Abstract
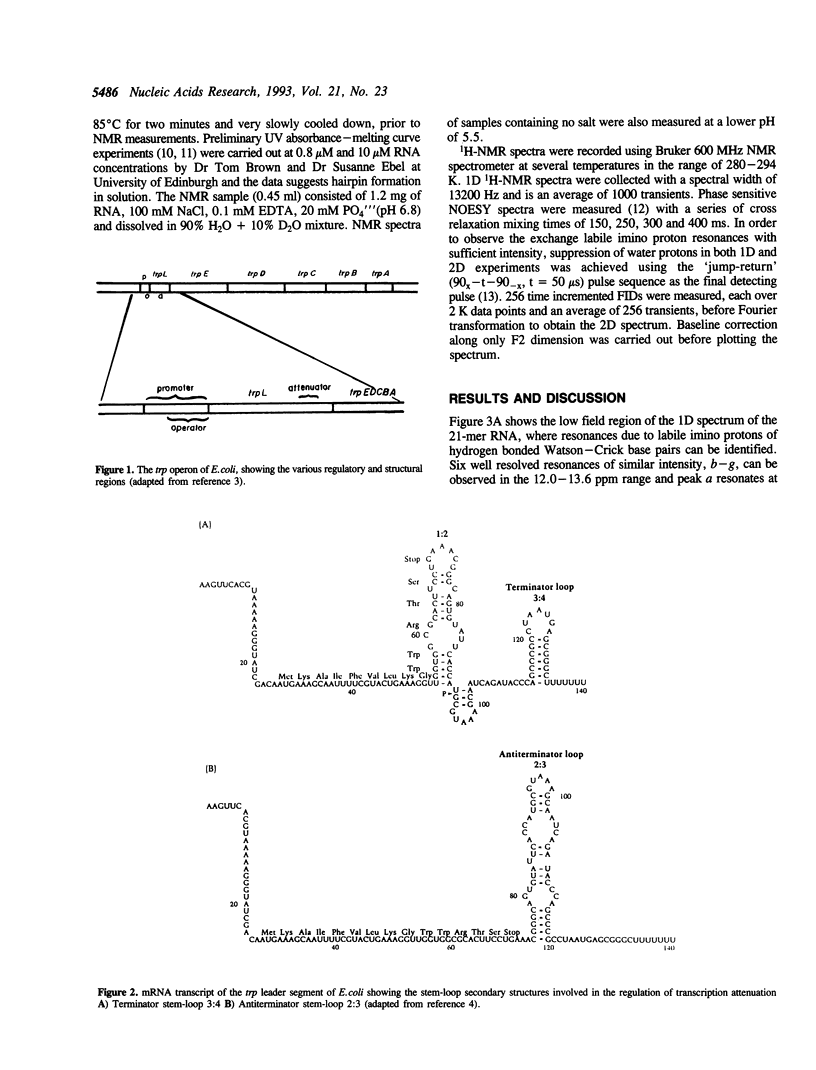
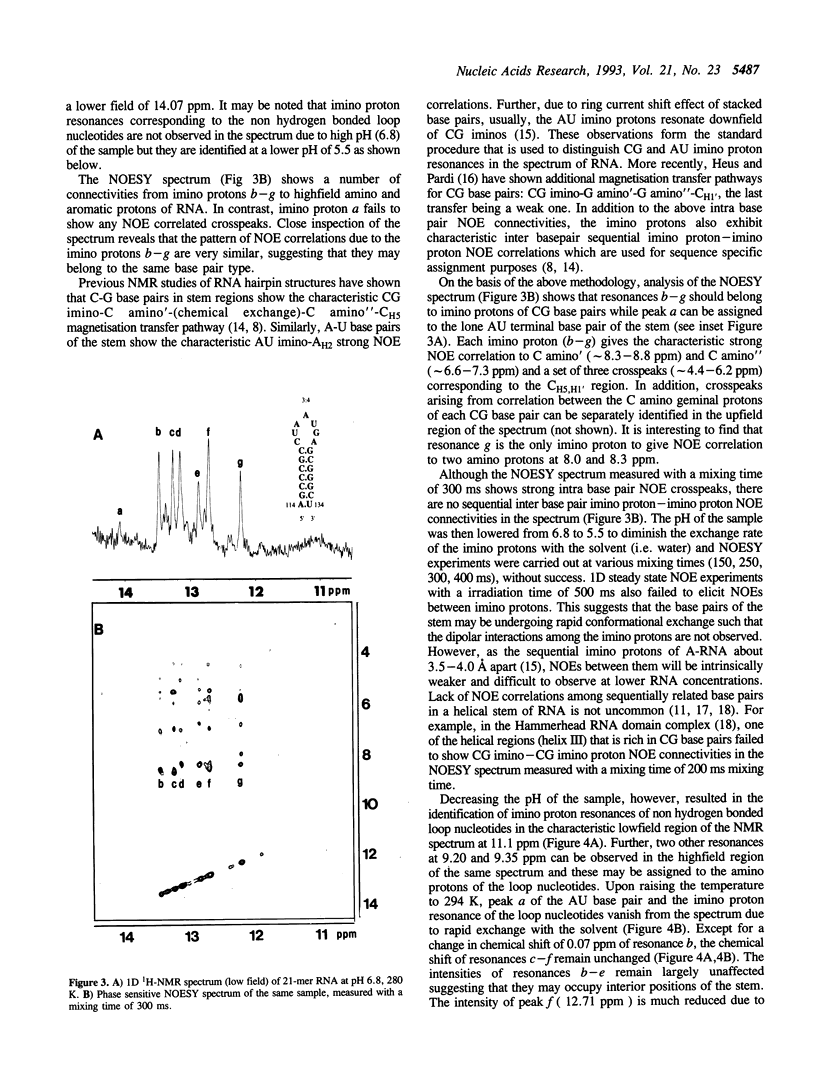
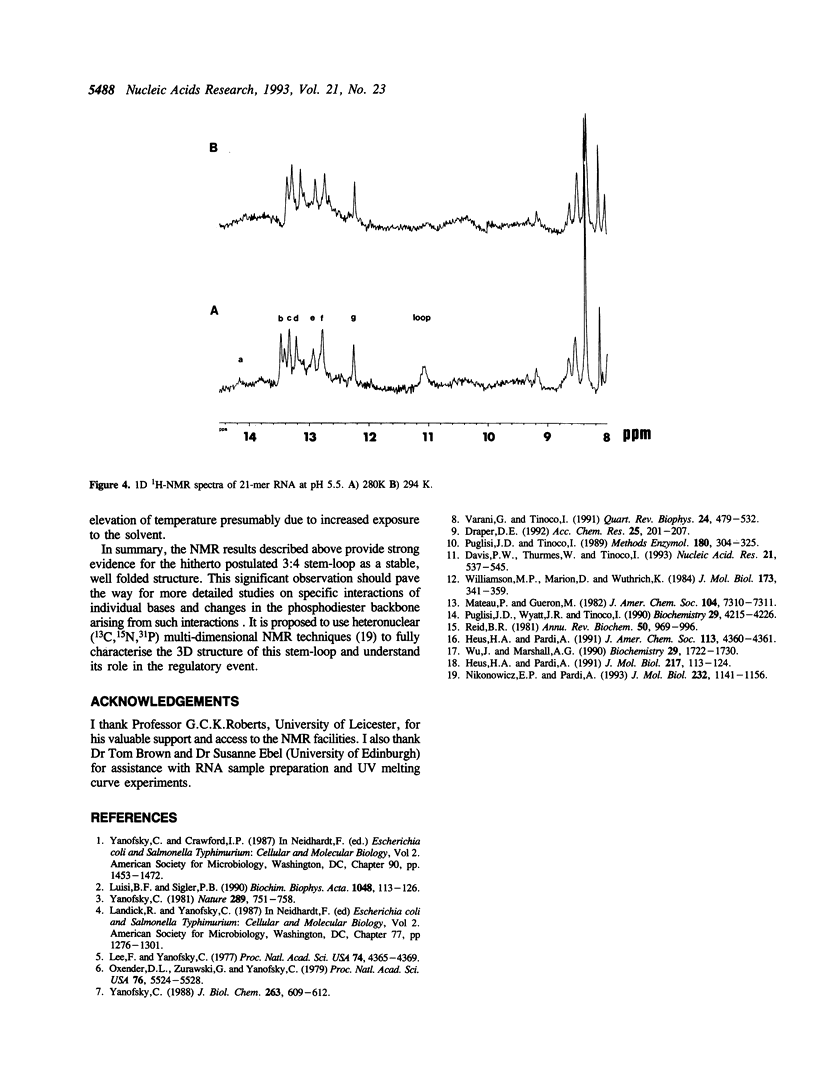
High field 1H-NMR studies of a synthetic 21-mer RNA fragment, corresponding to residues +114 to +134 within the trp leader mRNA transcript, have been carried out. Seven well resolved imino proton resonances corresponding to six C-G and one A-U hydrogen bonded base pairs, together with their characteristic NOE patterns can be identified in the NMR spectrum. This experimental result provides direct evidence for the postulated stem-loop secondary structure, 3:4, which has been reported to act as a transcription termination signal for RNA polymerase.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davis P. W., Thurmes W., Tinoco I., Jr Structure of a small RNA hairpin. Nucleic Acids Res. 1993 Feb 11;21(3):537–545. doi: 10.1093/nar/21.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Nuclear magnetic resonance studies of the hammerhead ribozyme domain. Secondary structure formation and magnesium ion dependence. J Mol Biol. 1991 Jan 5;217(1):113–124. doi: 10.1016/0022-2836(91)90615-d. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi B. F., Sigler P. B. The stereochemistry and biochemistry of the trp repressor-operator complex. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):113–126. doi: 10.1016/0167-4781(90)90047-6. [DOI] [PubMed] [Google Scholar]
- Nikonowicz E. P., Pardi A. An efficient procedure for assignment of the proton, carbon and nitrogen resonances in 13C/15N labeled nucleic acids. J Mol Biol. 1993 Aug 20;232(4):1141–1156. doi: 10.1006/jmbi.1993.1466. [DOI] [PubMed] [Google Scholar]
- Oxender D. L., Zurawski G., Yanofsky C. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Solution conformation of an RNA hairpin loop. Biochemistry. 1990 May 1;29(17):4215–4226. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- Reid B. R. NMR studies on RNA structure and dynamics. Annu Rev Biochem. 1981;50:969–996. doi: 10.1146/annurev.bi.50.070181.004541. [DOI] [PubMed] [Google Scholar]
- Varani G., Tinoco I., Jr RNA structure and NMR spectroscopy. Q Rev Biophys. 1991 Nov;24(4):479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- Williamson M. P., Marion D., Wüthrich K. Secondary structure in the solution conformation of the proteinase inhibitor IIA from bull seminal plasma by nuclear magnetic resonance. J Mol Biol. 1984 Mar 5;173(3):341–359. doi: 10.1016/0022-2836(84)90125-6. [DOI] [PubMed] [Google Scholar]
- Wu J. J., Marshall A. G. 500-MHz proton NMR evidence for two solution structures of the common arm base-paired segment of wheat germ 5S ribosomal RNA. Biochemistry. 1990 Feb 20;29(7):1722–1730. doi: 10.1021/bi00459a009. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Transcription attenuation. J Biol Chem. 1988 Jan 15;263(2):609–612. [PubMed] [Google Scholar]


