Abstract
Background
In ovo electroporation is a widely used technique to study gene function in developmental biology. Despite the widespread acceptance of this technique, no genome-wide analysis of the effects of in ovo electroporation, principally the current applied across the tissue and exogenous vector DNA introduced, on endogenous gene expression has been undertaken. Here, the effects of electric current and expression of a GFP-containing construct, via electroporation into the midbrain of Hamburger-Hamilton stage 10 chicken embryos, are analysed by microarray.
Results
Both current alone and in combination with exogenous DNA expression have a small but reproducible effect on endogenous gene expression, changing the expression of the genes represented on the array by less than 0.1% (current) and less than 0.5% (current + DNA), respectively. The subset of genes regulated by electric current and exogenous DNA span a disparate set of cellular functions. However, no genes involved in the regional identity were affected. In sharp contrast to this, electroporation of a known transcription factor, Dmrt5, caused a much greater change in gene expression.
Conclusions
These findings represent the first systematic genome-wide analysis of the effects of in ovo electroporation on gene expression during embryonic development. The analysis reveals that this process has minimal impact on the genetic basis of cell fate specification. Thus, the study demonstrates the validity of the in ovo electroporation technique to study gene function and expression during development. Furthermore, the data presented here can be used as a resource to refine the set of transcriptional responders in future in ovo electroporation studies of specific gene function.
Background
Determining the function of genes involved in embryonic development requires manipulation of gene expression in a spatially and temporally restricted manner. Whilst transgenesis in mice is used to this end, it is a time consuming and costly technique. Moreover, achieving the required spatiotemporal targeting via conditional transgenesis is not always achievable [1]. In contrast, the highly precise localisation and timing of expression construct insertion made possible by the in ovo electroporation technique provides an economical and efficient alternative to transgenesis [2]. These advantages make in ovo electroporation a widely used technique to study gene function during development.
In ovo electroporation is employed in both gain and loss of function studies and reporter studies to analyse gene function and regulation during development. This method has been applied to all areas of developmental biology, including neurogenesis and neural differentiation [3,4], axon outgrowth and guidance [5] axonal patterning in the developing limb [6], somitogenesis [7,8], skeletal muscle development [9] and eye development [10,11].
In ovo electroporation has been used to study development in mouse embryos [12] along with other vertebrates, including Danio rerio [13] and Xenopus laevis [13,14] and non-vertebrates such as Drosophila melanogaster [15] and Ascidiacea [16] (for reviews, see [17,18]). However, in ovo electroporation has been most widely applied to avian embryos because the avian embryo develops in ovo and has a planar topology. These characteristics greatly facilitate injection of the DNA construct, placement of electrodes and incubation of the electroporated embryo.
Independent of the experimental organism, electroporation involves microinjecting a gene expression construct, typically into a natural body cavity such as the neural tube. Subsequently, electrodes are placed flanking the site of injection and an electric current is applied across the embryo in the form of a rapid series of square wave pluses. This electric field transiently disrupts the stability of the plasma membrane, creating pores in the cell membrane. The negatively charged DNA constructs migrate towards the positive electrode and enter the cells in their path via these pores. The tissue adjacent to the negatively charged electrode remains untransfected, providing an internal control [19]. This method enables defined tissues to be targeted, by location of DNA injection and positioning of electrodes, at precise times of development. For more detailed information about specific parameters required refer to [2,20]. Further advances now enable focal electroporation and electroporation of different constructs in close proximity, increasing the precision and complexity of in ovo electroporation studies. This technique uses beads soaked in the DNA construct, which are microsurgically implanted into the embryo instead of microinjection [21].
In ovo electroporation is most commonly applied to Hamburger-Hamilton (HH) stage (st) 10 to 20 embryos. Embryos older than HH st20 have more compact tissues and increased tissue layers, making microinjection and in ovo electroporation more difficult. For older embryos ex ovo explant electroporation is an alternative method [22].
Typically, the construct used for in ovo electroporation contains the gene of choice along with green fluorescent protein (GFP), or another marker, in order to identify the cells that have taken up the construct. A popular construct to obtain transient expression is pCAβ-IRES-GFP containing the β-actin promoter, a cytomegalovirus (CMV) enhancer, a polylinker for inserting the desired gene followed by an internal ribosomal entry site (IRES) and GFP [23]. Expression of the translated product of the construct - for example, GFP - can be detected 2.5 hours after electroporation and peaks around 20 to 24 hours [24]. Expression of transient constructs can be maintained for 3 to 11 days [2,25]. Constitutive expression can be obtained by integrating plasmids into the genome using methods such as transposon-mediated gene transfer [26]. Using constructs with inducible promoters - for example, the tetracycline on tetracycline off system - enables further control over the timing of exogenous DNA expression [27]. It is also possible to use cell-type-specific enhancers [2].
In addition to overexpression studies, in ovo electroporation can also be used to carry out loss-of-function studies using RNA interference [28,29] or dominant negative constructs [30]. Constructs containing the gene of interest linked to either a repressor or an activator enable investigation of the transcriptional activity of genes in vivo [31]. As well as investigating gene function, in ovo electroporation can be used to study the activity of promoters during development in vivo using reporter constructs [32,33].
Traditionally, the downstream effects of in ovo electroporation of a gene of interest have been analysed using in situ hybridisation and antibody staining of a few select gene products. Parallel analysis of these genes in embryos electroporated with GFP alone is used as a control to identify any non-specific effects. This approach has the major disadvantage of surveying only a limited number of genes for qualitative transcriptional changes. To date, no effect of the in ovo electroporation technique on gene expression has been reported. However, evidence from other techniques, such as electrochemotherapy and electrogene therapy on malignant melanoma cells, indicates that both electric current and exogenous expression of DNA have a small but significant affect on endogenous gene expression [34].
The other potentially influential component of in ovo electroporation is the introduction of DNA constructs, which results in significant levels of exogenous DNA within the electroporated cells. The cellular response to expression of plasmid DNA has been analysed in several cell lines: Chinese hamster ovary epithelial cells (CHO-K1), mouse fibroblast cells (NIH3T3), human embryonic kidney cells (HEK293) and several melanoma lines [35]. Exogenous DNA expression in these cell lines was shown to have differing affects, triggering a DNA damage response in CHO-K1 and NIH3T3 cells but causing no cellular response in HEK293 cells or melanoma cell lines [35]. These differences in response to exogenous DNA expression are postulated to be due to species differences [35]. How chicken cells respond to exogenous DNA expression or indeed how multicellular living organisms respond is yet to be examined.
Despite these reports suggesting exposure to current and exogenous DNA within cells can cause changes in endogenous gene expression, there has been no systematic analysis of the effects of either of these components within the developing embryo. Understanding the experimentally induced changes caused by in ovo electroporation has recently become extremely pertinent since the sequencing of the chicken genome and the availability of a highly representative chicken genome microarray (Affymetrix GeneChip Chicken Genome Array) now enables the coupling of in ovo electroporation to genome-wide analysis. This strategy provides an opportunity to use in ovo electroporation to unravel the role of genes during the embryonic development in a high-throughput and genome-wide manner. If this strategy is to be used, a better understanding of the effects of this technique are required. To address this, we carried out microarray analysis of the effects of both the electric current and exogenous DNA expression associated with in ovo electroporation.
Results
Experimental strategy
To investigate the effects of the electric current and exogenous DNA expression associated with in ovo electroporation on endogenous gene expression of the developing chicken embryo, we used microarray analysis to compare the gene expression profile of four pooled samples of ventral lateral midbrain (VLM) tissue. The four samples were unelectroporated VLM (VLM), VLM exposed to electric current only (VLMi), VLM exposed to expression of GFP and electric current (VLMg) and VLM exposed to exogenous expression of a regulatory gene, Dmrt5 (Doublesex and Mab related transcription factor like 5) and electric current (VLMd) (Figure 1). The current was generated with an ECM830 Electro Square Porator (5 × 50-ms pulses of 12 V). The GFP construct used was pCAβ-IRES-GFP. Electroporation was carried out at HH st10, and embryos were collected 24 hours later. Thus, our experiment was specifically designed to interrogate the set of genes 'stably' regulated following electroporation, rather than those responding to the initial treatment. We reasoned that genes identified at this 24 hour time point would be more likely to have a significant contribution to altering long-term cell identity as opposed to those genes whose expression changed rapidly in the minutes following electrical insult.
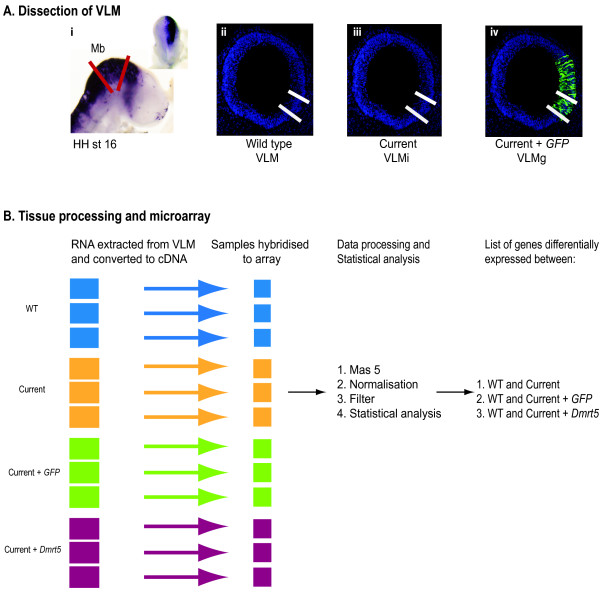
Figure 1.
Experimental strategy. (A) Dissection of the ventral lateral midbrain (VLM) region. (i) In situ sagital view of a HH st16 electroporated embryo expressing pCAβ-IRES-Dmrt5 construct (dorsal view in the inset). Red lines indicate the midbrain (Mb) region, which was dissected out. (ii-iv) Coronal sections of midbrain. White lines mark the VLM region. This region was isolated from (ii) control embryos (VLM), (iii) VLM exposed to current (VLMi), (iv) VLM exposed to current + GFP (VLMg), and VLM exposed to current + Dmrt5 (VLMd; image not shown) for investigation of transcriptional profiles by microarray analysis. (B) Tissue processing and microarray analysis. Six VLM tissues were pooled for each biological replicate, and three biological replicates were used for each condition. cDNA was isolated from these pools and hybridised to the Affymetrix Chicken Genome Array. Following MAS5, normalisation and filtering, genes whose expression differed significantly between the wild type (WT; VLM) and VLMi, VLMg and VLMd were identified by one-way ANOVA.
Effect of in ovo electroporation on endogenous gene expression
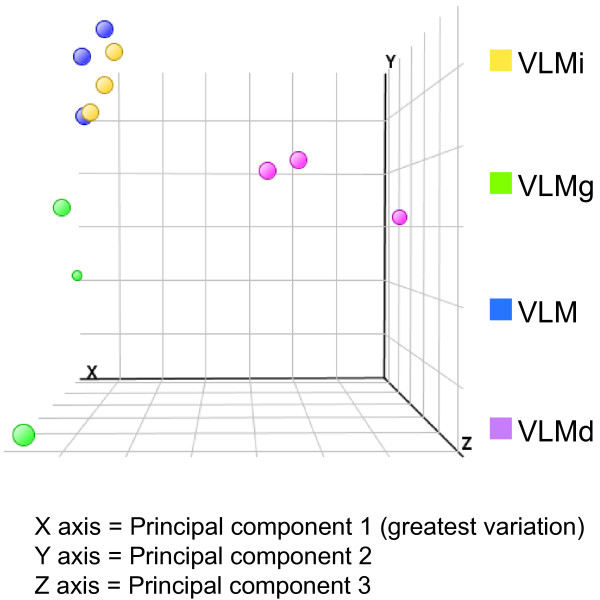
The distribution of gene expression values across the whole microarray was monitored before and after normalisation by quantile grouping (box plots) and was found to be similar for each set of biological replicates (data not shown). The variation in biological replicates was also analysed by principal component analysis (PCA), which measures the variation in expression levels between the microarrays. PCA showed all biological replicates cluster together, demonstrating that the biological replicate datasets are similar and reproducible. As well as biological replicates clustering together, three experimental conditions, VLM, VLMi and VLMg, cluster together, indicating there is little difference in gene expression between these three conditions. In contrast, exogenous expression of the regulatory gene Dmrt5 caused the VLMd samples to cluster separately from the other conditions (Figure 2). This preliminary analysis of global gene expression levels indicates that exposure to current or current + GFP has little effect on endogenous gene expression, whilst addition of a regulatory gene causes a much larger change in endogenous gene expression.
Figure 2.
Principal component analysis to identify genome-wide transcriptional variation caused by in ovo electroporation. This plot shows the variation between the samples; each dot represents the global gene expression of a single microarray. The greatest variation is measured on the x-axis, then the y- and z-axes, respectively. Wild-type VLM, VLMi and VLMg samples cluster together on the x-axis, indicating that there is little variation in the gene expression between these three conditions. VLMd, in which the regulatory gene Dmrt5 is exogenously expressed, clusters separately, indicating larger genome-wide variation between this sample and the others.
Analysis of the array data to identify genes that show significant differential expression (P > 0.05, >2.0-fold change) upon exposure to current, current + GFP or current + Dmrt5, when compared to the unelectroporated tissue, identified that only 41 of the 32,773 oligonucleotides spotted on the array were changed by exposure to current and 176 by current + GFP. This equates to a change in gene expression of 0.1% (current) and 0.5% (current + GFP), of the oligonucelotides found on the array, respectively. By comparison, exogenous expression of a regulatory gene, Dmrt5, led to a significant change (P > 0.05, >2.0-fold change) in the expression of 479 oligonucleotides or 1.5% of the oligonucleotides on the array. For further analysis, these oligonucleotide lists were processed to remove probe set duplicates and oligonucelotides that did not correspond to annotated genes (Table 1).
Table 1.
Differential expression of genes upon exposure to current, current + GFP and current + Dmrt5
| Condition | Number of oligonucleotides differentially expressed | Percentage of oligonucleotides showing differential expression | Number of annotated endogenous genes differentially expressed |
|---|---|---|---|
| Current | 41 | 0.13 | 21 |
| Current + GFP | 176 | 0.54 | 111 |
| Current + Dmrt5 | 479 | 1.46 | 309 |
Effect of current on endogenous gene expression
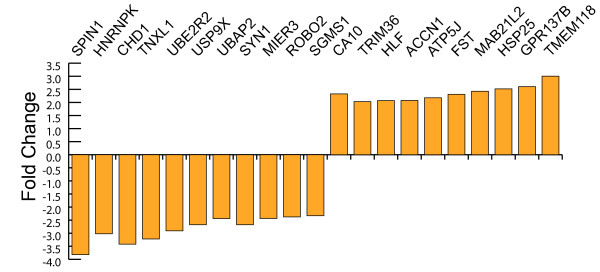
Only 41 of the 32,773 oligonucelotides were significantly differentially expressed 24 hours after exposure to current alone. This corresponds to 21 annotated endogenous genes (Figure 3). There was an equal proportion of up- and down-regulated genes (10 and 11, respectively) and the range of their fold changes was -3.8 to 3.0 (Table 2). Thus, the application of an electric current alone to developing neuronal cells within the ventral midbrain has a small effect on the constituents of the cell transcriptome. These findings are consistent with other in vitro studies that suggest current alone has a minimal impact on cellular identity and specification [34].
Figure 3.
Genes showing differential expression in response to current. Genes showing differential expression when exposed to current and their fold change.
Table 2.
Genes showing differential expression upon exposure to current and their fold change
| Gene name | Fold change |
|---|---|
| SPIN1 | -3.8 |
| HNRNPK | -3.0 |
| CHD1 | -3.4 |
| TXNL1 | -3.2 |
| UBE2R2 | -2.9 |
| USP9X | -2.6 |
| UBAP2 | -2.4 |
| SYN1 | -2.6 |
| MIER3 | -2.4 |
| ROBO2 | -2.3 |
| SGMS1 | -2.3 |
| CA10 | 2.3 |
| TRIM36 | 2.0 |
| HLF | 2.0 |
| ACCN1 | 2.0 |
| ATPJ5 | 2.1 |
| FST | 2.3 |
| MA21L2 | 2.4 |
| HSP25 | 2.5 |
| GPR137B | 2.6 |
| TMEM118 | 3.0 |
We next looked at the composition of the 21 responders in this group to determine the set of cellular activities affected. The 21 responders belong to a disparate array of biological functions (Table 3). The small number of genes meant it was not possible to identify biological processes that were significantly enriched in this population using Gene Ontology (GO) analysis.
Table 3.
Biological functions of genes that responded to current
| Biological category | Genes |
|---|---|
| Response to oxidative stress | Hnrnpk, Usp9x, Hlf, Txn1l, Hsp25 |
| Response to pH/redox changes | Ca10, Atp5j, Accn1, Txn1l |
| Regulation of apoptosis | Accn1, Hlf, Usp9x, Hnrnpk, Sgms1 |
Upregulation of Heat shock protein 25 (Hsp25) was observed. Two members of the ubiquitination pathway, uber2r and ubap2, also responded upon exposure to current. Five of the 21 responders are involved in response to oxidative stress. However, no major oxidative stress components show a change in expression. Four of the 21 genes are reported to respond to pH/redox changes. Five responders are involved in regulation of apoptosis, either to promote (two genes) or inhibit (three genes) this process. For all three of these biological functions, regulators of processes rather than determiners are affected, suggesting that the effect of the electric current on biological processes is minimal.
Effect of current and vector DNA on endogenous gene expression
The set of genes derived from the control tissues (VLM) was compared with their electroporated counterparts expressing GFP (VLMg). This analysis revealed that only 176 of the 32,773 oligonucleotides represented were significantly differentially expressed (P > 0.05, >2.0-fold change) which corresponds to less than 0.5% of the transcriptome. This corresponds to 111 endogenous genes (Table 4). Of these 111 responders to current + GFP, only 10 were upregulated, with the remaining 101 being downregulated. All of the upregulated genes showed a 2.0- to 3.3-fold change except for BSFB (beaded filament structural protein1, filensin), which was upregulated 7.8-fold. BSFB is a filament protein and forms a structural constituent of eye lens [36], and the significance of its upregulation is not known. All downregulated genes showed between a 2.0- and 3.6-fold change in expression. Thus, the application of an electric current in combination with expression of exogenous DNA in the form of GFP has a greater effect on neuronal cells than current alone. However, this is still a much smaller effect than exogenous expression of a regulatory gene, such as Dmrt5, which causes a change in gene expression of 309 endogenous genes.
Table 4.
Genes showing differential expression upon exposure to current + GFP and their fold change
| Gene name | Fold change |
|---|---|
| MYH10 | -2.0 |
| RCBTB1 | -2.0 |
| BAZ1B | -2.0 |
| ZFHX3 | -2.0 |
| EP400 | -2.0 |
| GSPT1 | -2.0 |
| YLPM1 | -2.0 |
| ANKHD1-EIF4EBP3 | -2.0 |
| SGMS1 | -2.0 |
| MLL | -2.1 |
| WAC | -2.1 |
| PITPNM3 | -2.1 |
| TNRC6B | -2.1 |
| SMPD3 | -2.1 |
| EPN2 | -2.1 |
| CCDC88A | -2.1 |
| LOC427360 | -2.1 |
| TLN2 | -2.1 |
| DGCR8 | -2.1 |
| LOC418437 | -2.1 |
| TMTC3 | -2.1 |
| NIPBL | -2.1 |
| PTCHD1 | -2.1 |
| BEND7 | -2.1 |
| GNPTAB | -2.1 |
| SENP5 | -2.1 |
| NKTR | -2.1 |
| FBXW8 | -2.1 |
| NISCH | -2.1 |
| ACPL2 | -2.1 |
| LOC429468 | -2.2 |
| KCNK5 | -2.2 |
| TBL1X | -2.2 |
| UBE4B | -2.2 |
| NTRK3 | -2.2 |
| MLL3 | -2.2 |
| ALS2 | -2.2 |
| PHIP | -2.2 |
| PPP6R3 | -2.2 |
| SPEN | -2.2 |
| ERC1 | -2.2 |
| TJP1 | -2.2 |
| ABCB11 | -2.2 |
| PRPF38B | -2.2 |
| LRRN1 | -2.2 |
| IL17RD | -2.2 |
| TXNL1 | -2.3 |
| CHD1 | -2.2 |
| USP40 | -2.3 |
| PLXNB2 | -2.3 |
| 18s rRNA | -2.3 |
| SPTBN1 | -2.3 |
| PCDH19 | -2.3 |
| PDCD6 | -2.3 |
| FZD7 | -2.3 |
| RASGEF1A | -2.3 |
| NAV2 | -2.3 |
| EP300 | -2.3 |
| GLS | -2.4 |
| ATRX | -2.4 |
| FAM130A2 | -2.4 |
| MDN1 | -2.4 |
| RPRD1A | -2.4 |
| VCAN | -2.4 |
| ARHGAP18 | -2.4 |
| JAG1 | -2.4 |
| BAZ2B | -2.4 |
| HNRNPK | -2.5 |
| CKAP5 | -2.5 |
| SYNE2 | -2.5 |
| ASPM | -2.5 |
| PLA2G2E | -2.6 |
| FIGN | -2.6 |
| HERC2 | -2.6 |
| TMEM121 | -2.6 |
| SALL3 | -2.6 |
| TRAF3 | -2.6 |
| AKAP13 | -2.6 |
| RBM25 | -2.7 |
| LOC776927 | -2.7 |
| SON | -2.7 |
| ARHGAP17 | -2.7 |
| AKAP9 | -2.8 |
| VGLL4 | -2.8 |
| ZEB2 | -2.8 |
| LRP2 | -2.8 |
| Ube2r2 | -2.9 |
| UBAP2 | -2.9 |
| YTHDC1 | -3.1 |
| TRIO | -3.1 |
| GUCY1A2 | -3.3 |
| LMO7 | -3.3 |
| PDZRN3 | -3.4 |
| SFRS18 | -3.4 |
| HMGA2 | -3.5 |
| PRTG | -3.5 |
| FHOD3 | -3.5 |
| VCL | -3.6 |
| SPIN | -3.6 |
| DLG1 | -3.7 |
| CACNG3 | 2.0 |
| DNTTIP1 | 2.0 |
| HUS1 | 2.0 |
| PSPH | 2.5 |
| KCTDC7 | 2.8 |
| WNT3A | 2.8 |
| EIF4A1 | 3.1 |
| EGFL7 | 2.9 |
| CIRBP | 3.3 |
| BFSP1 | 7.8 |
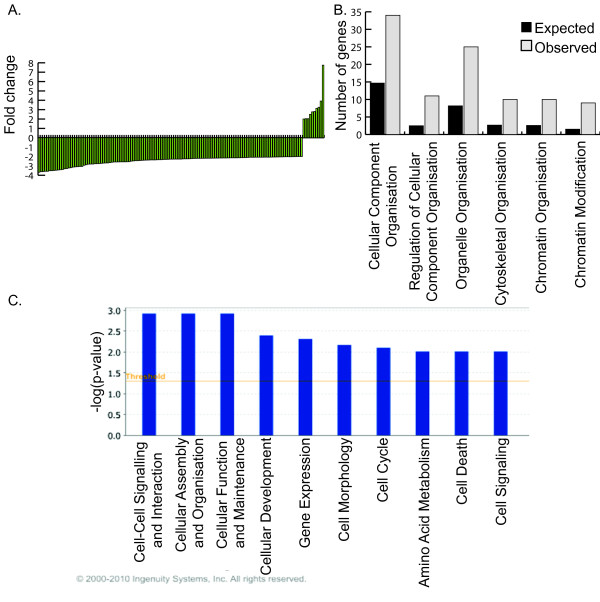
We next looked at the composition of the 111 responders in this group to determine the cellular activities affected. GO Tree Machine (GOTM) analysis was used to identify biological processes that were significantly enriched in this population. The biological function 'cellular component organisation' was significantly affected by exposure to current + GFP. Within this category 'regulation of cellular component organisation', 'organelle organisation' and 'cytoskeletal organisation' were significantly affected. These changes in 'organelle organisation' included the subterms 'chromatin organisation' and 'chromatin modification', which were significantly over-represented in the tissue exposed to current + GFP. This indicates that current + GFP may affect the cellular organisation (Figure 4A). This finding was also identified by Ingenuity Pathway Analysis (IPA). Using IPA, the most significant biological functions within the list of responders are 'cell-cell signalling and interaction', 'cellular assembly and organisation' and 'cellular function and maintenance' (Figure 4B). 'Cell death' is the ninth most significant pathway, with 9 out of the 111 genes found to be involved in this process. However, the changes in these apoptotic genes include ones that could both promote and reduce apoptosis (Figure 4B).
Figure 4.
Effect of electric current + GFP expression on endogenous gene expression of the VLM. (A) Genes showing differential expression when exposed to current + GFP and their fold changes. For gene names refer to Table 3. (B) GOTM analysis showing biological function and expected and observed number of genes in each category compared to the number expected from a random set of 111 genes. (C) IPA functional analysis showing the top ten biological functions enriched in the VLMg compared to VLM samples.
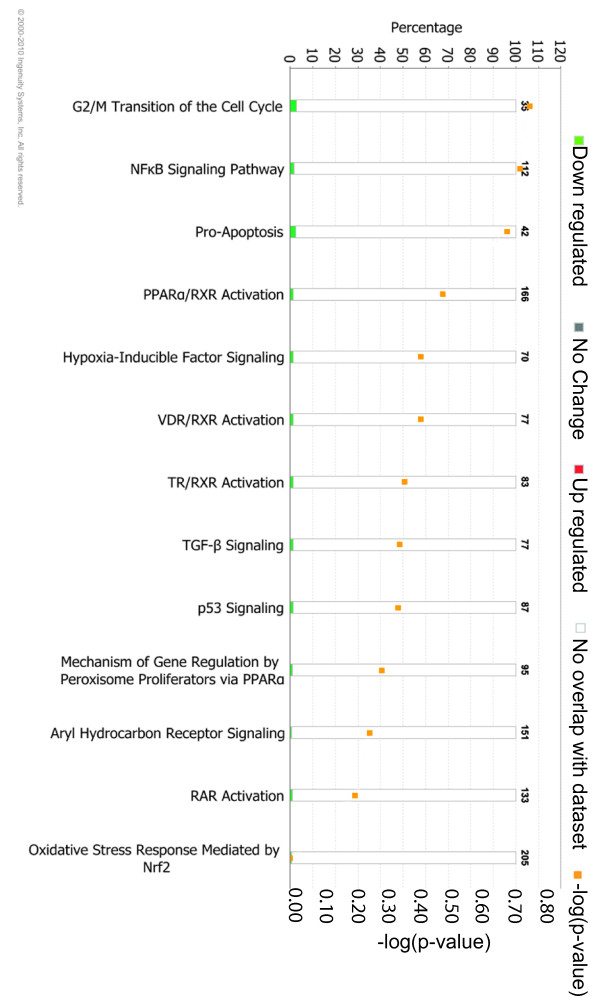
To identify if any of the changes in gene expression upon exposure to current + GFP elicit a toxic affect, such as oxidative stress or apoptosis, IPA toxicity analysis was used (Figure 5). This analysis found some genes involved in pro-apoptosis and p53 signalling in the set of 111 responders. However, the number found is less than would be expected to occur in a random set of 111 genes, suggesting this is not statistically significant. Thus, in ovo electroporation does not induce toxic effects within the embryo as defined by the pathway analysis software.
Figure 5.
Ingenuity Pathway Analysis toxicity analysis. The genes showing differential expression upon exposure to current + GFP are not significantly involved in any type of toxicity. The bar chart shows the percentage of genes involved in each type of toxicity (left axis). Numbers above the bars are the number of genes that would equate to 100% for each type of toxicity. The orange points show the -log(P-value) (right axis) for each category, and thus indicate whether a significant number of genes are found within a category to suggest an actual toxicity affect. A -log(P-value) of 1.3 would be considered significant; all -log(P-values) are lower than 1.3 and therefore not significant.
In ovo electroporation does not affect the regional identity of the VLM
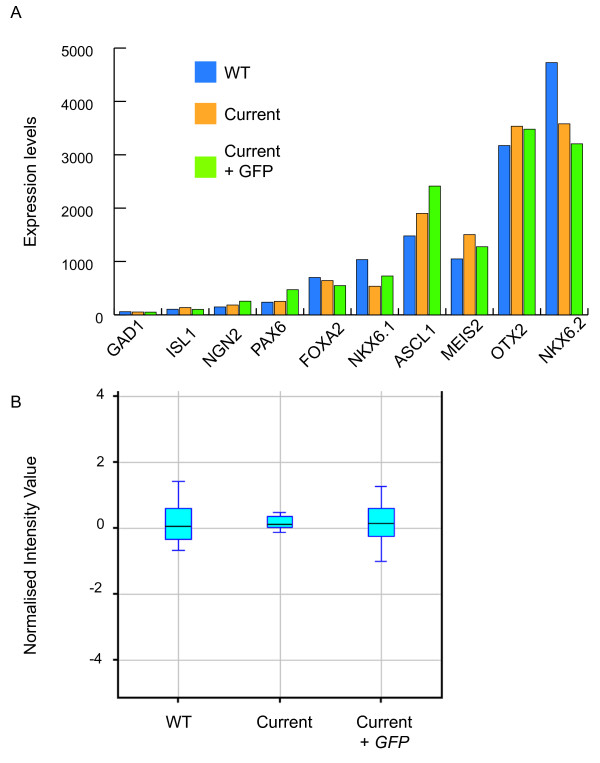
Given the high number of patterning marker genes that are well described for the VLM, we were able to analyse the effects of current and expression of GFP on patterning of this region. The VLM is lateral to the floor plate, in the Nkx6.2 expressing region; therefore, we expected markers such as Nkx6.2 to be present in the microarray. Analysis of the transcriptional profile obtained for the VLM was reflective of the known genetic profile of the region and the expression levels of the ten marker genes were similar in the VLM, VLMi and VLMg samples (Figure 6A).
Figure 6.
Regional identity of VLM unaffected by current alone or current + GFP. (A) Expression profile of ten marker genes of the VLM obtained from the microarray analysis of VLM (wild type (WT)), VLMi and VLMg. In all conditions, marker genes are seen at the expected levels, and there is little difference between the three conditions. (B) Box plot showing the normalised expression values for all ten marker genes. All conditions have the same median and error bars overlap, indicating that the ten marker genes are not significantly differentially expressed between the three conditions. This shows the regional identity is not significantly affected by exposure to current or current + GFP. Statistical analysis using one-way ANOVA also shows that these marker genes are not differentially expressed.
Indeed, statistical analysis using one-way analysis of variance (ANOVA) shows that these marker genes are not differentially expressed between the three samples to a P-value of 0.05. These data are illustrated in Figure 6B, which is a box plot of all ten marker genes. All the box plots have almost the same mean value and all overlap (Figure 6B). These data demonstrate that neither electric current nor current + GFP affect the regional identity of the tissue.
Discussion
In ovo electroporation is an extremely powerful technique to investigate the function of genes and regulatory regions during development. With advances in methods used to analyse electroporated embryos, such as microarray analysis and next generation sequencing, a better understanding of the effects of this technique are necessary. To this end we carried out a genome-wide analysis of the effect of this technique on endogenous gene expression.
Our analysis has established that the electric current used during electroporation (5 × 50-ms pulses of 12 V) has a minimal affect on gene expression, causing a change in expression of only 21 endogenous genes. The upregulation of Hsp25, which encodes a heat shock protein known to be upregulated 24 hours after heat shock in response to protein aggregation [37], combined with changes in two members of the ubiquitination pathway indicate that some protein denaturation may have occurred upon exposure to current.
We have also established that the current and expression of exogenous DNA, in the form of GFP, has a small but statistically significant effect on gene expression, with no toxicity pathways significantly activated. The expression of exogenous DNA has a greater affect on endogenous gene expression than exposure to current alone, with 111 genes affected by exposure to current + GFP.
Interestingly, exogenous DNA expression leads to the downregulation of 101 genes, in contrast to the upregulation of 10 genes, suggesting that exogenous DNA expression, in these conditions, results in a repression of endogenous gene expression.
There has been surprisingly little investigation into the cellular response to exogenous DNA expression. Hus1, a gene involved in the ataxia telangiectasia-mutated (ATM) and ATM and Rad3-related signalling network, has been shown to be upregulated in response to exogenous DNA expression in CHO-K1 and NI3T3 cells [35] and in response to current + GFP in this experiment. IPA and GOTM analysis indicate that the exogenous expression of DNA causes changes in 'cellular organisation' functions, including 'cytoskeletal organisation' and 'chromatin organisation'. Despite the downregulation of several genes involved in 'chromatin organisation and modification', this does not appear to have had an appreciable effect on gene expression since only 0.5% of the genes show a change in gene expression after exposure to current + GFP.
The absence of any effect on the regional identity of the VLM indicates that any changes seen were not sufficient to affect the specification of the tissue at the time point investigated here.
In contrast to the above findings, current + Dmrt5 has a far larger affect on gene expression, causing a change in expression of 309 genes. These changes in gene expression are associated with cell specification, a known function of Dmrt5 (unpublished data). Comparison of the range of fold changes seen in genes affected by current + Dmrt5 (-15 to +1,125), current (-3.8 to +3.0) or current + GFP (-3.7 to +7.8) further highlight the minimal affect that the current and exogenous DNA expression used in this technique have on endogenous gene expression when compared to the affect seen when a regulatory gene is exogenously expressed.
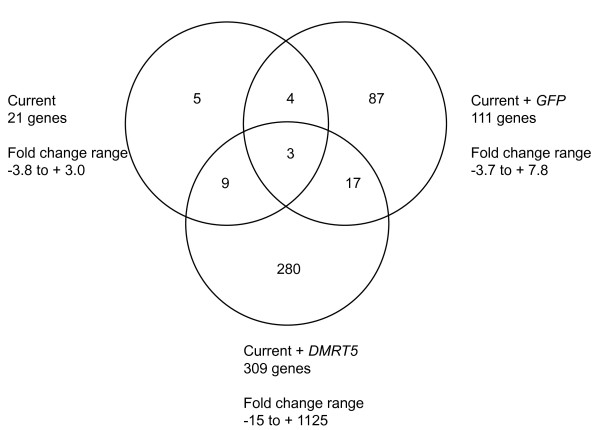
Three genes are affected in all experimental conditions, current, current + GFP and current + Dmrt5 (Figure 7). These are Hnrnpk (Heterogeneous nuclear ribonucleoprotein K), Chd1 (Chromodomain-helicase-DNA-binding protein 1) and Txnl1 (Thioredoxin-like protein 1). Hnrnpk is an RNA binding protein known to associate with pre-mRNAs in the nucleus and influences RNA processing, metabolism and transport. It is involved in maintaining cellular ATP levels in stress conditions, possibly by protecting its target mRNAs [38]. Chd1 is a chromatin remodelling enzyme that regulates transcription [39], and Txnl1 is a redox sensor [40].
Figure 7.
Venn diagram showing overlap between genes differentially expressed in all three conditions. Venn diagram showing annotated endogenous genes that are differentially expressed in all three conditions - current (VLMi), current + GFP (VLMg) and current + Dmrt5 (VLMd) - and the fold change range for each condition.
Of the 21 genes affected by current, 7 are also affected by current + GFP and 12 are also affected by current + Dmrt5 (Figure 7). This indicates that the changes identified in response to current are reproducible, but that the addition of DNA does alter the tissue's response to the current. Of the 111 genes affected by current + GFP, only 19 are affected by exogenous expression of Dmrt5 (Figure 7). This again indicates that the genes responding to the experimental conditions are affected in a combinatorial nature.
Conclusions
We have investigated the effects of both the current required for in ovo electroporation and the effect of exogenous DNA expression resulting from in ovo electroporation. The results of microarray analysis show that current and current + GFP have a small effect on endogenous gene expression, without affecting regional identity. This study demonstrates the validity of the in ovo electroporation technique to study gene function and expression within the developing embryo.
Materials and methods
In ovo electroporation
HH st10 embryos were windowed and electrodes place either side of the developing head (CUY610P1.5-1, Sonidel Ltd (Dublin, Ireland). A solution containing 7 μg/μl DNA construct pCAβ-IRES-GFP or pCAβ-IRES-Dmrt5, 2% polyvinyl alcohol, 0.05% Fast Green in water was injected into the developing midbrain. Five 12 V square wave pulses of 50 ms duration with an interval of 100 ms were applied across the electrodes using an ECM830 Electro-S Square Porator (BTX Inc. (Hawthorne, NY, USA). Following exposure to current, embryos were incubated at 37°C for 24 hours; embryos were collected at HH st16.
Tissue preparation
Collected embryos were washed in PBS and transferred to clean PBS for dissection of the midbrain region, as marked by red lines in Figure 1A. Dissected tissue was transferred to 2 U/ml dispase (Sigma (St Louis, MO, USA) for 10 to 15 minutes. Mechanical dissection was used to remove the neural tube from the mesoderm. VLM was dissected from wild-type embryos (VLM), embryos that had experienced electric current (VLMi), embryos exposed to current and GFP (VLMg) and embryos exposed to current and the regulatory gene Dmrt5 (VLMd); Figure 1A(ii-iv) shows schematics of the region isolated. Only embryos showing high expression of GFP were used; Figure 1A(i) shows an in situ of an electroporated embryo, and Figure 1A(ii-iv) show sections of electroporated embryos, highlighting the electroporation efficiency and region that was dissected from the embryos.
For each microarray, six VLM, six VLMi, six VLMg or six VLMd tissue samples were collected and pooled. Total RNA was isolated from each pool (Absolutely RNA Miniprep Kit, Stratagene (La Jolla, CA, USA). RNA was sent to UK Bioinformatics for processing and analysis. (London, UK) The procedure used was as follows. cDNA was biotin labelled and samples were hybridized to Affymetrix GeneChip Chicken Genome Array according to the manufacturer's protocol. For each condition three sets of pooled samples were collected and hybridised to three arrays to obtain three biological replicates for each condition.
Bioinformatics
Probe levels were calculated from raw data using the MAS5 algorithm embedded into the GCOS suite (version 1.2; Affymetrix). Data were analysed using the GeneSpring package (version GX7; Agilent Technologies, Wokingham, Berkshire, UK). The suitability of the expression data sets for inclusion in the analysis and the overall relationship between biological replicates was assessed using quantile plots and PCA. Samples were first normalised to the 50th percentile (median) across the entire expression data set. Genes were then filtered to remove any genes absent in all arrays. To identify genes showing differential expression between the three samples, a one-way ANOVA with no multiple testing correction was carried out. A P-value of 0.05 and a two-fold change in gene expression was used to determine genes showing differential expression between the samples.
Ingenuity network analysis
The differentially expressed genes were analysed using IPA v8.0-2602 (Ingenuity Systems, Redwood City, CA, USA). The Ingenuity Knowledge Base is the largest knowledge base of its kind, with millions of findings curated from the full text literature. For analysis, the Affymetrix Chicken Genome Array gene list was used as a reference set. All data sources, all species, and all tissues and cell lines were used for the analysis. IPA uses a Fisher's exact test to determine which toxicity pathways and biological functions were significantly enriched within the set of genes showing differential expression compared to the entire list of genes represented on the array.
Gene Ontology Tree Machine
GO provides a controlled vocabulary of about 20,000 terms in three independent hierarchies for cellular components, molecular functions and biological processes. The genes showing differential expression upon exposure to current or current + GFP were converted into mouse annotation and input into the GOTM program. As a reference set the entire mouse genome was chosen. The GOTM web-based tool carries out statistical analysis to identify enriched GO categories for the input gene sets and generates a GO tree to visualize GO terms that are enriched in the input gene list. Hypergeometric test was used to select enriched GO terms for each cluster compared to the GO terms of the entire mouse genome. A GO category was considered as enriched if the P-value was <0.01.
In situ hybridisation
Whole mounts were exposed to digoxigenin-tagged antisense RNA mouse Dmrt5 probe overnight at 70°C for hybridisation with the exogenous Dmrt5 following the protocol described in [41].
Abbreviations
CHO-K1: Chinese hamster ovary epithelial cells; Dmrt5: Double-sex and mab3-related transcription factor like 5; GFP: green fluorescent protein; GO: Gene Ontology; GOTM: Gene Ontology Tree Machine; HEK293: human embryonic kidney cells; HH: Hamburger-Hamilton; IPA: Ingenuity Pathway Analysis; IRES: internal ribosomal entry site; NIH3T3: mouse fibroblast cells; PBS: phosphate-buffered saline; PCA: principal component analysis; st: stage; VLM: ventral lateral midbrain; VLMd: ventral lateral midbrain exposed to current and Dmrt5; VLMg: ventral lateral midbrain exposed to current and GFP; VLMi: ventral lateral midbrain exposed to current.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
EKF, EG, and DC devised experiments. EKF carried out electroporation and isolation of VLM from experimental embryos, analysis of microarray data and wrote the manuscript. EG carried out isolation of VLM from experimental embryos and advised on the manuscript. DC carried out RNA extraction, microarray experiments, microarray analysis and advised on the manuscript. ML provided financial support for the project. All authors read and approved the final manuscript.
Contributor Information
Emma K Farley, Email: e.farley07@csc.mrc.ac.uk.
Emily Gale, Email: emily.gale@imperial.ac.uk.
David Chambers, Email: david.2.chambers@kcl.ac.uk.
Meng Li, Email: meng.li2@imperial.ac.uk.
Acknowledgements
We thank X Nan, I Jaeger, M Clements and M Chotalia for helpful comments on the manuscript. This work was supported by grants from the UK Medical Research Council, EU frame work 7 (Neurostemcell), and the UK Parkinson Disease Society.
References
- Bockamp E, Sprengel R, Eshkind L, Lehmann T, Braun JM, Emmrich F, Hengstler JG. Conditional transgenic mouse models: from the basics to genome-wide sets of knockouts and current studies of tissue regeneration. Regen Med. 2008;3:217–235. doi: 10.2217/17460751.3.2.217. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Bel-Vialar S, Krumlauf R. 'Shocking' developments in chick embryology: electroporation and in ovo gene expression. Nat Cell Biol. 1999;1:E203–207. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Lee JW, Lee SK. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron. 2009;62:641–654. doi: 10.1016/j.neuron.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam SM, Shinmyo Y, Okafuji T, Su Y, Naser IB, Ahmed G, Zhang S, Chen S, Ohta K, Kiyonari H, Abe T, Tanaka S, Nishinakamura R, Terashima T, Kitamura T, Tanaka H. Draxin, a repulsive guidance protein for spinal cord and forebrain commissures. Science. 2009;323:388–393. doi: 10.1126/science.1165187. [DOI] [PubMed] [Google Scholar]
- Luria V, Krawchuk D, Jessell TM, Laufer E, Kania A. Specification of motor axon trajectory by ephrin-B:EphB signaling: symmetrical control of axonal patterning in the developing limb. Neuron. 2008;60:1039–1053. doi: 10.1016/j.neuron.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sato Y, Saito D, Tadokoro R, Takahashi Y. EphrinB2 coordinates the formation of a morphological boundary and cell epithelialization during somite segmentation. Proc Natl Acad Sci USA. 2009;106:7467–7472. doi: 10.1073/pnas.0902859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaal M, Gros J, Lesbros C, Marcelle C. In ovo electroporation of avian somites. Dev Dyn. 2004;229:643–650. doi: 10.1002/dvdy.10433. [DOI] [PubMed] [Google Scholar]
- Gros J, Serralbo O, Marcelle C. WNT11 acts as a directional cue to organize the elongation of early muscle fibres. Nature. 2009;457:589–593. doi: 10.1038/nature07564. [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Chiodini F, Ebeling M, Alliod C, Kundzewicz A, Castro D, Ballivet M, Guillemot F, Matter-Sadzinski L, Matter JM. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136:3767–3777. doi: 10.1242/dev.033449. [DOI] [PubMed] [Google Scholar]
- Kubo F, Nakagawa S. Hairy1 acts as a node downstream of Wnt signaling to maintain retinal stem cell-like progenitor cells in the chick ciliary marginal zone. Development. 2009;136:1823–1833. doi: 10.1242/dev.029272. [DOI] [PubMed] [Google Scholar]
- Garcia-Moreno F, Pedraza M, Di Giovannantonio LG, Di Salvio M, Lopez-Mascaraque L, Simeone A, De Carlos JA. A neuronal migratory pathway crossing from diencephalon to telencephalon populates amygdala nuclei. Nat Neurosci. 2010;13:680–689. doi: 10.1038/nn.2556. [DOI] [PubMed] [Google Scholar]
- Hendricks M, Jesuthasan S. Electroporation-based methods for in vivo, whole mount and primary culture analysis of zebrafish brain development. Neural Dev. 2007;2:6. doi: 10.1186/1749-8104-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizenmann A, Brunet I, Lam JS, Sonnier L, Beurdeley M, Zarbalis K, Weisenhorn-Vogt D, Weinl C, Dwivedy A, Joliot A, Wurst W, Holt C, Prochiantz A. Extracellular Engrailed participates in the topographic guidance of retinal axons in vivo. Neuron. 2009;64:355–366. doi: 10.1016/j.neuron.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdar KP, Wagner TN, Finnerty V. Electroporation of Drosophila embryos. Methods Mol Biol. 1995;48:239–243. doi: 10.1385/0-89603-304-X:239. [DOI] [PubMed] [Google Scholar]
- Irvine SQ, Fonseca VC, Zompa MA, Antony R. Cis-regulatory organization of the Pax6 gene in the ascidian Ciona intestinalis. Dev Biol. 2008;317:649–659. doi: 10.1016/j.ydbio.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz M, Eberhart J, Mastick GS, Krull CE. Sparking new frontiers: using in vivo electroporation for genetic manipulations. Dev Biol. 2001;233:13–21. doi: 10.1006/dbio.2001.0181. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Analyzing gene regulation in ascidian embryos: new tools for new perspectives. Differentiation. 2002;70:132–139. doi: 10.1046/j.1432-0436.2002.700402.x. [DOI] [PubMed] [Google Scholar]
- Muramatsu T, Mizutani Y, Ohmori Y, Okumura J. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem Biophys Res Commun. 1997;230:376–380. doi: 10.1006/bbrc.1996.5882. [DOI] [PubMed] [Google Scholar]
- Krull CE. A primer on using in ovo electroporation to analyze gene function. Dev Dyn. 2004;229:433–439. doi: 10.1002/dvdy.10473. [DOI] [PubMed] [Google Scholar]
- Simkin JE, McKeown SJ, Newgreen DF. Focal electroporation in ovo. Dev Dyn. 2009;238:3152–3155. doi: 10.1002/dvdy.22142. [DOI] [PubMed] [Google Scholar]
- Pu HF, Young AP. Glucocorticoid-inducible expression of a glutamine synthetase-CAT-encoding fusion plasmid after transfection of intact chicken retinal explant cultures. Gene. 1990;89:259–263. doi: 10.1016/0378-1119(90)90014-I. [DOI] [PubMed] [Google Scholar]
- Yaneza M, Gilthorpe JD, Lumsden A, Tucker AS. No evidence for ventrally migrating neural tube cells from the mid- and hindbrain. Dev Dyn. 2002;223:163–167. doi: 10.1002/dvdy.1241. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Watanabe Y, Funahashi J. Misexpression of genes in brain vesicles by in ovo electroporation. Dev Growth Differ. 2000;42:199–201. doi: 10.1046/j.1440-169x.2000.00501.x. [DOI] [PubMed] [Google Scholar]
- Luo J, Redies C. Overexpression of genes in Purkinje cells in the embryonic chicken cerebellum by in vivo electroporation. J Neurosci Methods. 2004;139:241–245. doi: 10.1016/j.jneumeth.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Sato Y, Kasai T, Nakagawa S, Tanabe K, Watanabe T, Kawakami K, Takahashi Y. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev Biol. 2007;305:616–624. doi: 10.1016/j.ydbio.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Saito D, Tanabe K, Suetsugu R, Nakaya Y, Nakagawa S, Takahashi Y. Tet-on inducible system combined with in ovo electroporation dissects multiple roles of genes in somitogenesis of chicken embryos. Dev Biol. 2007;305:625–636. doi: 10.1016/j.ydbio.2007.01.042. [DOI] [PubMed] [Google Scholar]
- Pekarik V, Bourikas D, Miglino N, Joset P, Preiswerk S, Stoeckli ET. Screening for gene function in chicken embryo using RNAi and electroporation. Nat Biotechnol. 2003;21:93–96. doi: 10.1038/nbt770. [DOI] [PubMed] [Google Scholar]
- Sahin M, Greer PL, Lin MZ, Poucher H, Eberhart J, Schmidt S, Wright TM, Shamah SM, O'connell S, Cowan CW, Hu L, Goldberg JL, Debant A, Corfas G, Krull CE, Greenberg ME. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron. 2005;46:191–204. doi: 10.1016/j.neuron.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/S0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Araki I, Nakamura H. Engrailed defines the position of dorsal di-mesencephalic boundary by repressing diencephalic fate. Development. 1999;126:5127–5135. doi: 10.1242/dev.126.22.5127. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;4:509–519. doi: 10.1016/S1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Takemoto T, Kamachi Y, Kondoh H. Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech Dev. 2004;121:1145–1158. doi: 10.1016/j.mod.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Mlakar V, Todorovic V, Cemazar M, Glavac D, Sersa G. Electric pulses used in electrochemotherapy and electrogene therapy do not significantly change the expression profile of genes involved in the development of cancer in malignant melanoma cells. BMC Cancer. 2009;9:299. doi: 10.1186/1471-2407-9-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoucheva O, Alexeev V, Yoon K. Differential cellular responses to exogenous DNA in mammalian cells and its effect on oligonucleotide-directed gene modification. Gene Ther. 2006;13:266–275. doi: 10.1038/sj.gt.3302643. [DOI] [PubMed] [Google Scholar]
- Yoon KH, Blankenship T, Shibata B, Fitzgerald PG. Resisting the effects of aging: a function for the fiber cell beaded filament. Invest Ophthalmol Vis Sci. 2008;49:1030–1036. doi: 10.1167/iovs.07-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Fujimoto M, Nakamura K, Inouye S, Sugahara K, Izu H, Nakai A. Hsp25, a member of the Hsp30 family, promotes inclusion formation in response to stress. FEBS Lett. 2004;565:28–32. doi: 10.1016/j.febslet.2003.12.085. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Naiki T, Saito M, Irie K. hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions. Genes Cells. 2009;14:113–128. doi: 10.1111/j.1365-2443.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felberbaum-Corti M, Morel E, Cavalli V, Vilbois F, Gruenberg J. The redox sensor TXNL1 plays a regulatory role in fluid phase endocytosis. PLoS One. 2007;2:e1144. doi: 10.1371/journal.pone.0001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA, Patel K, Wilkinson DG. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]