Abstract
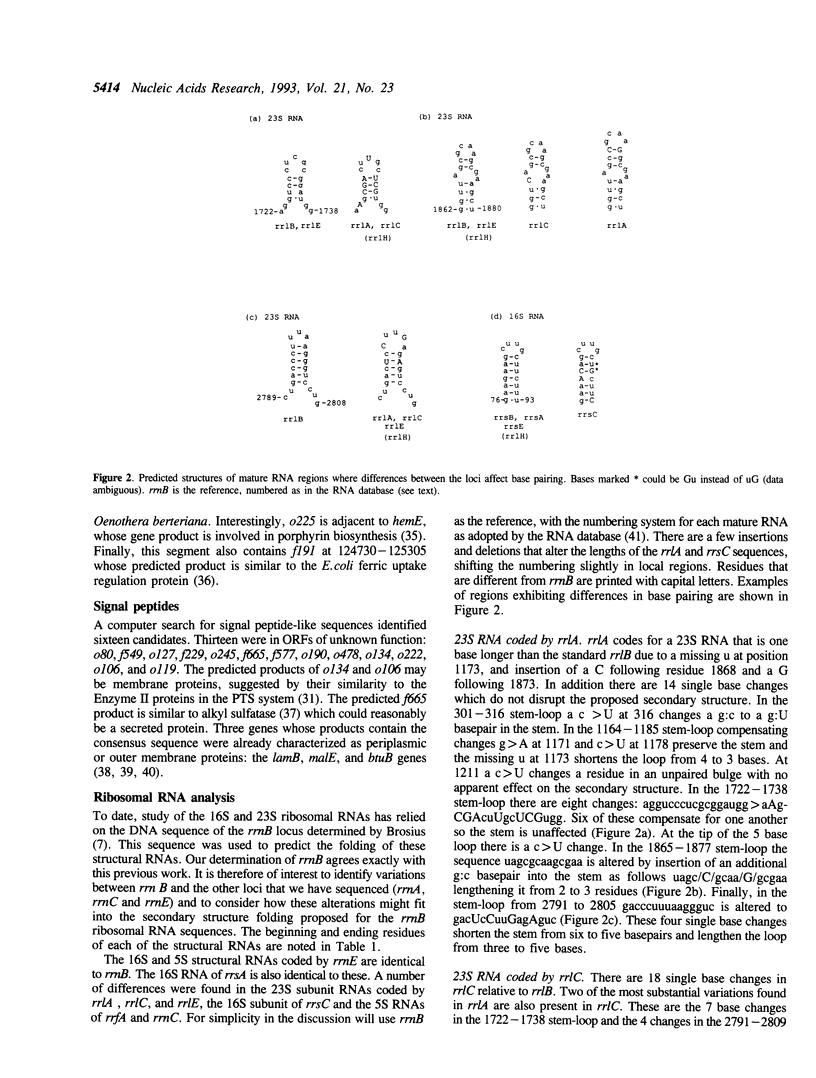
We present the sequence of 176 kilobases of the Escherichia coli K-12 genome, from katG at 89.2 to an open reading frame (ORF) of unknown function at 92.8 minutes on the genetic map. This brings the total of contiguous sequence from the E. coli genome project to 500 kb (81.5 to 92.8 minutes). This segment contains 134 putative coding genes (ORFs) of which 66 genes were previously identified. Eight new genes--acs, pepE, and nrfB-G--were identified as well as the previously mapped gldA and alr genes. Still, 58 ORFs remain unidentified despite literature and similarity searches. The arrangement of proposed genes relative to possible promoters and terminators suggests 55 potential transcription units. Other features include 13 REP elements, one IRU (ERIC) repeat, 59 computer-predicted bends, 42 Chi sites and one new grey hole. Sixteen signal peptides were found, including those of lamB, btuB, and malE. Two ribosomal RNA loci, rrnB and rrnE, are located in this segment, so we have now sequenced four of the seven E. coli rRNA loci. Comparison of the rRNA loci reveals some differences in the ribosomal structural RNAs which are generally compatible with the proposed secondary structures.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman D. L., Trawick D. R., Kranz R. G. Bacterial cytochromes c biogenesis. Genes Dev. 1992 Feb;6(2):268–283. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- Benson S. A., Hall M. N., Silhavy T. J. Genetic analysis of protein export in Escherichia coli K12. Annu Rev Biochem. 1985;54:101–134. doi: 10.1146/annurev.bi.54.070185.000533. [DOI] [PubMed] [Google Scholar]
- Berlyn M. B., Letovsky S. Genome-related datasets within the E. coli Genetic Stock Center database. Nucleic Acids Res. 1992 Dec 11;20(23):6143–6151. doi: 10.1093/nar/20.23.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Ouzounis C., Sander C., Scharf M., Schneider R., Sonnhammer E. What's in a genome? Nature. 1992 Jul 23;358(6384):287–287. doi: 10.1038/358287a0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Burland V., Daniels D. L., Plunkett G., 3rd, Blattner F. R. Genome sequencing on both strands: the Janus strategy. Nucleic Acids Res. 1993 Jul 25;21(15):3385–3390. doi: 10.1093/nar/21.15.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The complete nucleotide sequence of the ribosomal 16-S RNA from Excherichia coli. Experimental details and cistron heterogeneities. Eur J Biochem. 1979 Oct 15;100(2):399–410. doi: 10.1111/j.1432-1033.1979.tb04183.x. [DOI] [PubMed] [Google Scholar]
- Chopra A. K., Peterson J. W., Prasad R. Cloning and sequence analysis of hydrogenase regulatory genes (hydHG) from Salmonella typhimurium. Biochim Biophys Acta. 1991 Dec 2;1129(1):115–118. doi: 10.1016/0167-4781(91)90224-a. [DOI] [PubMed] [Google Scholar]
- Condon C., Philips J., Fu Z. Y., Squires C., Squires C. L. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 1992 Nov;11(11):4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerton I. F., Fincham J. R., Sandeman R. A., Hynes M. J. Comparison and cross-species expression of the acetyl-CoA synthetase genes of the Ascomycete fungi, Aspergillus nidulans and Neurospora crassa. Mol Microbiol. 1990 Mar;4(3):451–460. doi: 10.1111/j.1365-2958.1990.tb00611.x. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Daniels D. L. The complete AvrII restriction map of the Escherichia coli genome and comparisons of several laboratory strains. Nucleic Acids Res. 1990 May 11;18(9):2649–2651. doi: 10.1093/nar/18.9.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin A., Hussain H., Griffiths L., Grove J., Sambongi Y., Busby S., Cole J. Regulation and sequence of the structural gene for cytochrome c552 from Escherichia coli: not a hexahaem but a 50 kDa tetrahaem nitrite reductase. Mol Microbiol. 1993 Sep;9(6):1255–1265. doi: 10.1111/j.1365-2958.1993.tb01255.x. [DOI] [PubMed] [Google Scholar]
- Davison J., Brunel F., Phanopoulos A., Prozzi D., Terpstra P. Cloning and sequencing of Pseudomonas genes determining sodium dodecyl sulfate biodegradation. Gene. 1992 May 1;114(1):19–24. doi: 10.1016/0378-1119(92)90702-q. [DOI] [PubMed] [Google Scholar]
- De Rijk P., Neefs J. M., Van de Peer Y., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1992 May 11;20 (Suppl):2075–2089. doi: 10.1093/nar/20.suppl.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G. P., Rudd K. E., Morgan M. K., Bayat H., Ames G. F. Physical mapping of repetitive extragenic palindromic sequences in Escherichia coli and phylogenetic distribution among Escherichia coli strains and other enteric bacteria. J Bacteriol. 1992 Jul;174(14):4583–4593. doi: 10.1128/jb.174.14.4583-4593.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood M., Nomura M. Deletion of a ribosomal ribonucleic acid operon in Escherichia coli. J Bacteriol. 1980 Aug;143(2):1077–1080. doi: 10.1128/jb.143.2.1077-1080.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R. MacPattern: protein pattern searching on the Apple Macintosh. Comput Appl Biosci. 1991 Jan;7(1):105–106. doi: 10.1093/bioinformatics/7.1.105. [DOI] [PubMed] [Google Scholar]
- Galakatos N. G., Daub E., Botstein D., Walsh C. T. Biosynthetic alr alanine racemase from Salmonella typhimurium: DNA and protein sequence determination. Biochemistry. 1986 Jun 3;25(11):3255–3260. doi: 10.1021/bi00359a026. [DOI] [PubMed] [Google Scholar]
- Galinier A., Bleicher F., Nègre D., Perrière G., Duclos B., Cozzone A. J., Cortay J. C. Primary structure of the intergenic region between aceK and iclR in the Escherichia coli chromosome. Gene. 1991 Jan 2;97(1):149–150. doi: 10.1016/0378-1119(91)90024-6. [DOI] [PubMed] [Google Scholar]
- Gilson E., Saurin W., Perrin D., Bachellier S., Hofnung M. Palindromic units are part of a new bacterial interspersed mosaic element (BIME). Nucleic Acids Res. 1991 Apr 11;19(7):1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S- and 23S-like) ribosomal RNA structures. Nucleic Acids Res. 1992 May 11;20 (Suppl):2095–2109. doi: 10.1093/nar/20.suppl.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller K., Mann B. J., Kadner R. J. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol. 1985 Mar;161(3):896–903. doi: 10.1128/jb.161.3.896-903.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F. The Escherichia coli K-12 "wild types" W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993 Jun;175(11):3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R. Z., Tang J. C., Lin E. C. Experimental evolution of a novel pathway for glycerol dissimilation in Escherichia coli. J Mol Evol. 1983;19(6):429–436. doi: 10.1007/BF02102318. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Wuebbens M. M., Rajagopalan K. V. The structure of a molybdopterin precursor. Characterization of a stable, oxidized derivative. J Biol Chem. 1989 Aug 15;264(23):13440–13447. [PubMed] [Google Scholar]
- Kellermann O., Szmelcman S. Active transport of maltose in Escherichia coli K12. Involvement of a "periplasmic" maltose binding protein. Eur J Biochem. 1974 Aug 15;47(1):139–149. doi: 10.1111/j.1432-1033.1974.tb03677.x. [DOI] [PubMed] [Google Scholar]
- Knappe J., Sawers G. A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):383–398. doi: 10.1111/j.1574-6968.1990.tb04108.x. [DOI] [PubMed] [Google Scholar]
- Lilley P. E., Stamford N. P., Vasudevan S. G., Dixon N. E. The 92-min region of the Escherichia coli chromosome: location and cloning of the ubiA and alr genes. Gene. 1993 Jul 15;129(1):9–16. doi: 10.1016/0378-1119(93)90690-5. [DOI] [PubMed] [Google Scholar]
- Mallinder P. R., Pritchard A., Moir A. Cloning and characterization of a gene from Bacillus stearothermophilus var. non-diastaticus encoding a glycerol dehydrogenase. Gene. 1992 Jan 2;110(1):9–16. doi: 10.1016/0378-1119(92)90438-u. [DOI] [PubMed] [Google Scholar]
- Meadow N. D., Fox D. K., Roseman S. The bacterial phosphoenolpyruvate: glycose phosphotransferase system. Annu Rev Biochem. 1990;59:497–542. doi: 10.1146/annurev.bi.59.070190.002433. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Overbeek R., Larsen N., Marsh T. L., McCaughey M. J., Maciukenas M. A., Kuan W. M., Macke T. J., Xing Y., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1992 May 11;20 (Suppl):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett G., 3rd, Burland V., Daniels D. L., Blattner F. R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993 Jul 25;21(15):3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J. L., Blattner F. R. Formal description of a DNA oriented computer language. Nucleic Acids Res. 1982 Jan 11;10(1):69–84. doi: 10.1093/nar/10.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger G. A., Hammer B. A., Johnson E. A., Lin E. C. Anaerobic growth of Escherichia coli on glycerol by importing genes of the dha regulon from Klebsiella pneumoniae. J Gen Microbiol. 1989 May;135(5):1255–1262. doi: 10.1099/00221287-135-5-1255. [DOI] [PubMed] [Google Scholar]
- Stan-Lotter H., Clarke D. M., Bragg P. D. Isolation of a fourth cysteinyl-containing peptide of the alpha-subunit of the F1 ATPase from Escherichia coli necessitates revision of the DNA sequence. FEBS Lett. 1986 Mar 3;197(1-2):121–124. doi: 10.1016/0014-5793(86)80310-6. [DOI] [PubMed] [Google Scholar]
- Stoker K., Reijnders W. N., Oltmann L. F., Stouthamer A. H. Initial cloning and sequencing of hydHG, an operon homologous to ntrBC and regulating the labile hydrogenase activity in Escherichia coli K-12. J Bacteriol. 1989 Aug;171(8):4448–4456. doi: 10.1128/jb.171.8.4448-4456.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmelcman S., Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975 Oct;124(1):112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Horn P. B., Backstrom A. D., Stewart V., Begley T. P. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J Bacteriol. 1993 Feb;175(4):982–992. doi: 10.1128/jb.175.4.982-992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman S. A., Daub E., Grisafi P., Botstein D., Walsh C. T. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence, enzyme purification, and characterization. Biochemistry. 1984 Oct 23;23(22):5182–5187. doi: 10.1021/bi00317a015. [DOI] [PubMed] [Google Scholar]
- Wijsman H. J. The characterization of an alanine racemase mutant of Escherichia coli. Genet Res. 1972 Dec;20(3):269–277. doi: 10.1017/s001667230001380x. [DOI] [PubMed] [Google Scholar]
- Wild J., Hennig J., Lobocka M., Walczak W., Kłopotowski T. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol Gen Genet. 1985;198(2):315–322. doi: 10.1007/BF00383013. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Williams H. D., Zamanian M., Gibson F., Poole R. K. Isolation and characterization of Escherichia coli mutants affected in aerobic respiration: the cloning and nucleotide sequence of ubiG. Identification of an S-adenosylmethionine-binding motif in protein, RNA, and small-molecule methyltransferases. J Gen Microbiol. 1992 Oct;138(10):2101–2112. doi: 10.1099/00221287-138-10-2101. [DOI] [PubMed] [Google Scholar]
- Wu L. F., Tomich J. M., Saier M. H., Jr Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the fruB(HI) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J Mol Biol. 1990 Jun 20;213(4):687–703. doi: 10.1016/S0022-2836(05)80256-6. [DOI] [PubMed] [Google Scholar]


