ET using DNA-modified electrodes on gold [87–93]. While these experiments are interesting for many reasons, perhaps the most important question regarding DNA-mediated ET is whether the mechanism of this process differs in any way from that of DNA-mediated HT. Unfortunately, ET rates in these electrochemical constructs are limited by slow transfer through the thiol linker that connects the DNA to the gold surface [94]. Complexes such as (mes)2Pt(dppz)2+, which have been used both to oxidize CPG and to reduce CPC, are promising probes for solution state studies of DNA HT and ET, but these complexes are difficult to tether to DNA, making comparative studies of the distance dependence of HT and ET untenable [95].
To this end, our lab has developed an iridium complex that is amenable to functionalization and acts as both a photooxidant and a photoreductant in the presence of DNA [96]. The complex, Ir(ppy)2(dppz’)+ (ppy = phenylpyridine), contains a dppz ligand modified with a carboxylic acid functionality which enables covalent tethering of the complex to the 5′ end of a DNA single strand via a C6 alkyl chain. We envisage the intercalation of this complex as though the dppz ligand were threaded through the DNA, with the tether on one side of the duplex and the metal center and ancillary ligands on the other. Such a binding mode could easily be achieved during annealing of the DNA single strands to create the duplex. The excited state oxidation and reduction potentials of the complex are estimated to be 1.7 and −0.9 V vs. NHE, respectively, indicating that it is competent for both oxidative HT to guanine and reductive ET to thymine or cytosine. Because this single complex can be used to probe both DNA HT and DNA ET, the mechanisms and efficiencies of these processes can be directly compared in the same duplex.
We conducted these studies by taking advantage of the fast ring-opening kinetics of cyclopropylamine-modified nucleobases. When non-covalent Ir complex was added to duplexes containing CPG, only ten minutes of irradiation were needed to achieve complete degradation of the CP rings. The reaction was less efficient in the case of CPC: after twenty minutes of irradiation, the yield of ring opening was 86% [96]. The mechanism of CPG ring-opening in this construct is oxidative, while that of CPC ring-opening is reductive. If the CPC ring-opening reaction were to occur by an oxidative mechanism, substitution of inosine for the guanine opposite CPC should result in more efficient damage due to the decreased competition for holes. This effect was not observed. Further experiments involving covalently tethered Ir-DNA conjugates support these observations [97]. When CPG was incorporated into an adenine tract several bases away from the Ir complex intercalation site, the CPG ring-opening yield after one hour of irradiation was 46%. When CPC was incorporated into the duplex at the same site, the ring-opening yields were 31% when CPC was base paired with inosine and only 10% when CPC was base paired with guanine. These results suggest that within an adenine tract, CPC decomposition is an oxidative process, and that HT through an adenine tract is preferred over ET. When the modified bases were incorporated into thymine tracts instead, the results turned out differently. In these duplexes, the CPG ring-opening yield was very similar, at 55% after one hour of irradiation. However, the ring-opening yields for CPC embedded within thymine tracts were much lower, at 5% when CPC was paired with guanine and only 2% when CPC was paired with inosine. In this case, the yield was lower for the inosine-containing duplex, suggesting that CPC had been reduced and that ET is the preferred mechanism of charge transport through pyrimidines. Thus, the mechanism of CPC ring-opening depends strongly on the sequence context.
By systematically varying the distance between the intercalation site of the tethered Ir complex and the electron or hole acceptor in these assemblies, it is possible to compare the distance dependences of hole and electron transport directly, from the same probe and within the same sequence context. Experiments of this type were carried out utilizing CPA as a kinetically fast hole trap and 5-bromouridine (BrU) as a fast, irreversible electron trap [98]. In order to reduce BrU, excited Ir was first reduced by ascorbate via the flash-quench technique. The distance dependencies of the two processes were characterized by the parameter β, which serves as a proxy for the resistivity of the sequence and is a measure of the exponential decay in CT yield with distance. For HT, β = 0.05, while for ET, β = 0.10 or 0.12, depending on whether BrU was embedded within an adenine tract or a thymine tract, respectively. The shallow distance dependence observed in both cases suggests that HT and ET occur by similar mechanisms. Importantly, the amount of attenuation in CT yield upon the incorporation of a mismatch or abasic site at the position in the bridge neighboring the CT trap was identical for the CPA and BrU strands, indicating that successful charge migration along the duplex is less strongly affected by a change in the redox potential of the bridge as it is by perturbations in base stacking.
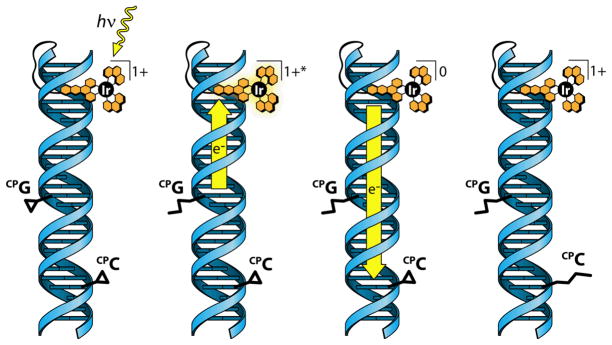
The ability of Ir(ppy)2(dppz’)+ to participate in both electron and hole transfer within DNA allows for a two-step CT process, dubbed the “ping pong” reaction, shown in Figure 5, in which the complex is first reduced by DNA-mediated HT, then subsequently reoxidized by DNA-mediated ET, following a single photoexcitation event. This reaction was utilized in a series of experiments involving HT to CPA followed by ET to either BrU or CPC in order to understand more fully the similarities and differences between DNA HT and ET [99]. In one experiment, the distance between the CPA and the Ir binding site was increased while the distance between the BrU and the Ir binding site remained the same. As the CPA was moved farther from the Ir binding site, both the CPA ring-opening yield and the BrU decomposition yield decreased, but the ratio of the decomposition yield to the ring-opening yield remained at about 40%. Importantly, very little BrU decomposition was observed in the absence of either CPA or the Ir complex. Strikingly, when CPC was substituted for BrU, although CPA and CPC ring-opening yields decreased as the CPA distance increased, CPC ring-opening was stoichiometric with CPA ring-opening. Base pairing CPC with inosine rather than guanine had no effect, suggesting that CPC is opened reductively in this sequence context. These results show that the ping-pong reaction is generalizable and very efficient. Importantly, by using a single probe to trigger both HT and ET under the same experimental conditions, we have shown that the two mechanisms have very similar characteristics, and that DNA CT, whether reductive or oxidative, is a general reporter for the integrity of the DNA base stack.
Figure 5.
Ping-pong electron transfer. From left to right: photoexcitation of the Ir complex results in DNA-mediated ET from the CPG base. Subsequent ET from the Ir complex reduces the CPC base.
3. DNA CHARGE TRANSPORT IN A BIOLOGICAL CONTEXT
Given this remarkable CT chemistry that can lead to redox reactions over long molecular distances and that is exquisitely sensitive to perturbations in the base-paired structure, we have begun to explore the biological implications of this chemistry. Below, we consider several examples where DNA-mediated CT may be powerfully utilized within the cell. Could oxidative radical damage be funneled to particular sites in the genome, and could other sites be insulated? Moreover, can we consider metalloproteins as participates in DNA CT? Indeed, could DNA-mediated CT be used for long-range signaling across the genome?
3.1. Generation of Mitochondrial DNA Mutations
DNA-mediated CT in vitro experiments have revealed that one-electron oxidation reactions, initiated using covalently tethered or intercalative metal complexes, can occur over distances as large as 200 Å [63]. This then leads to consideration of DNA CT in a physiological context, where cells must utilize a variety of pathways to protect the genome from incessant oxidative stress. Rh(phi)2(bpy)3+, a metal-based photooxidant, has emerged as a tool to probe hole migration over long distances in DNA [63]. Using ligation-mediated PCR to identify lesions, we have also utilized Rh(phi)2(bpy)3+ to probe DNA CT in nuclei isolated from HeLa cells. Damage in these systems occurs at the 5′-G of GG sites, even at sites with constitutively bound proteins, suggesting that oxidation occurs via DNA-mediated CT [55,56,100].
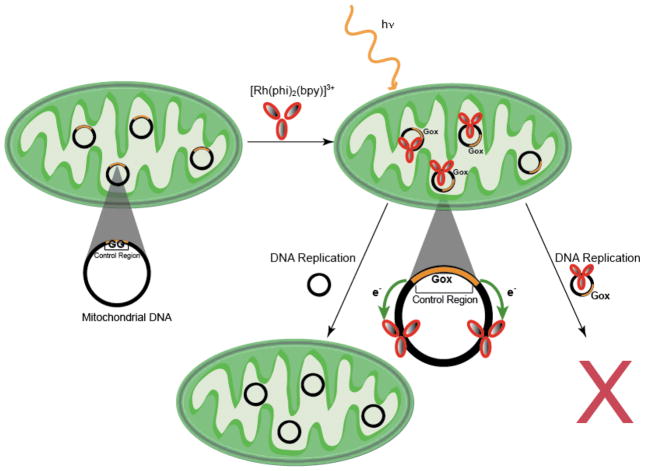
Mitochondria, cellular sites of respiration, accumulate ROS resulting from oxidative phosphorylation [5,101,102]. These organelles also contain their own DNA, making them particularly interesting systems to examine with respect to DNA CT. Photoactivation of Rh(phi)2(bpy)3+ at long wavelengths (365 nm) in the presence of DNA generates base mutations, such as 8-oxo-2′-deoxyguanosine (8-oxodG), identical to naturally occurring oxidative lesions. Irradiation at shorter wavelengths (313 nm) results in direct strand cleavage, marking the specific site of complex intercalation [103,104]. HeLa cells incubated with Rh(phi)2(bpy)3+, which is readily absorbed, and then irradiated, can also be analyzed for DNA mutations and oxidative damage [105]. Rh complex binding sites can be directly compared to sites of guanine oxidation with a primer extension assay [103]. Base oxidation resulting from DNA CT hole migration in vitro or in HeLa cells primarily occurs in conserved sequence block II, nucleotides 303–315, of mitochondrial DNA (mtDNA) [104]. Furthermore, this sequence is located within mitochondrial control region, which regulates primer formation for mtDNA replication [106,107]. Mutations within this region of mtDNA are associated with tumor formation and other disease conditions [108].
Remarkably, sites of DNA oxidation via CT within the mitochondria occur as far as 70 bases from the intercalated metal complex, suggesting that DNA CT plays a role in mtDNA oxidative damage [104]. Conserved sequence block II, which contains seven consecutive guanines, is a site of very low oxidation potential, and could therefore act as a sink for lesions such as 8-oxodG [100]. Extensive damage to this region through DNA-mediated funneling would save the cell from further error propagation by preventing replication of the damaged mitochondrial genome (Figure 6). Nevertheless, replication of DNA containing lesions ultimately leads to mutations such as dG to dT transversions, and these mutations also occur within conserved sequence block II. This ultimately reduces the amount of guanine in the regulatory region, eliminating the checkpoint and funneling site for oxidative stress, and limiting mutant mitochondrial function. Tumor cells, which do not necessarily depend on respiration, could then survive by utilizing alternative energy pathways independent of mitochondria [105].
Figure 6.
Multiple copies of mitochondrial DNA (black) are found in mitochondria (green) organelles within the cell. Irradiation of Rh(phi)2(bpy)3+ photooxidant results in oxidation of sites with low oxidation potential (Gox). Damage in the genome is funneled (green arrows), via DNA-mediated electron transfer (e−) to the control region (orange), preventing replication of the lesion-filled plasmid (bottom right). DNA replication of undamaged DNA occurs, aiding in the survival functional error-free mitochondria (bottom left).
3.2. DNA Charge Transport with Metalloproteins: Establishing DNA-bound Redox Potentials
3.2.1 Base Excision Repair Enzymes
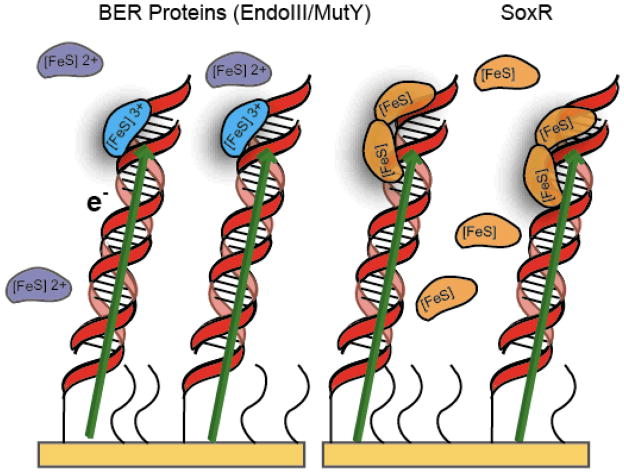
Several DNA repair proteins contain redox-active [FeS] clusters. However, a distinct role for these moieties in these proteins been investigated only recently. In a range of electrochemical studies, we have observed that all of the common DNA base lesions attenuate CT. In fact, all of the single base mismatches in DNA, irrespective of sequence context, inhibit DNA CT [88,109,110]. We have thus proposed, given the remarkable sensitivity of DNA CT to damaged bases, that repair proteins could use DNA CT to search for lesion-containing sites in DNA. Thus, many repair proteins containing [4Fe4S] clusters have been examined in our laboratory in this context [111–114]. MutY and endonuclease III (EndoIII), for example, highly homologous glycosylases from Escherichia coli, both contain a [4Fe4S]2+ cluster within a Cys-X6-Cys-X2- Cys-X5-Cys loop near the C terminus of the protein [115–120]. A vast number of DNA repair proteins are known to contain this loop, defined by the first two ligating cysteines, a proposed structural element [119–121]. Although the [4Fe4S] cluster of MutY is positioned within this loop, the protein is capable of folding without the cluster [122]. There is no increased structural stability due to the [4Fe4S] cluster, yet it is required for DNA binding. In the case of EndoIII, the [4Fe4S]2+ cluster is solvent-accessible and undergoes decomposition with ferricyanide. The protein is resistant to reduction, having an estimated [4Fe4S]2+/1+ couple midpoint potential of less than −600 mV [118,121]. Notably, initial measurements of the redox potentials of these proteins were performed in the absence of DNA. Since these are DNA-binding proteins, we then wondered whether a DNA environment would alter the redox properties of the [4Fe4S] cluster. Measurements of DNA-bound potentials were then made using DNA-modified electrodes to examine this question (Figure 7).
Figure 7.
Examples of surfaces used for DNA/protein electrochemistry of BER proteins (left) or SoxR (right). DNA duplexes are attached to the gold via a 5′ thiol linker. Mercaptohexanol (curved lines) is used to backfill so that the duplexes are in an upright position. Electrons travel (arrow) from the gold surface to the bound protein. EndoIII and MutY are bound when the [4Fe4S] cluster is in the 3+ oxidation state, and SoxR binds as a dimer.
MutY-bound DNA was probed electrochemically on loosely packed DNA gold electrode films [111]. A thiol-terminated duplex with a non-specific DNA binding site was attached to a gold surface and passivated with mercaptohexanol. Cyclic voltammetry of MutY at the DNA-modified surface yielded a midpoint potential of +90 mV vs. NHE, an appropriate potential for a physiologically active redox switch. In the absence of DNA, examination of a surface coated with mercaptohexanol yielded no electrochemical signal. Therefore, DNA was required to observe reduction of the [4Fe4S] cluster-containing protein. When the electrode was modified with duplex DNA containing an abasic site, no signal was observed, proving that electron transfer occurs through the DNA to the bound MutY protein. The redox reaction of MutY is DNA-mediated.
Redox potentials of +100 mV vs. NHE are common for high-potential iron proteins, and in the case of MutY (+90 mV vs. NHE) this redox potential indicated a shift in the oxidation state of the [4Fe4S] cluster from 2+ to 3+ as the protein binds to the DNA [112,123]. EndoIII was also examined bound to a DNA-modified electrode. As with MutY, when the potential was held at +50 mV, thus promoting oxidation of the cluster, the signal increased. Quite remarkably, both proteins display signals with midpoint potentials of ~100 mV vs. NHE in the presence of DNA. Both also display a much smaller signal when the intervening duplex contains an abasic site. We can then attribute oxidation of the cluster to DNA-mediated CT from the electrode through the base pair stack to the bound protein.
Building upon these studies, we then examined differences in the redox potential of EndoIII bound to DNA versus free using highly oriented pyrolytic graphite (HOPG) electrodes [113]. In these experiments, graphite electrodes were modified with pyrenated DNA, and both cyclic voltammetry and square wave voltammetry data were collected. A midpoint potential of ~20 mV vs. NHE was observed in the presence of well-matched DNA and EndoIII. In contrast, in the absence of DNA, oxidative scans of EndoIII yielded an irreversible anodic peak at ~250 mV and loss of the yellow solution color, indicating degradation of the [4Fe4S] cluster [113]. Consecutive positive scans showed broad, irregular signals at −80 and −710 mV vs. NHE, representative of clusters that have lost an iron atom to become [3Fe4S]. The 2+/3+ EndoIII [4Fe4S] cluster redox couple was found to have a positive potential (~250 mV). These data taken together indicate that EndoIII in the 3+ oxidation state binds DNA at least a thousand-fold more tightly than the reduced 2+ form. This characteristic shift in binding affinity between the reduced and oxidized forms of MutY and EndoIII has been an important element in our proposed mechanism for how these repair proteins scan the genome using DNA CT to find lesions [112,113,124,125].
3.2.2. The SoxR Transcription Factor
The SoxR transcription factor regulates the response to superoxide within the cell. SoxR binds DNA as a dimer and contains two [2Fe2S] clusters (one in each monomer) that are not required for protein folding [126–129]. The oxidation of SoxR leads to the expression of the SoxS transcription factor, which controls expression of genes involved in protecting the cell from stress. Interestingly, the oxidized and reduced formed of the protein show equal affinities for the SoxR promoter [114]. Redox potentials for SoxR without DNA, however, are about −290 mV, providing somewhat of a conundrum since that would suggest that at cellular potentials, SoxR is already oxidized; this is not an optimum situation for a sensor of oxidation [128,130,131]. Determination of the DNA-bound redox potential of SoxR provides insight into the activation mechanism of the protein. Well-matched DNA duplexes, attached to an HOPG electrode via pyrene modification, were self-assembled to form loosely packed DNA monolayers. Redmond Red was used as the redox-active probe in these studies, with a midpoint potential of −160 mV vs. NHE [132]. A quasi-reversible stable electrochemical signal was found for the [2Fe2S] cluster of SoxR at +200 mV vs. NHE [114]. Unlike MutY and EndoIII, SoxR shows asymmetry in the reduction and oxidation waves, displaying a better resolved anodic peak. Measurements of SoxR extracted from different organisms yield similar potentials. Significantly, then, there is ~500 mV positive shift in redox potential of SoxR upon DNA binding. It is likely that this shift in potential provides the energy for the torquing of the DNA by oxidized SoxR, activating transcription.
Critically, we see the importance of performing redox measurements of DNA binding proteins in the presence of DNA, as the DNA polyanion alters the protein environment and potential. MutY, EndoIII, and SoxR show redox activity in a physiologically relevant regime only when bound to DNA. These redox-active proteins, then, may use DNA CT as a sensor for oxidative damage and to locate lesions awaiting repair.
3.3. DNA-mediated Cross-linking and Oxidation of MutY
Methods used to probe long-range DNA-mediated oxidation of 5′-GG-3′ have aided in the study of DNA/protein interactions, allowing us to gain insight into how these metalloproteins might take advantage of the unique property of DNA to conduct charge. Experiments harnessing DNA-mediated CT to generate oxidized guanine, leading to DNA-protein cross-links, have provided a more detailed look at the amino acid/DNA interface [133–141]. Noncovalent ruthenium intercalators, upon photoactivation, have been used to generate guanine radicals, and these radicals can promote adducts with histone proteins.
DNA/MutY cross-linking experiments were performed using covalently tethered Rh(phi)2(bpy’)3+ to generate oxidative damage [141]. MutY was chosen for initial studies, since base flipping was thought to be involved in lesion detection. It is worth noting that lysine 142 in MutY was thought to play a large mechanistic role during the repair of 8-oxodG:A lesions based on early cross-linking and NMR investigations [50,134,142]. In our experiments, interestingly, there was no apparent change in distal/proximal guanine ratios upon DNA-mediated guanine oxidation in the presence of up to 200 nM of protein. This result indicated that MutY binding does not perturb long-range DNA CT, and thus might use a mechanism other than base flipping to search for damage. Oligonucleotides with an 8-oxodG:7-deaza-A MutY binding site 20 Å away from the tethered Rh photooxidant were then used to probe DNA CT-mediated cross-link formation between MutY and 8-oxodG. A covalent cross-link of 8-oxodG with MutY was identified involving lysine 142. This provided an example, then, of long range DNA-mediated CT providing a route to a protein/DNA redox reaction [141].
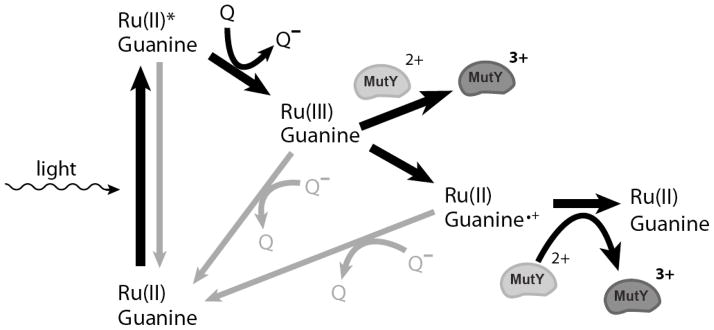
With reliable methods to generate oxidative damage and the knowledge that MutY is redox-active based electrochemical studies, we have shown that MutY itself can be activated in a DNA-mediated redox dependent manner [124]. Ru(phen)2dppz2+ binds to DNA by intercalation and is excited in the presence of visible light. This excited state is quenched by an electron acceptor to form a Ru(III) species in situ that can then oxidize guanines from a distance, and the guanine radical, once formed, can oxidize MutY (Scheme 4) [16,133]. To test this, we irradiated poly(dGC) and poly(dAT) duplexes in the presence of Ru(phen)2dppz2+ and a quencher, Co(NH3)5Cl2+, with or without MutY. EPR spectra at 10 K revealed a primary g value of 2.02 which was attributed to the [3Fe4S]1+ cluster, an oxidative degradation product of the [4Fe4S]2+ cluster [124]. Traces showed an additional signal at g values of 2.08 and 2.06 for both poly(dGC) and poly(dAT) duplexes in the presence of MutY, assigned to the oxidized [4Fe4S]3+ cluster [124,143]. Notably, with poly(dAT), these signals were significantly lower in intensity than with poly(dGC). Thus, the [4Fe4S] cluster of MutY can become oxidized to the 3+ state with or without intervening guanine radicals. The guanine radical, owing to its relatively long lifetime, can, however, increase the yield of protein oxidation.
Scheme 4.
The flash-quench technique used to generate Ru(III) and subsequently oxidize DNA-bound MutY. Following photoexcitation, Ru(II)* is quenched forming powerful ground state oxidant Ru(III) which can then proceed to oxidize guanine within the base stack. The guanine radical not only forms oxidative products but also oxidizes proteins, such as MutY. Back electron transfer reactions are shown in gray.
DNA-mediated oxidation of MutY was investigated on a faster time scale using transient absorption measurements in the presence of Ru(phen)2dppz2+, poly(dGC) DNA, and a Ru(NH3)63+ quencher [124]. As the DNA-bound metal complex was photoactivated, transient absorption decays were observed at several wavelengths. Transient absorption signals fit to a biexponential function indicated two phases: a fast phase corresponding to the presence of guanine radical in duplex DNA that decreased in the presence of MutY and a slow phase with a spectral profile characteristic of the [4Fe4S]3+/2+ difference spectrum. Importantly, these experiments established that guanine radical formation could directly lead to oxidation of bound protein. The intervening guanine radicals may allow MutY more time to undergo oxidation.
Biochemical studies examining 42-mer DNA duplexes that contain a 5′-GG-3′ doublet and that are covalently linked to a ruthenium intercalator also showed inhibition of 5′-G damage in the presence of MutY. This system differs from covalently tethered Rh(phi)2(bpy’)3+ used to generate oxidative damage in previous experiments, owing to its ability to generate guanine radicals through the flash-quench pathway. Guanine has an oxidation potential of −1.29 V vs. NHE, so it is easily oxidized in the presence of Ru(III) complex, which has a reduction potential of 1.5 V vs. NHE [55,58]. Importantly, the midpoint potential of DNA-bound MutY [4Fe4S]2+/3+ is about +0.1 V vs. NHE. Thus, instead of generating oxidative DNA damage, MutY can intercept the holes from the guanine radicals, becoming oxidized itself [111]. This explains the inhibition of damage observed in the denaturing PAGE gel experiments tracking 5′-G damage. EPR measurements further confirmed this hypothesis, displaying strong signals attributed to the oxidized [4Fe4S]3+ cluster in samples of Ru complex tethered to the 5′-GG-3′-containing 42-mer duplex in the presence of MutY. Collectively, transient absorption, EPR, and biochemical experiments indicate that MutY can be oxidized in a DNA-mediated fashion. Moreover, the implications of these findings for DNA repair by redox-active proteins are critical, since within the cell, oxidative stress may lead to this chemistry as a means to activate repair.
We have thus proposed a mechanism to activate repair using DNA CT. As we have shown, binding to DNA shifts the potential of the MutY [4Fe4S] cluster toward oxidation, activating it when repair is required. The BER proteins all have similar DNA-bound redox potentials and contain a reduced [4Fe4S]2+ in solution in the absence of oxidative stress [124]. Guanine radicals, forming endogenously under conditions of oxidative stress, could initiate binding of protein to DNA, oxidizing the [4Fe4S] cluster from 2+ to 3+, and increasing protein affinity for the duplex. As this occurs, an electron is transferred through the DNA to a distally bound BER protein. This distant protein is then reduced, loses its affinity for DNA, and is free to relocate to another site. The process comprises a scan of the region of DNA between the proteins as long as CT occurs efficiently. In the presence of a lesion known to attenuate charge transfer, such as a mismatch or oxidized base, however, DNA CT does not occur between bound proteins, and thus the proteins remain in the vicinity of the damage, slowly proceeding to the site of damage. In this model, proteins are expected to redistribute onto regions of DNA containing lesions. This mechanism is currently being investigated both in vitro and in vivo [125].
3.4. DNA-mediated Oxidation of SoxR Activates Transcription
Applying our findings on DNA-mediated CT involvement in BER pathways, we have examined SoxR, which contains a metallo transcription factor. SoxR is activated in the presence of oxidative stress; however, the specific oxidant is unknown. The protein is reversibly inactivated in vitro by reduction of its [2Fe2S] clusters using dithionite [128]. In vivo studies, using redox-cyclers such as paraquat to induce oxidative stress, show that superoxide is not the direct activator of SoxR. Rather, the redox-cyclers deplete cellular NADPH, which is normally required to keep SoxR in a reduced form. The redox-cyclers then undergo autooxidation and produce superoxide by losing an electron to dioxygen. Plumbagin and phenazine methosulfate, notably, have yielded reversible oxidation of the [2Fe2S] clusters [128]. Electrochemistry of SoxR in our lab showed a redox-active signal for the [2Fe2S] cluster of SoxR at +200 mV vs. NHE, indicating that the protein undergoes one-electron oxidation when bound to DNA [114]. Due to the redox shift we observed upon DNA binding, we questioned whether the DNA-bound form of SoxR might be the missing oxidative switch. As DNA is subjected to oxidative damage, guanine radicals capable of migrating through the base pair stack are generated. These guanine radicals can then serve as the oxidant for SoxR. Once oxidized, SoxR can promote transcription under conditions when the cell needs it the most. The oxidative signal then would be the guanine radical.
We have learned that the electronic coupling of photooxidants used to probe DNA-mediated CT is an important factor in observing this process. Therefore, SoxR studies were recently conducted in our lab using a strong DNA intercalator, Ru(phen)(dppz)(bpy’)2+ to generate guanine radicals resulting in irreversible oxidative damage [144]. Co(NH3)5Cl2+ was used as the quencher in these studies, eliminating back electron transfer, so that the major pathway in the reaction yields oxidative damage. Similar to DNA-bound MutY experiments described earlier in which MutY oxidation reduced the extent of guanine damage, the yield of 5′-GG-3′ oxidation products was measured with and without oxidized or reduced SoxR. Upon reduction of SoxR with sodium dithionite in the presence of DNA-bound Ru, quencher, and light, damage at guanine sites was attenuated. This indicates that SoxR is able to donate an electron to the oxidized guanine, filling the radical hole with its own lost electron, and being converted from a reduced to an oxidized form. Conversely, there was no attenuation in damage when oxidized SoxR was used in the experiment. These results clearly show that SoxR is able to interact with the DNA base stack, thus participating in DNA-mediated CT. The SoxR response to guanine radicals generated in DNA suggests that oxidative damage can initiate protein activation.
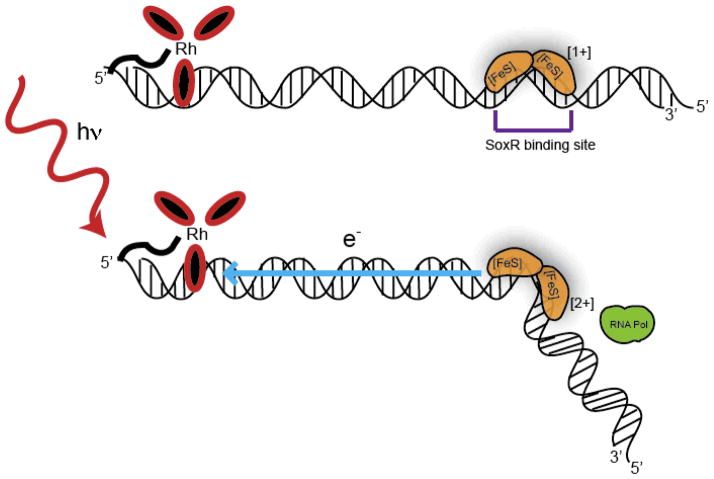
In order to examine the SoxR response to DNA damage in its native cellular environment, E. coli cultures were treated with Rh(phi)2bpy3+. This intercalator, upon photoactivation, was previously shown to generate guanine damage in mitochondria and HeLa cells [103–105]. In the experiment, transcription of the soxS RNA product, observed using reverse transcription PCR, indicated activation of SoxR. Bacteria irradiated in the presence of the Rh photooxidant showed much greater soxS product levels than those without irradiation. Since the complex strictly induces DNA damage and cannot oxidize SoxR directly without DNA, SoxR must be activated by guanine radicals in these experiments. Expression levels of soxS in the presence of Rh(phi)2bpy3+ were similar to those measured when methyl viologen, a redox-cycler, was used, with both systems activating SoxR to the same extent. But is SoxR oxidized from a distance through DNA-mediated CT [61,144]? To test this idea, Rh(phi)2bpy3+ was tethered to a 180-mer duplex of DNA containing the SoxR binding site and the −10 and −35 promoter regions of soxS (Figure 8), and gene products were monitored with an anaerobic abortive transcription assay following irradiation [127]. The abortive transcription product, a 4-mer, was only observed in samples containing reduced SoxR and Rh-tethered DNA. Importantly, there was no direct photooxidation of SoxR by Rh(phi)2bpy3+ without DNA. Remarkably, then, DNA-mediated transcriptional activation of SoxR was indeed demonstrated over a 270 Å distance. DNA CT has emerged not only as an indicator of oxidative damage, but also as a signal to metalloproteins in the genome to initiate repair.
Figure 8.
Rh(phi2)(bpy’)3+ is tethered to a duplex of DNA containing the SoxR binding site. SoxR is bound in the reduced (+1) state (yellow protein). Photoactivation of the metal complex triggers electron transfer, oxidizing SoxR (2+), which may then kink the DNA and recruit transcription machinery such as RNA polymerase (green).
4. CONCLUSIONS
Initially used as general charge donors and acceptors to probe DNA CT, the utility of metal complexes have come a long way. Precise tuning of reactivity and specificity has resulted in complexes now capable of generating specific DNA damage in vivo. Probe design, as discussed, takes into account properties including ligand sterics, binding mode, coupling to the base pair stack, and stereospecificity. Metallointercalators, such as Rh(phi)2bpy3+, initiate electron transfer in DNA upon photoactivation, resulting in observable damage to guanine, which is the most easily oxidized nucleobase. Charge transport through DNA can occur over long distances, and it is because of metal complexes used as probes that we are able to establish this unique property of DNA. The photophysical properties of these complexes in particular have facilitated spectroscopic characterization. Recently, iridium complexes have also aided in elucidation of the hole transfer mechanism in DNA and allowed for direct comparison between hole and electron transfer.
Significantly, in vivo studies depend on generation of DNA damage by photooxidants to elucidate the role of CT within cells. Utilizing metal complexes as photooxidants, the funneling of damage to specific regulatory sites in the mitochondrial genome has been identified. Moreover, CT can occur not just to generate guanine damage, but also to activate repair proteins and transcription. Here too we have seen DNA-mediated redox chemistry, but now with metalloproteins. Using reactions with simple coordination complexes to guide our questions, our goal now shifts to discovery of various endogenous “metal complex” systems in which a metalloprotein couples with DNA using DNA-mediated CT to activate signaling across the genome.
Acknowledgments
We are grateful to NIH for their support of this research. We also thank our many colleagues for their efforts and insights, especially Harry Gray, whose seminal work on protein electron transfer, especially that using our favorite flash-quench methodology, inspired many experiments.
References
- 1.van Loon B, Markkanen E, Hübscher U. DNA Repair. 2010;9:604. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 2.David SS, O’Shea VL, Kundu S. Nature. 2007;477:941. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Møller P, Folkmann JK, Forchhammer L, Bräuner EV, Danielsen PH, Risom L, Loft S. Cancer Lett. 2008;266:84. doi: 10.1016/j.canlet.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 4.Cooke MS, Evans MD, Dizdaroglu M. J Lunec FASEB J. 2003;17:1195. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 5.Nishikawa M. Cancer Lett. 2008;266:53. doi: 10.1016/j.canlet.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 6.Klaunig JE, Kamendulis LM. Annu Rev Pharmacol Toxicol. 2004;44:239. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 7.Kopelevich Y, Esquinazi P. Adv Mater. 2007;19:4559. [Google Scholar]
- 8.Lerman LS. J Mol Biol. 1961;3:18. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- 9.Berman HM, Young PR. Ann Rev Biophys Bioeng. 1981;10:87. doi: 10.1146/annurev.bb.10.060181.000511. [DOI] [PubMed] [Google Scholar]
- 10.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Nat Struct Biol. 2000;7:117. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 11.Pierre VC, Kaiser JT, Barton JK. Proc Natl Acad Sci USA. 2007;104:429. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeglis BM, Pierre VC, Barton JK. Chem Commun. 2007;44:4565. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeglis BM, Pierre VC, Kaiser JT, Barton JK. Biochemistry. 2009;48:4247. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barton JK, Kumar CV, Turro NJ. J Am Chem Soc. 1986;108:6391. [Google Scholar]
- 15.Murphy CJ, Barton JK. Methods Enzymol. 1993;226:576. doi: 10.1016/0076-6879(93)26027-7. [DOI] [PubMed] [Google Scholar]
- 16.Friedman AE, Chambron J-C, Sauvage J-P, Turro NJ, Barton JK. J Am Chem Soc. 1990;112:4960. [Google Scholar]
- 17.Hartshorn RM, Barton JK. J Am Chem Soc. 1992;114:5919. [Google Scholar]
- 18.Jenkins Y, Friedman AE, Turro NJ, Barton JK. Biochemistry. 1992;31:10809. doi: 10.1021/bi00159a023. [DOI] [PubMed] [Google Scholar]
- 19.Turro C, Bossman SH, Jenkins Y, Barton JK, Turro NJ. J Am Chem Soc. 1995;117:9026. [Google Scholar]
- 20.Olson EJC, Hu D, Hörmann A, Jonkman AM, Arkin MR, Stemp EDA, Barton JK, Barbara PF. J Am Chem Soc. 1997;119:11458. [Google Scholar]
- 21.Chang I-J, Gray HB, Winkler JR. J Am Chem Soc. 1991;113:7056. [Google Scholar]
- 22.Barton JK, Dannenberg JJ, Raphael AL. J Am Chem Soc. 1982;104:4967. [Google Scholar]
- 23.Barton JK, Basile LA, Danishefsky A, Alexandrescu A. Proc Natl Acad Sci USA. 1984;81:1961. doi: 10.1073/pnas.81.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton JK, Danishefsky AT, Goldberg JM. J Am Chem Soc. 1984;106:2172. [Google Scholar]
- 25.Barton JK, Raphael AL. J Am Chem Soc. 1984;106:2466. [Google Scholar]
- 26.Barton JK, Lolis E. J Am Chem Soc. 1985;107:708. [Google Scholar]
- 27.Kumar CV, Barton JK, Turro NJ. J Am Chem Soc. 1985;107:5518. [Google Scholar]
- 28.Westerlund F, Pierard F, Eng MP, Nordén B, Lincoln P. J Am Chem Soc. 2005;109:17327. doi: 10.1021/jp0517091. [DOI] [PubMed] [Google Scholar]
- 29.Barton JK. Science. 1986;233:727. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- 30.Barton JK, Goldberg JM, Kumar CV, Turro NJ. J Am Chem Soc. 1986;108:2081. [Google Scholar]
- 31.Jenkins Y, Barton JK. J Am Chem Soc. 1992;114:8736. [Google Scholar]
- 32.Holmlin RE, Dandliker PJ, Barton JK. Bioconj Chem. 1999;10:1122. doi: 10.1021/bc9900791. [DOI] [PubMed] [Google Scholar]
- 33.Zeglis BM, Barton JK. J Am Chem Soc. 2006;128:5654. doi: 10.1021/ja061409c. [DOI] [PubMed] [Google Scholar]
- 34.Puckett CA, Barton JK. J Am Chem Soc. 2009;131:8738. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puckett CA, Barton JK. Bioorg Med Chem. 2010;18:3564. doi: 10.1016/j.bmc.2010.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krotz AH, Kuo LY, Shields TP, Barton JK. J Am Chem Soc. 1993;115:3877. [Google Scholar]
- 37.Krotz AH, Hudson BP, Barton JK. J Am Chem Soc. 1993;115:12577. [Google Scholar]
- 38.Sitlani A, Barton JK. Biochemistry. 1994;33:12100. doi: 10.1021/bi00206a013. [DOI] [PubMed] [Google Scholar]
- 39.Shields TP, Barton JK. Biochemistry. 1995;34:15037. doi: 10.1021/bi00046a009. [DOI] [PubMed] [Google Scholar]
- 40.Hudson BP, Barton JK. J Am Chem Soc. 1998;120:6877. [Google Scholar]
- 41.Sardesai NY, Zimmermann K, Barton JK. J Am Chem Soc. 1994;116:7502. [Google Scholar]
- 42.Sardesai NY, Barton JK. J Biol Inorg Chem. 1997;2:762. [Google Scholar]
- 43.Hastings CA, Barton JK. Biochemistry. 1999;38:10042. doi: 10.1021/bi982039a. [DOI] [PubMed] [Google Scholar]
- 44.Terbrueggen RH, Barton JK. Biochemistry. 1995;34:8227. doi: 10.1021/bi00026a003. [DOI] [PubMed] [Google Scholar]
- 45.Terbrueggen RH, Johann TW, Barton JK. Inorg Chem. 1998;37:6874. doi: 10.1021/ic980837j. [DOI] [PubMed] [Google Scholar]
- 46.Franklin SJ, Barton JK. Biochemistry. 1998;37:16093. doi: 10.1021/bi981798q. [DOI] [PubMed] [Google Scholar]
- 47.Odom DT, Parker CS, Barton JK. Biochemistry. 1999;38:5155. doi: 10.1021/bi9827969. [DOI] [PubMed] [Google Scholar]
- 48.Fitzsimons MP, Barton JK. J Am Chem Soc. 1997;119:3379. [Google Scholar]
- 49.Purugganan MD, Kumar CV, Turro NJ, Barton JK. Science. 1988;241:1645. doi: 10.1126/science.241.4873.1645. [DOI] [PubMed] [Google Scholar]
- 50.Murphy CJ, Arkin MR, Ghatlia ND, Bossmann S, Turro NJ, Barton JK. Proc Natl Acad Sci USA. 1994;91:5315. doi: 10.1073/pnas.91.12.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy CJ, Arkin MR, Jenkins Y, Ghatlia ND, Bossmann SH, Turro NJ, Barton JK. Science. 1993;262:1025. doi: 10.1126/science.7802858. [DOI] [PubMed] [Google Scholar]
- 52.Stemp EDA, Arkin MR, Barton JK. J Am Chem Soc. 1995;117:2375. [Google Scholar]
- 53.Holmlin RE, Stemp EDA, Barton JK. J Am Chem Soc. 1996;118:5236. [Google Scholar]
- 54.Holmlin RE, Barton JK. Inorg Chem. 1995;34:7. doi: 10.1021/ic970869r. [DOI] [PubMed] [Google Scholar]
- 55.Steenken S, Jovanovic SV. J Am Chem Soc. 1997;119:617. doi: 10.1021/ja003823x. [DOI] [PubMed] [Google Scholar]
- 56.Saito I, Takayama M, Sugiyama H, Nakatani K. J Am Chem Soc. 1995;117:6406. [Google Scholar]
- 57.Kelley SO, Barton JK. Science. 1999;283:375. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 58.Stemp EDA, Arkin MR, Barton JK. J Am Chem Soc. 1997;119:2921. [Google Scholar]
- 59.Candeias LP, Steenken S. J Am Chem Soc. 1989;111:1094. [Google Scholar]
- 60.Shibutani S, Takeshita M, Grollman AP. Nature. 1991;349:431. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 61.Hall DB, Holmlin RE, Barton JK. Nature. 1996;382:731. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 62.Arkin MR, Stemp EDA, Pulver SC, Barton JK. Chem Biol. 1997;4:389. doi: 10.1016/s1074-5521(97)90129-0. [DOI] [PubMed] [Google Scholar]
- 63.Núñez ME, Hall DB, Barton JK. Chem Biol. 1999;6:85. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 64.Williams TT, Odom DT, Barton JK. J Am Chem Soc. 2000;122:9048. [Google Scholar]
- 65.Bhattacharya PK, Barton JK. J Am Chem Soc. 2001;123:8649. doi: 10.1021/ja010996t. [DOI] [PubMed] [Google Scholar]
- 66.Hall DB, Kelley SO, Barton JK. Biochemistry. 1998;37:15933. doi: 10.1021/bi981750c. [DOI] [PubMed] [Google Scholar]
- 67.Kelley SO, Holmlin RE, Stemp EDA, Barton JK. J Am Chem Soc. 1997;119:9861. [Google Scholar]
- 68.Kelley SO, Barton JK. Chem Biol. 1998;5:413. doi: 10.1016/s1074-5521(98)90158-2. [DOI] [PubMed] [Google Scholar]
- 69.Wan C, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc Natl Acad Sci USA. 1999;96:6014. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanvah S, Schuster GB. Pure Appl Chem. 2006;78:2297. [Google Scholar]
- 71.Williams TT, Dohno C, Stemp EDA, Barton JK. J Am Chem Soc. 2004;126:8148. doi: 10.1021/ja049869y. [DOI] [PubMed] [Google Scholar]
- 72.O’Neill MA, Barton JK. J Am Chem Soc. 2002;124:13053. doi: 10.1021/ja0208198. [DOI] [PubMed] [Google Scholar]
- 73.O’Neill MA, Barton JK. Proc Natl Acad Sci USA. 2002;99:16543. doi: 10.1073/pnas.012669599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Neill MA, Becker H-C, Wan C, Barton JK, Zewail AH. Angew Chem Int Ed. 2003;42:5896. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 75.Genereux JC, Barton JK. Chem Rev. 2010;110:1642. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakatani K, Dohno C, Saito I. J Am Chem Soc. 2001;123:9681. doi: 10.1021/ja010479a. [DOI] [PubMed] [Google Scholar]
- 77.Shao F, O’Neill MA, Barton JK. Proc Natl Acad Sci USA. 2004;101:17914. doi: 10.1073/pnas.0408128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dohno C, Ogawa A, Nakatani K, Saito I. J Am Chem Soc. 2003;125:10154. doi: 10.1021/ja035887o. [DOI] [PubMed] [Google Scholar]
- 79.Musa OM, Horner JH, Shahin H, Newcomb M. J Am Chem Soc. 1996;118:3862. [Google Scholar]
- 80.Shaffer CL, Morton MD, Hanzlik RP. J Am Chem Soc. 2001;123:349. doi: 10.1021/ja003048l. [DOI] [PubMed] [Google Scholar]
- 81.O’Neill MA, Dohno C, Barton JK. J Am Chem Soc. 2004;126:1316. doi: 10.1021/ja037802p. [DOI] [PubMed] [Google Scholar]
- 82.O’Neill MA, Barton JK. J Am Chem Soc. 2004;126:11471. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 83.Shao F, Augustyn K, Barton JK. J Am Chem Soc. 2005;127:17445. doi: 10.1021/ja0563399. [DOI] [PubMed] [Google Scholar]
- 84.Augustyn KE, Genereux JC, Barton JK. Angew Chem Int Ed Eng. 2007;46:5731. doi: 10.1002/anie.200701522. [DOI] [PubMed] [Google Scholar]
- 85.Genereux JC, Augustyn KE, Davis ML, Shao F, Barton JK. J Am Chem Soc. 2008;130:15150. doi: 10.1021/ja8052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Genereux JC, Wuerth SM, Barton JK. submitted for publication. [Google Scholar]
- 87.Kelley SO, Barton JK. Bioconj Chem. 1997;8:31. doi: 10.1021/bc960070o. [DOI] [PubMed] [Google Scholar]
- 88.Kelley SO, Jackson NM, Hill MG, Barton JK. Angew Chem Int Ed. 1999;38:941. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 89.Boon EM, Salas JE, Barton JK. Nat Biotech. 2002;20:282. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 90.Boon EM, Barton JK. Bioconj Chem. 2003;14:1140. doi: 10.1021/bc034139l. [DOI] [PubMed] [Google Scholar]
- 91.Boal AK, Barton JK. Bioconj Chem. 2005;16:312. doi: 10.1021/bc0497362. [DOI] [PubMed] [Google Scholar]
- 92.Gorodetsky AA, Buzzeo MC, Barton JK. Bioconj Chem. 2008;19:2285. doi: 10.1021/bc8003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Genereux JC, Barton JK. Nat Chem. 2009;1:106. doi: 10.1038/nchem.188. [DOI] [PubMed] [Google Scholar]
- 94.Drummond TG, Hill MG, Barton JK. J Am Chem Soc. 2004;126:15010. doi: 10.1021/ja044910i. [DOI] [PubMed] [Google Scholar]
- 95.Lu W, Vicic DA, Barton JK. Inorg Chem. 2005;44:7970. doi: 10.1021/ic051124l. [DOI] [PubMed] [Google Scholar]
- 96.Shao F, Elias B, Lu W, Barton JK. Inorg Chem. 2007;46:10187. doi: 10.1021/ic7014012. [DOI] [PubMed] [Google Scholar]
- 97.Shao F, Barton JK. J Am Chem Soc. 2007;129:14733. doi: 10.1021/ja0752437. [DOI] [PubMed] [Google Scholar]
- 98.Elias B, Shao F, Barton JK. J Am Chem Soc. 2008;130:1152. doi: 10.1021/ja710358p. [DOI] [PubMed] [Google Scholar]
- 99.Elias B, Genereux JC, Barton JK. Angew Chem Int Ed Eng. 2008;47:9067. doi: 10.1002/anie.200803556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Merino EJ, Boal AK, Barton JK. Curr Opin Chem Biol. 2008;12:229. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wallace DC. Annu Rev Genet. 2005;39:359. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cadenas E, Davies KJA. Free Radical Biol Med. 2000;29:222. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 103.Merino EJ, Barton JK. Biochemistry. 2007;46:2805. doi: 10.1021/bi062024+. [DOI] [PubMed] [Google Scholar]
- 104.Merino EJ, Barton JK. Biochemistry. 2008;47:1511. doi: 10.1021/bi701775s. [DOI] [PubMed] [Google Scholar]
- 105.Merino EJ, Davis ML, Barton JK. Biochemistry. 2009;48:660. doi: 10.1021/bi801570j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu B, Clayton DA. EMBO J. 1996;15:3135. [PMC free article] [PubMed] [Google Scholar]
- 107.Pham XH, Farge G, Shi YH, Gaspari M, Gustafsson CM, Falkenberg M. J Biol Chem. 2006;281:24647. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 108.Tan DJ, Bai RK, Wong LJC. Cancer Res. 2002;62:972. [PubMed] [Google Scholar]
- 109.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2000;18:1096. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 110.Kelley SO, Boon EM, Barton JK, Jackson NM, Hill MG. Nucleic Acids Res. 1999;27:4830. doi: 10.1093/nar/27.24.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boon EM, Livingston AL, Chmiel NH, David SS, Barton JK. Proc Natl Acad Sci. 2003;100:12543. doi: 10.1073/pnas.2035257100. 101 (2004) 4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boal AK, Yavin E, Lukianova OA, O’Shea VL, David SS, Barton JK. Biochemistry. 2005;44:8397. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 113.Gorodetsky AA, Boal AK, Barton JK. J Am Chem Soc. 2006;128:12082. doi: 10.1021/ja064784d. [DOI] [PubMed] [Google Scholar]
- 114.Gorodetsky AA, Dietrich LEP, Lee PE, Demple B, Newman DK, Barton JK. Proc Natl Acad Sci USA. 2008;105:3684. doi: 10.1073/pnas.0800093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.David SS, Williams SD. Chem Rev. 1998;98:1221. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 116.Michaels ML, Pham L, Nghiem Y, Cruz C. JH Miller Nucleic Acids Res. 1990;18:3841. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S. JA Tainer Nat Struct Biol. 1998;5:1058. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 118.Cunningham RP, Asahara H, Bank JF, Scholes CP, Slerno JC, Surerus K, Munck E, McCracken J, Peisach J, Emptage MH. Biochemistry. 1989;28:4450. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- 119.Kou C-F, McRee DE, Fischer CL, O’Handley SF, Cunningham RP, Tainer JA. Science. 1992;258:434. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 120.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. EMBO J. 1995;14:4108. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fu W, O’Handley S, Cunningham RP, Johnson MK. J Biol Chem. 1992;267:16135. [PubMed] [Google Scholar]
- 122.Porello SL, Cannon MJ, David SS. Biochemistry. 1998;37:6465. doi: 10.1021/bi972433t. [DOI] [PubMed] [Google Scholar]
- 123.Cowan JA, Lui SM. Adv Inorg Chem. 1998;45:313. [Google Scholar]
- 124.Yavin E, Boal AK, Stemp EDA, Boon EM, Livingston AL, O’Shea VL, David SS, Barton JK. Proc Natl Acad Sci. 2005;102:3546. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. Proc Natl Acad Sci. 2009;106:15237. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pomposiello PJ, Demple B. Trends Biotechnol. 2001;19:109. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 127.Hidalgo E, Demple B. EMBO J. 1994;13:138. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gaudu P, Weiss B. Proc Natl Acad Sci USA. 1996;93:10094. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ding H, Hidalgo E, Demple B. J Biol Chem. 1996;271:33173. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 130.Ding H, Demple B. Proc Natl Acad Sci USA. 1997;94:8445. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kobayashi K, Tagawa S. J Biochem. 2004;136:607. doi: 10.1093/jb/mvh168. [DOI] [PubMed] [Google Scholar]
- 132.Bard AJ, Faulkner LR. Electrochemical Methods. 2 Wiley; New York: 2001. [Google Scholar]
- 133.Burrows CJ, Muller JG. Chem Rev. 1998;98:1109. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 134.Hickerson RP, Chepanoske CL, Williams SD, David SS. J Am Chem Soc. 1999;121:9901. [Google Scholar]
- 135.Chakrabarti SK, Bai CJ, Subramanian KS. Toxicol Appl Pharmacol. 1999;154:245. doi: 10.1006/taap.1998.8584. [DOI] [PubMed] [Google Scholar]
- 136.Gavin IM, Melnick SM, Yurina NP, Khabarova MI, Bavykin SG. Anal Biochem. 1998;263:26. doi: 10.1006/abio.1998.2827. [DOI] [PubMed] [Google Scholar]
- 137.Zitkovich A, Lukanova A, Popov T, Taioli E, Cohen H, Costa M, Honiolo P. Biomarkers. 1996;1:86. doi: 10.3109/13547509609088675. [DOI] [PubMed] [Google Scholar]
- 138.Izzotti A, Bagnasco M, Camoirano A, Orlando M, De Flora S. Mut Res. 1998;400:233. doi: 10.1016/s0027-5107(98)00028-1. [DOI] [PubMed] [Google Scholar]
- 139.Villanueva A, Canete M, Trigueros C, Rodriguez-Blorlado L, Juarranz A. Biopolymers. 1993;33:239. doi: 10.1002/bip.360330206. [DOI] [PubMed] [Google Scholar]
- 140.Nguyen KL, Steryo M, Kurbanyan K, Nowizki KM, Butterfield SM, Ward SR, Stemp EDA. J Am Chem Soc. 2000;122:3585. [Google Scholar]
- 141.Boon EM, Pope MA, Williams SD, David SS, Barton JK. Biochemistry. 2002;41:8464. doi: 10.1021/bi012068c. [DOI] [PubMed] [Google Scholar]
- 142.House PG, Volk DE, Thiviyanathan V, Raymond RC, Luxon BA, Gorenstein DG, Lloyd SR. Prog Nucleic Acid Res Mol Biol. 2001;68:349. doi: 10.1016/s0079-6603(01)68111-x. [DOI] [PubMed] [Google Scholar]
- 143.Dilg AWE, Mincione G, Achterhold K, Iakovleva O, Mentler M, Luchinat C, Bertini I, Parak FG. J Biol Inorg Chem. 1999;4:727. doi: 10.1007/s007750050345. [DOI] [PubMed] [Google Scholar]
- 144.Lee PE, Demple B, Barton JK. Proc Natl Acad Sci. 2009;106:13164. doi: 10.1073/pnas.0906429106. [DOI] [PMC free article] [PubMed] [Google Scholar]