Abstract
The bacterial determinants of pulmonary Francisella induced inflammatory responses and their interaction with host components are not clearly defined. In this study, proteomic and immunoblot analyses showed presence of a cytoplasmic protein elongation factor Tu (EF-Tu) in the membrane fractions of virulent F. novicida, LVS and SchuS4, but not in an attenuated F. novicida mutant. EF-Tu was immunodominant in mice vaccinated and protected from virulent F. novicida. Moreover, recombinant EF-Tu induced macrophages to produce inflammatory cytokines in a TLR4 dependent manner. This study shows immune stimulatory properties of a cytoplasmic protein EF-Tu expressed on the membrane of virulent Francisella strains.
Keywords: Francisella, tularemia, Elongation factor-Tu, Toll-like receptor, innate immune response, inflammation
1. Introduction
Francisella tularensis is a Gram negative intracellular coccobacillus and the causative agent of tularemia. The pneumonic infections with this organism present the most debilitating form of the disease resulting in severe sepsis [1; 2]. The ease of bacterial transmission via aerosol routes and extremely low infectious doses has led to the CDC classification of Type A strains as Category A bioterrorism agents [1]. Out of the four subsp. currently identified for F. tularensis, intranasal infections with lethal doses of human pathogenic Type A strains of the subsp. tularensis and the murine model strain novicida cause a rapid death of mice within 3–5 days before the onset of adaptive immunity [2; 3; 4]. Thus innate immune responses play an important role in mediating the outcome of infection with this organism. However, the bacterial components stimulating these responses and the mechanisms of interaction between the bacterial and host cell components are only beginning to emerge [5; 6].
Toll-Like receptors (TLRs) are germ line encoded transmembrane receptors expressed by innate immune cells, mainly macrophages and dendritic cells. So far thirteen TLRs (10 in humans and 12 in mice) have been identified which recognize distinct mutation resistant microbial components causing activation of a variety of signal transduction pathways [7]. This results in production of cytokines and chemokines presenting the first line of defense against invading pathogens. Thus recognition of pathogen associated molecules by TLRs on innate immune cells is crucial for the generation of protective response.
Recent studies from our laboratory have shown that a transposon mutant of F. novicida strain U112 lacking a 58 kDa protein is attenuated and elicits a much reduced inflammatory response in comparison with the fully virulent wild-type strain [2]. Moreover, intranasal inoculation with this mutant protected the mice from an otherwise lethal challenge with the wild-type bacteria [3]. As the bacterial proteins present on the membrane are the most likely candidates to interact with innate immune components, we hypothesized that comparative proteomic analyses of the membranes of attenuated mutant and the virulent wild-type strain would yield information about virulence attributes and /or protective antigens of this pathogen.
In this study, we compared the membrane proteins isolated from the attenuated mutant and the wild-type F. novicida strain U112 and characterized the innate immune responses of bone-marrow derived macrophages towards putative virulence factors thus identified. We report that Francisella Elongation factor Tu (EF-Tu), a cytosolic protein normally involved in translational machinery is present in the membrane fractions of F. novicida, LVS as well as the fully virulent Type A strain SchuS4, and appears to act as a novel virulence factor capable of eliciting an inflammatory cytokine response in macrophages in a Toll-like receptor 4 (TLR4) dependent manner.
2. Materials and Methods
2.1. Bacterial strains and mice
The wild type F. novicida strain U112 and the transposon mutants lacking the 58kDa protein (locus tag FTN_0444) were kindly provided by Dr. L. Gallagher [8]. The Live Vaccine Strain (LVS) and Francisella tularensis Type A strain SchuS4 was kindly provided by Dr. Karl E. Klose (University of Texas at San Antonio). The bacteria were cultivated and stored as previously described [2]. For the expression of recombinant EF-Tu, the E. coli strains XL1-Blue (Invitrogen) and MKV15 (DE3), a triple mutant lacking secondary acyltransferases lpxL, lpxM and lpxP (kindly provided by Dr. Cheung Y. Hung, University of Texas at San Antonio, originally from Dr. Stephen Trent, University of Texas at Austin) were grown in Luria Bertani (LB) broth at 37 °C.
For protection studies and isolation of bone marrows, female mice aged 6–8 weeks were used. TLR2−/− and TLR4−/− mice in C57BL/6 background (originally from S. Akira, Osaka University, Osaka, Japan) were obtained from Dr. Michael Berton (Department of Microbiology and Immunology, University of Texas Health Science Center at San Antonio (UTHSCSA), TX). The wild-type C57BL/6 mice were obtained from the National Cancer Institute animal program (Bethesda, MD). All mouse strains were bred in the University of Texas at San Antonio (UTSA) animal facility. Experiments were conducted under the guidelines of the IACUC, UTSA, University of Texas System, the U.S. Department of Agriculture, and the National Institutes of Health.
2.2. Vaccination of mice with the attenuated mutant
Wild-type C57BL/6 mice were vaccinated with the 58 kDa mutant and challenged with the virulent F. novicida strain U112 as described in our previous study [3]. Sera from vaccinated and protected mice were collected 3 weeks after challenge and were titrated using U112 bacterial lysate by western blot analysis. Sera showing positive reactions at dilutions ≥ 1:2000 were used for further experiments described below.
2.3. Isolation of sarkosyl-insoluble membrane proteins of Francisella
The sarkosyl insoluble membrane fractions of the F. novicida 58 kDa mutant as well as the wild-type strains U112, Type B strain LVS and Type A strain SchuS4 were isolated using previously described methods with modifications [9]. Briefly, bacterial cultures grown overnight on agar plates were sonicated in 20mM Tris-HCl (pH 7.5) along with DNAse and protease inhibitor cocktail (Roche, Mannheim, Germany) on ice. The samples were incubated at room temperature (RT) for 30 min followed by centrifugation at 7500 g (20 min, 4 °C). An aliquot of the supernatant was used as a total protein fraction and the remainder was centrifuged at 200,000 g (1h, 4 °C) to pellet cell membranes. The sediment was suspended in Tris-HCl, treated for 30 min at RT with 2% (w / v) sarkosyl (Sigma) and then centrifuged at 200,000 g (1h). This step was repeated 3X and the last pellet was suspended in Tris, aliquoted and stored at −80 °C until further use. The protein concentration in the samples was estimated using a BCA protein estimation kit (Pierce, IL).
2.4. 2-DE western blot and identification of protein spots
The total and membrane protein fractions were resolved by 2-DE using the Bio-Rad IEF system by methods previously described [10]. For the first dimension 100µg of protein was resolved on pH 3–10 IPG strips (11cm) followed by the second dimension in 12.5% acrylamide gels. The proteins were transferred onto polyvinylidene polypyrrolidone fluoride (PVDF) membranes, and western blots were performed as previously described [3]. Sera from mice vaccinated with the 58 kDa mutant or the polyclonal antibodies raised against the recombinant EF-Tu were used as primary antibodies. Based on the western blot results, the protein spots were cut from coomassie stained 2-DE gels and were identified by MALDI-TOF/MS analysis in the Institutional Mass Spectrometry Facility at UTHSCSA as described [10].
2.5. Expression and purification of recombinant EF-Tu
The gene encoding Francisella EF-Tu (F. tularensis ORF FTT_0137) was amplified using SchuS4 genomic DNA as template, cloned into pGEX-6p vector (Amersham Biosciences Corp., Piscataway, NJ) and purified as described before [11] using E. coli XL1-Blue or MKV15 (DE3) strains. The purified protein was passed through Detoxi-Gel endotoxin removing columns per manufacturer’s instructions (Pierce, Rockford, IL) to remove contaminating LPS. The levels of LPS in purified EF-Tu preparations were checked by Limulus Amoebocyte Lysate (LAL) assay (Cambrex Bioscience) and was found to be <2.0 ng/mg protein.. As a control for endotoxin contamination, the detoxified EF-Tu or Ultrapure LPS was boiled for 45 min at 100 °C and used to stimulate cells. The concentration of EF-Tu was determined by Bicinchoninic acid (BCA) assay kit (Pierce, Rockford, IL).
2.6. Stimulation of bone-marrow derived macrophages (BMDMs) with recombinant EF-Tu and cytokine analysis
Bone marrows were isolated from wild-type C57BL/6, TLR2−/− or TLR4−/− mice and the cells were differentiated to macrophages as previously described [12]. On day 6 of culture cells were plated at 0.08×106 cells per well in 96-well flat-bottom plates and stimulated with the indicated concentrations of recombinant EF-Tu purified from XL1-Blue or MKV15(DE3). Ultrapure LPS (Escherichia coli serotype 0111:B4; Sigma-Aldrich, St. Louis, MO) and Pam3Cys4 or FSL-1 were used as known ligands for TLR4 and TLR2 respectively. Measurement of IL-6 and TNF-α in the supernatants at 24h of culture was done by sandwich ELISA (BD Pharmingen) as per the manufacture’s instructions.
3. Results
3.1. Francisella EF-Tu is present in the membrane fraction of the virulent Francisella strains U112, LVS and SchuS4 but not on attenuated F. novicida mutant and is highly immunodominant
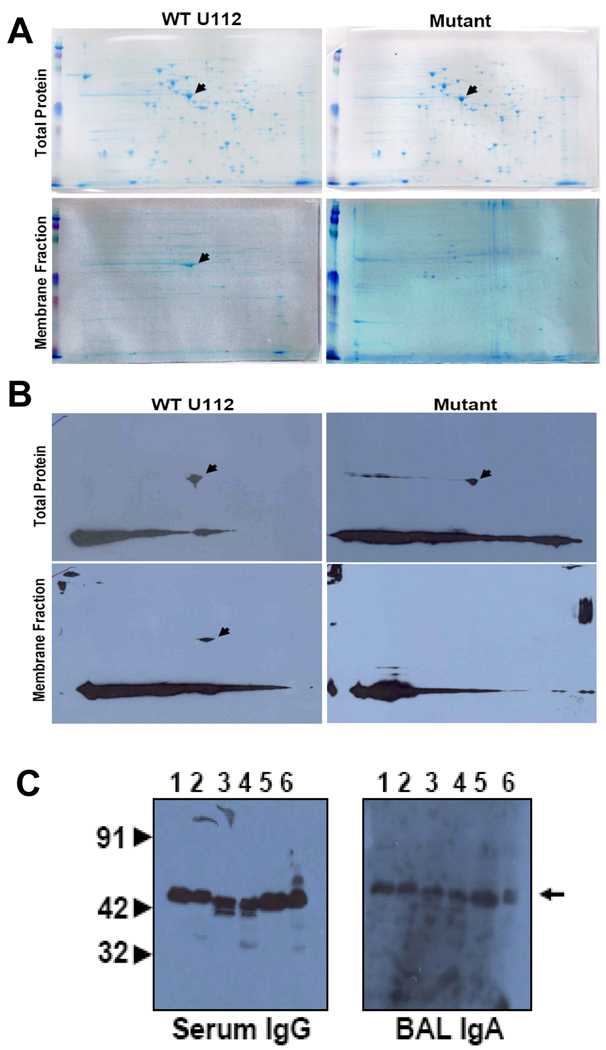
Our previous studies showed that the F. novicida transposon mutant lacking a 58kDa protein was highly attenuated in mice via intranasal infection route but protected the mice from a subsequent challenge with the wild-type strain U112 [2; 3]. In order to identify putative virulence factors and/or protective antigens, we compared the sarkosyl insoluble membrane fraction of the mutant and wild-type strain U112 of F. novicida. A 2-DE analysis of the fractions showed the presence of a major 42 kDa protein in the total protein fractions of both mutant and WT bacteria but was present in the membrane fraction of only the WT and not the mutant (Fig.1 A). A MALDI-TOF/MS analysis identified the protein as elongation factor Tu (EF-Tu). Western blot analysis of the total and membrane protein fractions of the wild type U112 and the attenuated strain using sera from mice vaccinated with the mutant and challenged with the wild-type strain (Mut/WT mice) showed that EF-Tu was highly immunodominant and induced a strong humoral response in the protected mice (Fig 1B). Moreover, EF-Tu was found to be present in the membrane fractions of LVS as well as the Type A strain SchuS4 and was highly immunoreactive to serum IgG and bronchoalveolar lavage (BAL) IgA antibodies from Mut/WT mice (Fig. 1C). The presence of EF-Tu in the membrane fractions of all the three virulent strains of Francisella (SchuS4, U112 and LVS) was confirmed by probing the protein fractions with a mouse antibody raised against purified recombinant Francisella EF-Tu (data not shown). These results demonstrate that EF-Tu, a protein normally present in the cytoplasm as part of the translational machinery of the cell, is expressed on the membranes of all the three strains of Francisella that are virulent in animals and/or humans. The absence of EF-Tu in the membrane fraction of the attenuated strain of F. novicida indicates that this protein may be acting as a virulence factor. Additionally, mucosal IgA and serum IgG and IgA antibodies directed against EF-Tu were produced abundantly in mice protected from an otherwise lethal challenge with F. novicida following vaccination with the attenuated mutant strain. This suggests that EF-Tu is immunodominant at local as well as systemic levels in the host during infection.
Fig. 1.
Fig. 1. Francisella EF-Tu is expressed on membrane of virulent but not on an attenuated strain and is highly immunodominant. (A) Total and sarkosyl-insoluble membrane fractions of wild-type and attenuated strains of F. novicida were fractionated on IEF strips of pH 4–7. The second dimension was run on 12.5 % SDS gels and stained with Coomassie. Arrow heads indicate EF-Tu. (B) Total and sarkosyl-insoluble membrane fractions of wild-type and attenuated strains of F. novicida separated on 2-DE gels and probed with pooled sera from mice vaccinated with the attenuated mutant and challenged with the wild-type F. novicida strain U112 (Mut/WT sera). The arrow heads indicate EF-Tu. (C) Total (Lanes 1, 3 and 5) and sarkosyl-insoluble membrane fractions (2, 4 and 6) of LVS, Type A strain SchuS4 and F. novicida strain U112 respectively probed with Mut/WT sera or bronchoalveoplar lavage (BAL) followed by horseradish peroxidase (HRP) conjugated anti-mouse IgG or anti-mouse IgA as secondary antibodies. The arrow indicates EF-Tu.
3.2. Recombinant EF-Tu elicits inflammatory cytokine production from bone-marrow macrophages in a TLR-4 dependent manner
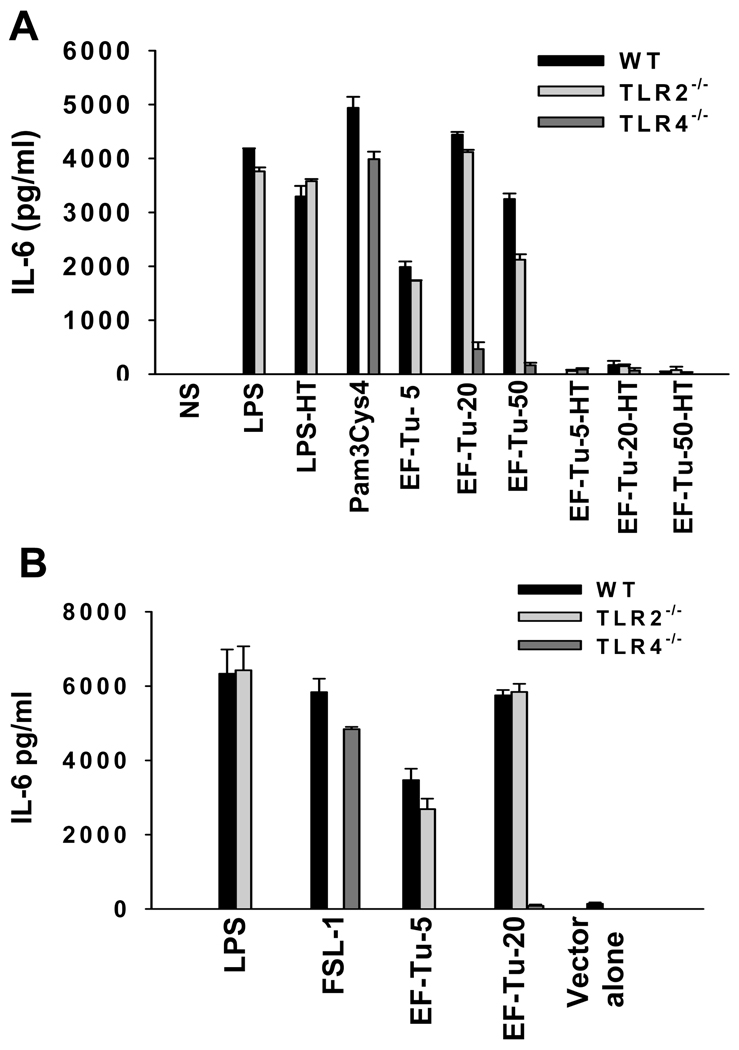
To determine the significance of membrane expression of EF-Tu that is normally expressed in cell cytosol, we tested if this protein has involved in the activation of macrophages, a cell-type central to initiation of innate immune responses. The ORF representing the Francisella EF-Tu (FTT_0137) was cloned using the genomic DNA of SchuS4 as template, and the protein was expressed in N-terminal GST-tag fusion form in E. coli strain XL1-Blue as well as MKV15 (DE3), a strain with a tetra-acylated LPS due to triple mutation in lpxL, lpxM and lpxP rendering its LPS non-stimulatory. Bone marrow derived macrophages (BMDMs) from wild-type C57BL/6 mice, were stimulated with 5–50ug/ml of recombinant EF-Tu for 24h. The culture supernatants were then tested for the levels of IL-6 and TNF-α by ELISA. As shown in Figure 2A, the wild-type BMDMs produced substantial amounts of IL-6 upon stimulation with all of the concentrations tested. Similar results were obtained with TNF-α also (data not shown). As TLR2 and TLR4 are the major PRRs of macrophages involved in recognition of PAMPs, we determined if either of these receptors could be involved in Francisella EF-Tu recognition. Whereas the stimulation with EF-Tu produced comparable amounts of IL-6 from wild-type and TLR2−/− BMDMs, the stimulatory effect was abolished in macrophages lacking TLR4 (Fig 2A). The levels of IL-6 induced by EF-Tu were comparable to those observed with the known ligands for TLR2 (Pam3Cys4, FSL1) and TLR4 (LPS). To ascertain that this effect was unrelated to trace amounts of LPS despite passing recombinant EF-Tu through EndoFree columns, the purified protein was boiled before stimulating the macrophages (Fig. 2A). As expected, the heat treatment abolished the stimulatory effect of purified EF-Tu but not LPS, confirming that the stimulatory effect was due to protein antigen and not contaminating LPS (Fig. 2A). Furthermore, the purified EF-Tu expressed in E. coli MKV15 with mutated LPS showed a similar stimulatory effect which was TLR4 dependent (Fig 2B). Taken together, these results show that Francisella EF-Tu is able to induce inflammatory cytokine production from macrophages in a TLR4 dependent manner.
Fig. 2.
Recombinant EF-Tu of Francisella stimulates bone-marrow derived macrophages to produce IL-6 in a TLR4 dependent manner. (A) Bone marrow derived macrophages from wild-type C57BL/6, TLR2−/− or TLR4−/− mice were stimulated with various concentrations (5–50ug/ml) of purified recombinant Francisella EF-Tu. Ultra-pure LPS (10ng/ml) or Pam3Cys4 (10ug/ml) was used as a control for TLR4 or TLR2 respectively. The amount of IL-6 in culture supernatants at 24h post-stimulation was measured by sandwich ELISA. LPS-HT and EF-Tu-HT denote heat treated LPS and EF-Tu respectively. (B) Francisella EF-Tu expressed and purified from E. coli triple mutant MKV15(DE3) with mutated LPS was used to stimulate macrophages derived from wild-type C57BL/6, TLR2−/− or TLR4−/− mice. As a negative control, bacterial lysates obtained from MKV15(DE3) bacteria transformed with empty vector were processed the same way. Ultra-pure LPS (10ng/ml) or FSL-1 (10ug/ml) was used as a control for TLR4 or TLR2 respectively. The amount of IL-6 in culture supernatants at 24h post-stimulation was measured by sandwich ELISA.
4. Discussion
Virulence determinants of pulmonary Francisella induced inflammatory response are largely unknown. In this study we identified elongation factor-Tu (EF-Tu), a protein normally present in cell cytoplasm to be present on the membrane of virulent Francisella strains U112, LVS and SchuS4. Our data also showed that this protein elicited inflammatory cytokine production in macrophages and this stimulatory effect was TLR4 dependent.
Recent studies have shown that several bacterial pathogens of plants and animals express EF-Tu, a prokaryotic cytoplasmic protein with a “house-keeping” function in peptide elongation during protein synthesis process, on their surface which plays a key role in facilitating the pathogenesis of these microbes by interaction with a variety of host molecules. For example, EF-Tu expressed on the membrane of P. aeruginosa and Mycoplasma pneumoniae acts as a virulence factor by binding to complement and fibronectin in order to facilitate the infection process [13; 14]. Recently, Francisella Type A strain SchuS4 has also been reported to bind to serine protease pasmin to degrade opsonizing antibody in order to evade host immunity [15]. It is tempting to speculate that EF-Tu may constitute one of the bacterial factors involved in this process, which needs to be examined experimentally. In addition, a recent study reported expression of EF-Tu on surface of LVS which facilitated the entry of the bacteria into THP1 cells [16]. Our study reported here showed that EF-Tu was expressed on membrane of F. novicida as well as the Type A strain SchuS4. Importantly, our comparative analysis of showed the presence of this protein in the membrane fraction of virulent strains of Francisella but not in an attenuated strain, which is defective in causing the disease in mice [2]. As the infection with this mutant results in lower bacterial burdens in systemic organs of infected mice, compared to the wild-type strain [2; 3], it is possible that absence of EF-Tu on membrane of this mutant strains contributes to this. This suggests a role of this protein as a virulence factor facilitating the pulmonary infection with Francisella in-vivo.
A strong mucosal and systemic antibody response to EF-Tu was observed in mice challenged with wild-type F. novicida strain U112 following intranasal vaccination with the attenuated mutant. As these mice are protected from an otherwise lethal infection with F. novicida, and other studies have shown an important role of antibodies in clearance of bacteria as well as beneficial effects of B cells on early innate immune responses [17], it is likely that this humoral response against EF-Tu contributes to the protection of mice observed in our studies. Additionally, human sera collected from tularemia patients have been shown to be highly immunoreactive to EF-Tu [18]. Furthermore, serological immunodominance of this protein has been shown in human patients as well as experimental animals infected with other pathogens such as Chlamydia and Anaplasma marginale [19; 20]. Thus, EF-Tu can not only serve as a diagnostic marker for seropositivity but also can be explored as a protective vaccine candidate. Further studies in this regard are underway in our laboratory.
The innate immune system of plants and animals is armed with a variety of pattern recognition receptors (PRRs) such as TLRs which specifically recognize pathogen associated molecular patterns (PAMPs) to initiate immune responses. In order to maintain the functionality of this system, these receptors have evolved to recognize PAMPs which are essential for normal function of the pathogen and which the pathogen cannot afford to mutate in order to escape the recognition. In this study, we show that EF-Tu, a protein essential for protein synthesis in Francisella, acts as a stimulator of innate immune cells in a TLR4 dependent manner. The immune stimulatory properties of this protein as a PAMP have been observed in bacterial infections of plants [21; 22]. Our study is the first report of Francisella EF-Tu eliciting an innate immune response in a TLR dependent manner. In toto, our findings reported here, present EF-Tu as a bacterial factor capable of activating macrophages in a TLR4 dependent manner as well as humoral immune response associated with protection from the disease. The immune stimulatory potential of this protein makes it an attractive candidate for devising protective measures against this deadly infection.
Acknowledgements
The authors thank Drs. Collin Manoil and Larry Gallagher, University of Washington for kindly providing the F. novicida mutant strain; and Dr. Derek P. Thomas, University of Texas at San Antonio for technical help with 2DE analysis. This work was supported by National Institute of Health Grants 1P01A10157986, NS35974, AI 59703 to JMT and American Heart Association Grant 10BGIA4300041 to JS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Tonat K. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 2.Sharma J, Li Q, Mishra BB, Pena C, Teale JM. Lethal pulmonary infection with Francisella novicida is associated with severe sepsis. J Leukoc Biol. 2009;86:491–504. doi: 10.1189/jlb.1208728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma J, Li Q, Mishra BB, Georges MJ, Teale JM. Vaccination with an attenuated strain of Francisella novicida prevents T-cell depletion and protects mice infected with the wild-type strain from severe sepsis. Infect Immun. 2009;77:4314–4326. doi: 10.1128/IAI.00654-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson RV, Crane DD, Bosio CM. Long lived protection against pneumonic tularemia is correlated with cellular immunity in peripheral, not pulmonary, organs. Vaccine. 2010;28:6562–6572. doi: 10.1016/j.vaccine.2010.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noah CE, Malik M, Bublitz DC, Camenares D, Sellati TJ, Benach JL, Furie MB. GroEL and lipopolysaccharide from Francisella tularensis live vaccine strain synergistically activate human macrophages. Infect Immun. 2010;78:1797–1806. doi: 10.1128/IAI.01135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vezzulli L, Pezzati E, Repetto B, Stauder M, Giusto G, Pruzzo C. A general role for surface membrane proteins in attachment to chitin particles and copepods of environmental and clinical vibrios. Lett Appl Microbiol. 2008;46:119–125. doi: 10.1111/j.1472-765X.2007.02269.x. [DOI] [PubMed] [Google Scholar]
- 10.Thomas DP, Lopez-Ribot JL, Lee SA. A proteomic analysis of secretory proteins of a pre-vacuolar mutant of Candida albicans. J Proteomics. 2009;73:342–351. doi: 10.1016/j.jprot.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma J, Zhong Y, Dong F, Piper JM, Wang G, Zhong G. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect Immun. 2006;74:1490–1499. doi: 10.1128/IAI.74.3.1490-1499.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunert A, Losse J, Gruszin C, Huhn M, Kaendler K, Mikkat S, Volke D, Hoffmann R, Jokiranta TS, Seeberger H, Moellmann U, Hellwage J, Zipfel PF. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J Immunol. 2007;179:2979–2988. doi: 10.4049/jimmunol.179.5.2979. [DOI] [PubMed] [Google Scholar]
- 14.Balasubramanian S, Kannan TR, Baseman JB. The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin. Infect Immun. 2008;76:3116–3123. doi: 10.1128/IAI.00173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crane DD, Warner SL, Bosio CM. A novel role for plasmin-mediated degradation of opsonizing antibody in the evasion of host immunity by virulent, but not attenuated, Francisella tularensis. J Immunol. 2009;183:4593–4600. doi: 10.4049/jimmunol.0901655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barel M, Hovanessian AG, Meibom K, Briand JP, Dupuis M, Charbit A. A novel receptor - ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: interaction between surface nucleolin and bacterial elongation factor Tu. BMC Microbiol. 2008;8:145. doi: 10.1186/1471-2180-8-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirimanjeswara GS, Olmos S, Bakshi CS, Metzger DW. Humoral and cell-mediated immunity to the intracellular pathogen Francisella tularensis. Immunol Rev. 2008;225:244–255. doi: 10.1111/j.1600-065X.2008.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havlasova J, Hernychova L, Halada P, Pellantova V, Krejsek J, Stulik J, Macela A, Jungblut PR, Larsson P, Forsman M. Mapping of immunoreactive antigens of Francisella tularensis live vaccine strain. Proteomics. 2002;2:857–867. doi: 10.1002/1615-9861(200207)2:7<857::AID-PROT857>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Bunk S, Susnea I, Rupp J, Summersgill JT, Maass M, Stegmann W, Schrattenholz A, Wendel A, Przybylski M, Hermann C. Immunoproteomic identification and serological responses to novel Chlamydia pneumoniae antigens that are associated with persistent C. pneumoniae infections. J Immunol. 2008;180:5490–5498. doi: 10.4049/jimmunol.180.8.5490. [DOI] [PubMed] [Google Scholar]
- 20.Lopez JE, Siems WF, Palmer GH, Brayton KA, McGuire TC, Norimine J, Brown WC. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect Immun. 2005;73:8109–8118. doi: 10.1128/IAI.73.12.8109-8118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]