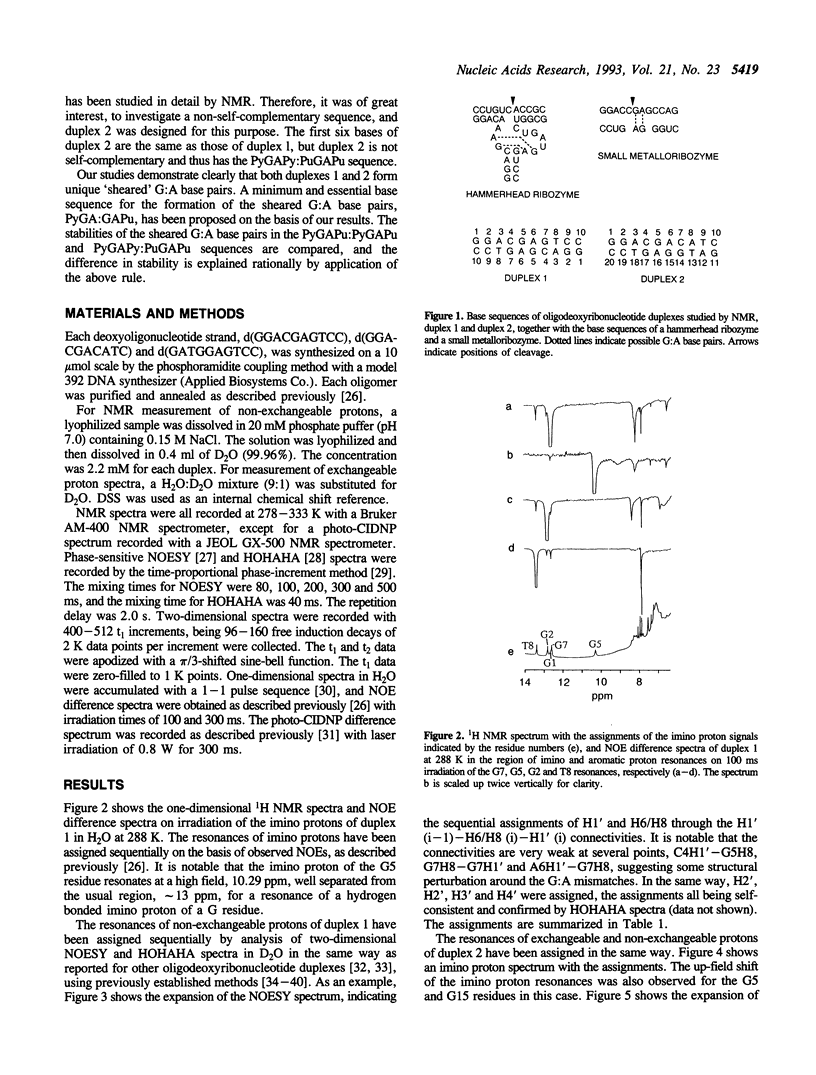
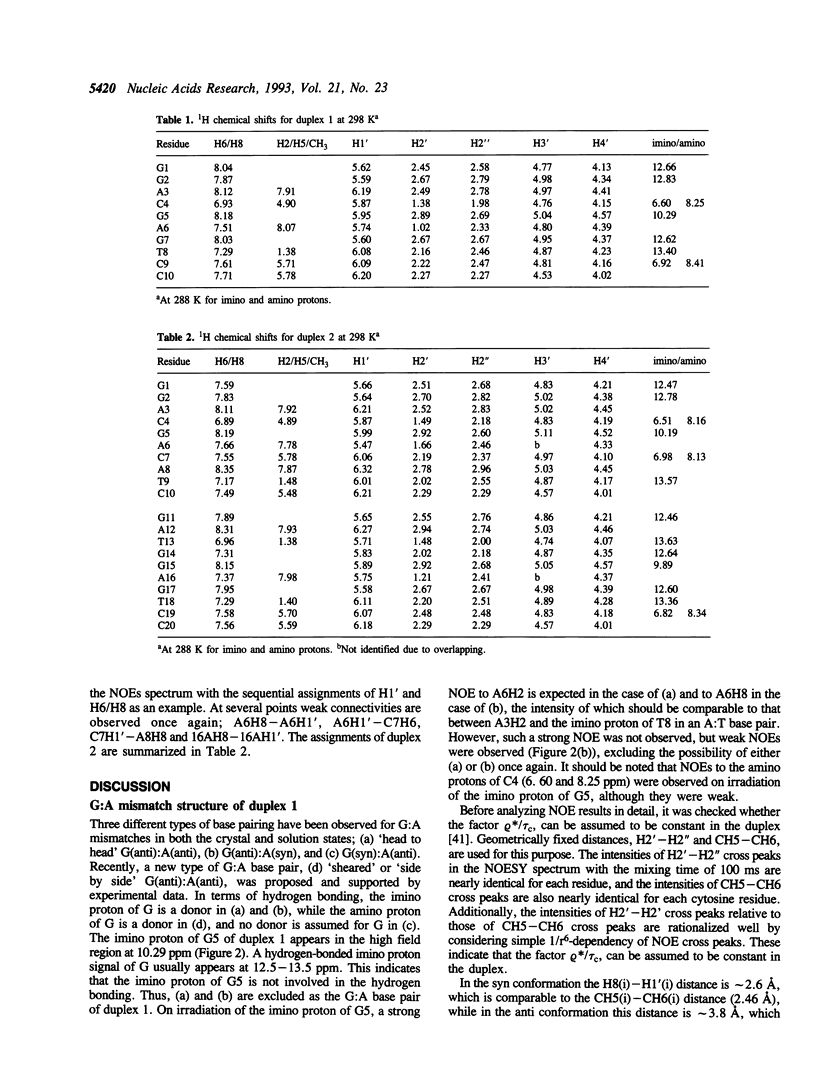
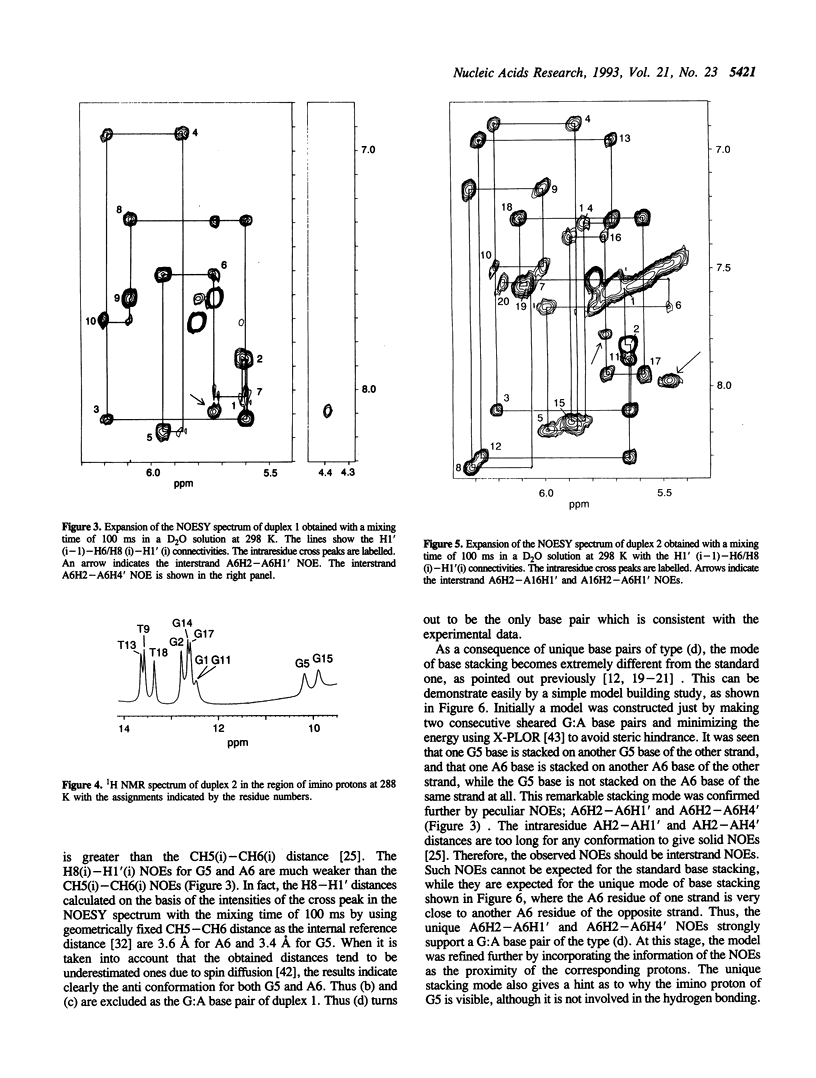
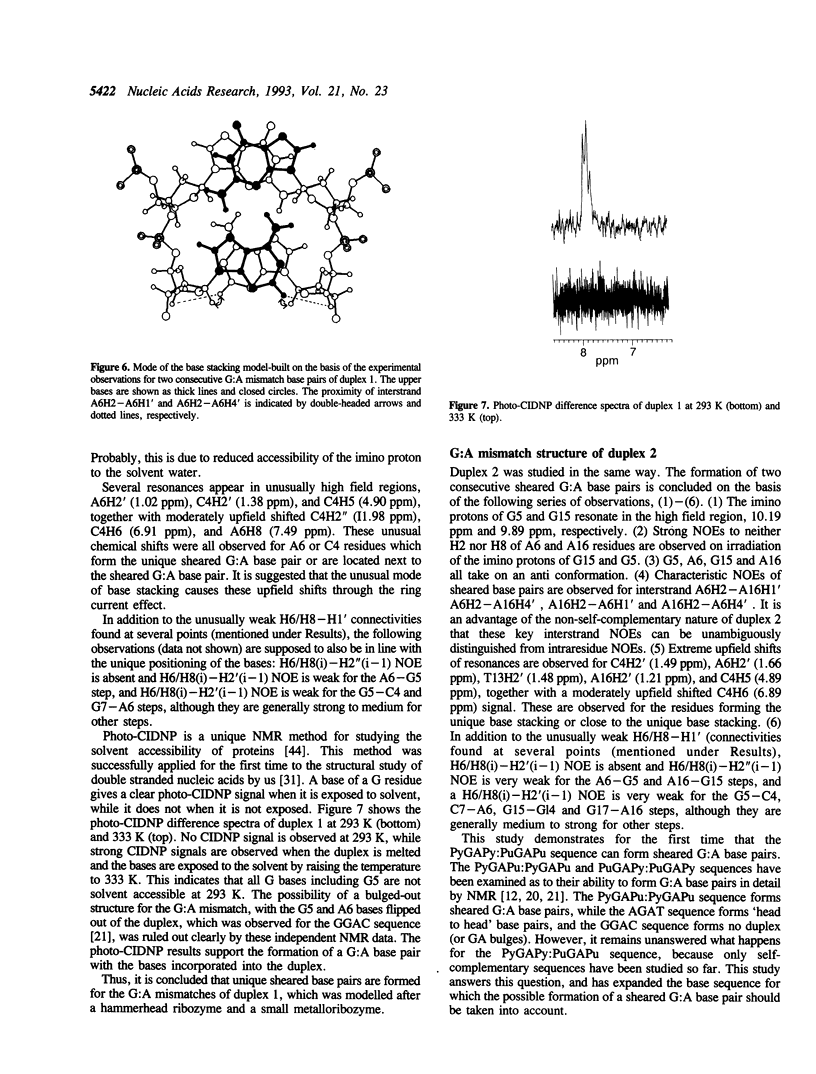
Abstract
The structures of two oligodeoxyribonucleotide duplexes, the base sequences of which were modelled after both a hammerhead ribozyme and a small metalloribozyme, were studied by NMR. Both duplexes contain adjacent G:A mismatches; one has a PyGAPu:PyGAPu sequence and the other a PyGAPy:PuGAPu sequence. It is concluded on the basis of many characteristic NOEs that in both duplexes G:A base pairs are formed in the unique 'sheared' form, where an amino proton instead of an imino proton of G is involved in the hydrogen bonding, and G and A bases are arranged 'side by side' instead of 'head to head'. A photo-CIDNP experiment, which gives unique and independent information on the solvent accessibility of nucleotide bases, also supports G:A base pairing rather than a bulged-out structure of G and A residues. This is the first demonstration that not only the PyGAPu:PyGAPu sequence but also the PyGAPy:PuGAPu sequence can form the unique sheared G:A base pairs. Taking the previous studies on G:A mismatches into account, the idea is suggested that a PyGA:GAPu sequence is a minimum and essential element for the formation of the sheared G:A base pairs. The sheared G:A base pairs in the PyGAPu:PyGAPu sequence are suggested to be more stable than those in the PyGAPy:PuGAPu sequence. This is explained rationally by the idea proposed above.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown T., Hunter W. N., Kneale G., Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Leonard G. A., Booth E. D., Chambers J. Crystal structure and stability of a DNA duplex containing A(anti).G(syn) base-pairs. J Mol Biol. 1989 May 20;207(2):455–457. doi: 10.1016/0022-2836(89)90268-4. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987 Jun 19;236(4808):1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- Cheng J. W., Chou S. H., Reid B. R. Base pairing geometry in GA mismatches depends entirely on the neighboring sequence. J Mol Biol. 1992 Dec 20;228(4):1037–1041. doi: 10.1016/0022-2836(92)90312-8. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Cheng J. W., Reid B. R. Solution structure of [d(ATGAGCGAATA)]2. Adjacent G:A mismatches stabilized by cross-strand base-stacking and BII phosphate groups. J Mol Biol. 1992 Nov 5;228(1):138–155. doi: 10.1016/0022-2836(92)90497-8. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M. Sequence-dependent structural variations in two right-handed alternating pyrimidine-purine DNA oligomers in solution determined by nuclear Overhauser enhancement measurements. EMBO J. 1983;2(12):2109–2115. doi: 10.1002/j.1460-2075.1983.tb01710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G. M., Lauble H., Frenkiel T. A., Gronenborn A. M. A two-dimensional NMR study of the solution structure of a DNA dodecamer comprising the concensus sequence for the specific DNA-binding sites of the glucocorticoid receptor protein. Eur J Biochem. 1984 Dec 17;145(3):629–636. doi: 10.1111/j.1432-1033.1984.tb08603.x. [DOI] [PubMed] [Google Scholar]
- Feigon J., Denny W. A., Leupin W., Kearns D. R. Proton nuclear magnetic resonance investigation of the conformation and dynamics in the synthetic deoxyribonucleic acid decamers d(ATATCGATAT) and d(ATATGCATAT). Biochemistry. 1983 Dec 6;22(25):5930–5942. doi: 10.1021/bi00294a037. [DOI] [PubMed] [Google Scholar]
- Fu D. J., McLaughlin L. W. Importance of specific purine amino and hydroxyl groups for efficient cleavage by a hammerhead ribozyme. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3985–3989. doi: 10.1073/pnas.89.9.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Uhlenbeck O. C., Pardi A. Sequence-dependent structural variations of hammerhead RNA enzymes. Nucleic Acids Res. 1990 Mar 11;18(5):1103–1108. doi: 10.1093/nar/18.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira M., Sugeta H., Kyogoku Y. A new model for the bending of DNAs containing the oligo(dA) tracts based on NMR observations. Nucleic Acids Res. 1990 Feb 11;18(3):613–618. doi: 10.1093/nar/18.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira M., Sugeta H., Kyogoku Y. Determination of the conformation of d(GGAAATTTCC)2 in solution by use of 1H NMR and restrained molecular dynamics. Biochemistry. 1990 Aug 7;29(31):7214–7222. doi: 10.1021/bi00483a008. [DOI] [PubMed] [Google Scholar]
- Katahira M., Sugeta H., Kyogoku Y., Fujii S., Fujisawa R., Tomita K. One- and two-dimensional NMR studies on the conformation of DNA containing the oligo(dA)oligo(dT) tract. Nucleic Acids Res. 1988 Sep 12;16(17):8619–8632. doi: 10.1093/nar/16.17.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Construction of a series of several self-cleaving RNA duplexes using synthetic 21-mers. FEBS Lett. 1988 Feb 15;228(2):228–230. doi: 10.1016/0014-5793(88)80004-8. [DOI] [PubMed] [Google Scholar]
- Li Y., Zon G., Wilson W. D. NMR and molecular modeling evidence for a G.A mismatch base pair in a purine-rich DNA duplex. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):26–30. doi: 10.1073/pnas.88.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zon G., Wilson W. D. Thermodynamics of DNA duplexes with adjacent G.A mismatches. Biochemistry. 1991 Jul 30;30(30):7566–7572. doi: 10.1021/bi00244a028. [DOI] [PubMed] [Google Scholar]
- Maltseva T., Sandström A., Ivanova I. M., Sergeyev D. S., Zarytova V. F., Chattopadhyaya J. Structural studies of the 5'-phenazinium-tethered matched and G-A-mismatched DNA duplexes by NMR spectroscopy. J Biochem Biophys Methods. 1993 May;26(2-3):173–236. doi: 10.1016/0165-022x(93)90046-q. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- McCall M. J., Hendry P., Jennings P. A. Minimal sequence requirements for ribozyme activity. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5710–5714. doi: 10.1073/pnas.89.13.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L., Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Structure refinement of oligonucleotides by molecular dynamics with nuclear Overhauser effect interproton distance restraints: application to 5' d(C-G-T-A-C-G)2. J Mol Biol. 1986 Apr 5;188(3):455–475. doi: 10.1016/0022-2836(86)90168-3. [DOI] [PubMed] [Google Scholar]
- Odai O., Kodama H., Hiroaki H., Sakata T., Tanaka T., Uesugi S. Synthesis and NMR study of ribo-oligonucleotides forming a hammerhead-type RNA enzyme system. Nucleic Acids Res. 1990 Oct 25;18(20):5955–5960. doi: 10.1093/nar/18.20.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O. C. A small metalloribozyme with a two-step mechanism. Nature. 1992 Aug 13;358(6387):560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Wu T. F., Cousineau B., Ogilvie K. K., Cedergren R. Mixed deoxyribo- and ribo-oligonucleotides with catalytic activity. Nature. 1990 Apr 5;344(6266):565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- Pieken W. A., Olsen D. B., Benseler F., Aurup H., Eckstein F. Kinetic characterization of ribonuclease-resistant 2'-modified hammerhead ribozymes. Science. 1991 Jul 19;253(5017):314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Heinemann U., Chandrasegaran S., Kan L. S., Kopka M. L., Dickerson R. E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987 Oct 23;238(4826):498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- Reid D. G., Salisbury S. A., Bellard S., Shakked Z., Williams D. H. Proton nuclear Overhauser effect study of the structure of a deoxyoligonucleotide duplex in aqueous solution. Biochemistry. 1983 Apr 12;22(8):2019–2025. doi: 10.1021/bi00277a044. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Taylor N. R., Kaplan B. E., Swiderski P., Li H., Rossi J. J. Chimeric DNA-RNA hammerhead ribozymes have enhanced in vitro catalytic efficiency and increased stability in vivo. Nucleic Acids Res. 1992 Sep 11;20(17):4559–4565. doi: 10.1093/nar/20.17.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Patel D. J., Sauer R. T., Karplus M. Two-dimensional 1H NMR study of the lambda operator site OL1: a sequential assignment strategy and its application. Proc Natl Acad Sci U S A. 1984 Jan;81(1):130–134. doi: 10.1073/pnas.81.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. M., Pieken W. A., Eckstein F. Function of specific 2'-hydroxyl groups of guanosines in a hammerhead ribozyme probed by 2' modifications. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):918–921. doi: 10.1073/pnas.89.3.918. [DOI] [PMC free article] [PubMed] [Google Scholar]



