Abstract
Hepatitis E virus (HEV) is an important human pathogen. At least four recognized and two putative genotypes of mammalian HEV have been reported: genotypes 1 and 2 are restricted to humans whereas genotypes 3 and 4 are zoonotic. The current experimental vaccines are all based on a single strain of HEV, even though multiple genotypes of HEV are co-circulating in some countries and thus an individual may be exposed to more than one genotype. Genotypes 3 and 4 swine HEV is widespread in pigs and known to infect humans. Therefore, it is important to know if prior infection with a genotype 3 swine HEV will confer protective immunity against subsequent exposure to genotypes 3 and 4 human and swine HEV. In this study, specific-pathogen-free pigs were divided into 4 groups of 6 each. Pigs in the three treatment groups were each inoculated with a genotype 3 swine HEV, and 12 weeks later, challenged with the same genotype 3 swine HEV, a genotype 3 human HEV, and a genotype 4 human HEV, respectively. The control group was inoculated and challenged with PBS buffer. Weekly sera from all pigs were tested for HEV RNA and IgG anti-HEV, and weekly fecal samples were also tested for HEV RNA. The pigs inoculated with swine HEV became infected as evidenced by fecal virus shedding and viremia, and the majority of pigs also developed IgG anti-HEV prior to challenge at 12 weeks post-inoculation. After challenge, viremia and fecal virus shedding of challenge viruses were not detected, suggesting that prior infection with a genotype 3 swine HEV prevented pigs from developing viremia and fecal virus shedding after challenges with homologous and heterologous genotypes 3 and 4 HEV. The results from this study have important implications for future development of an effective HEV vaccine.
Keywords: hepatitis E virus (HEV), swine HEV, cross-protection, prior infection, pigs
1. Introduction
Hepatitis E virus (HEV), the causative agent of human hepatitis E, is an important pathogen worldwide (Meng, 2010a, 2010b; Purcell and Emerson, 2008). Although the mortality rate is generally low, it can reach up to 28% in HEV-infected pregnant women (Bhatia et al., 2008; Jilani et al., 2007). As a fecal-orally transmitted virus, contaminated water is the most common source of infection for large outbreaks in developing countries due to poor sanitation conditions (Emerson and Purcell, 2003; Meng, 2010b). In some industrialized countries, sporadic cases of acute hepatitis E that are likely caused by zoonotic transmission have also been reported. Cases of autochthonous HEV infections from industrialized countries are increasing (Aikawa et al., 2002; Colson et al., 2010; Jameel, 1999; Legrand-Abravanel et al., 2010; Mizuo et al., 2005).
At least four recognized genotypes of mammalian HEV have been identified worldwide: genotypes 1 and 2 strains of HEV have a limited host range and are restricted to humans, whereas genotypes 3 and 4 have an expanded host range and are zoonotic (Meng, 2010a, 2010b; Pavio et al., 2010; Pavio and Mansuy, 2010). The first animal strain of HEV, swine HEV, was identified from pigs in the United States (Meng et al., 2007). Thus far, all viruses identified from pigs worldwide belong to either genotype 3 or genotype 4 (Meng, 2010a, 2010b; Okamoto, 2007). Besides pigs and humans, HEV has also been genetically identified from chickens (Haqshenas et al., 2001), rats (Johne et al., 2010), mongoose (Nakamura et al., 2006), deer (Takahashi et al., 2004; Tei et al., 2003), and rabbits (Zhao et al., 2009). Two putative new genotypes of mammalian HEV, rat HEV (Johne et al., 2010) and a novel wild boar HEV (Takahashi et al., 2011), have recently been identified. It appears that all mammalian HEV genotypes identified thus far may belong to a single serotype (Emerson and Purcell, 2003; Meng, 2010b), since antigenic cross-reactivity of the capsid protein among mammalian HEV strains and between avian HEV and mammalian HEV have been reported (Haqshenas et al., 2002). The avian HEV from chickens likely belongs to a separate genus (Bilic et al., 2009).
Hepatitis E is a recognized zoonotic disease, and several animal species such as domestic and wild pigs and deer can serve as reservoirs (Meng, 2010b; Pavio et al., 2010). Swine HEV is enzootic in domestic and wild pigs essentially in all swine-producing countries worldwide and has a very high incidence in swine herds (Bouwknegt et al., 2008; Casas et al., 2009; De Deus et al., 2008a, 2008b; Sonoda et al., 2004; Van der Poel et al., 2001). It has been reported that humans in regular contact with pigs such as veterinarians and other pig handlers appear to be at higher risk of HEV infection (Drobeniuc et al., 2001; Meng et al., 2003). Under experimental conditions, genotypes 3 and 4 strains of human HEV infected pigs, and conversely genotypes 3 and 4 strains of swine HEV infected non-human primates (Arankalle et al., 2006; Halbur et al., 2001; Meng et al., 1998). Strains of HEV recovered from human hepatitis E patients are genetically indistinguishable from swine HEV recovered from pig livers in grocery stores, and zoonotic human infections with genotypes 3 and 4 swine HEV through direct contact with infected animals or via consumption of undercooked or raw animal meats have also been reported (Colson et al., 2010; Rolfe et al., 2010; Takahashi et al., 2004; Yazaki et al., 2003).
Currently, there is very limited data regarding the cross-protection between different HEV strains belonging to different genotypes. The objective of this study was to evaluate the cross-protective ability of prior genotype 3 swine HEV infection in pigs against subsequent challenge with homologous swine HEV strain and heterologous human HEV strains of different genotypes.
2. Materials and Methods
2.1. Virus Stocks
The HEV infectious stocks used in this study were generated from previous studies, including a genotype 3 swine HEV with an infectious titer of 104.5 50% pig infectious dose (PID50) (Halbur et al., 2001; Meng et al., 1988), a genotype 3 U.S. strain of human HEV (Feagins et al., 2008; Halbur et al., 2001; Meng et al., 1988), and a genotype 4 Taiwanese strain of human HEV each diluted to approximately 3.4 × 108 genomic equivalent (GE) titer per ml of 10% fecal suspension in PBS buffer (Feagins et al., 2008; Wu et al., 2000). These virus stocks are known to be infectious when intravenously inoculated into pigs (Feagins et al., 2008; Halbur et al., 2001; Meng et al., 1988).
2.2. Experimental Design
Twenty-four, 8-week-old, cross-bred specific-pathogen-free (SPF) pigs that were tested negative for IgG anti-HEV were randomly divided into 4 groups of 6 pigs each. Pigs in each group were housed in separate rooms within a climate controlled Biosafety Level 2 (BSL-2) facility. A strict biosecurity protocol was followed for feeding and sample collection. Group 1 negative control pigs were inoculated intravenously (IV) with sterile PBS buffer and subsequently challenged with PBS buffer at 12 week post-inoculation (wpi). Pigs in groups 2, 3, and 4 were all inoculated IV with 104.5 PID50 of a genotype 3 strain of swine HEV, and subsequently challenged, at 12 wpi, with 3.4 × 108 GE titer of a genotype 3 US-2 strain of human HEV, 104.5 PID50 titer of the same genotype 3 strain of swine HEV used for initial inoculation, and 3.4 × 108 GE titer of a genotype 4 Taiwanese strain of human HEV, respectively. At 4 weeks post-challenge (wpc), all pigs were necropsied. Serum and fecal samples were collected prior to inoculation and weekly thereafter, and tested for the presence of HEV RNA by nested RT-PCR. The weekly serum samples were also tested for IgG anti-HEV with an ELISA.
2.3. ELISA for the detection of IgG anti-HEV
The ELISA was performed essentially as previously described (Feagins et al., 2007, 2008; Halbur et al., 2001; Meng et al., 1997, 1998) with the exception that the antigen used in this study was a recombinant protein expressed and purified from E.Coli, representing amino acids 452-617 of the HEV capsid protein (GenWay, Inc, San Diego, CA). The ELISA cutoff was set at 3 standard deviations above the mean OD values of the pre-inoculation samples (Feagins et al., 2007, 2008; Halbur et al., 2001; Meng et al., 1997, 1998).
2.4. Nested RT-PCR to detect HEV RNA in fecal and serum samples
The nested RT-PCR assays used for the detection of HEV RNA from serum and fecal samples were essentially the same as previously described (Feagins et al., 2007, 2008) with slight modifications. Strain-specific primers to amplify the ORF2 region of each virus were designed to detect the genotype 3 swine HEV, genotype 3 US-2 human HEV, and genotype 4 Taiwanese human HEV, respectively. The nested RT-PCR primer sequences for the detection of genotype 3 swine HEV are: first round, forward [5’-AGCTCCTGTACCTGATGTTGACTC-3’], reverse [5’-CTACAGAGCGCCAGCCTTGATTGC-3’]; second round, forward [5’-GCTCACGTCATCTGTCGCTGCTGG-3’], and reverse [5’-GGGCTGAACCAAAATCCTGACATC-3’]. The primer sequences for the detection of genotype 3 US-2 strain of human HEV include: first round, forward [5’-GTTGCTCTTCGTGCTTTTGCCTATG-3’], and reverse [5’-CCAGCGGGCGATAACGGATTGTAGC-3’]; second round, forward [5’-GGCGGTGGTTTCTGGGGTGAC-3’], and reverse [5’-GTACCCGAAGCGACAGATGACG-3’]. The primers for the detection of genotype 4 Taiwanese strain of human HEV consist of: first round, forward [5’-CGCAGGTTTATGGTGTTAGCC-3’], and reverse [5’-GTCGGGGTGGTGGTTGTTTGGG-3’]; second round, forward [5’-GCTCCTCCTGCTTTTGCCTATGC-3’], and reverse [5’-CGGTGGCGATGGTGGATGTGAGTG-3’].
Total RNAs were extracted with Trizol Reagent (Life Technologies, Carlsbad, CA) from 150 μl of serum or 10% fecal suspension. Reverse transcription and cDNA synthesis were performed at 42°C for 1 hr with 1 μl (10 μM) of the first round reverse primer specific for each virus, 1 μl (200 units/μl) of Superscript II reverse transcriptase (Life Technologies, Carlsbad, CA), 1 μl of 0.1M dithiothreitol, 4 μl of 5× RT buffer, 0.5 μl (40 units/μl) of RNasin ribonuclease inhibitor (Promega, Madison, WI), and 1 μl of 10 mM deoxynucleoside triphosphates. Nested PCR using AmpliTaq Gold DNA polymerase (Applied Biosystems, Carlsbad, CA) and genotype-specific primers was then performed on each sample [34]. The cycling parameters included an initial denaturation step at 95°C for 9 min, followed by 39 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 52°C, extension for 1 min at 72°C, and a final extension step at 72°C for 7min. The expected final PCR products of the amplified ORF2 gene region for genotype 3 swine HEV, genotype 3 human HEV, and genotype 4 human HEV are 680 bp, 779 bp, and 508 bp, respectively.
3. Results
3.1. Fecal virus shedding in pigs inoculated with genotype 3 swine HEV prior to challenge
Rectal temperature, body weight or feed intake was not recorded, since HEV infection in pigs does not cause clinical sign of diseases (Halbur et al., 2001). Three pigs in the negative control group 1 and one pig in group 4 died of over-eating prior to challenge, and were excluded from the study. Prior to challenge, the weekly fecal samples from each pig were tested for the presence of the genotype 3 swine HEV by a nested RT-PCR. Fecal virus shedding started at 1 wpi with five positive pigs in group 2, three positive pigs in group 3, and 2 positive pigs in group 4 (Table 1). Genotype 3 swine HEV RNA was thereafter detected in the feces of all pigs in groups 2, 3 and 4 prior to challenge at 12 wpi (Table 1). Fecal virus shedding was not detectable in any of the group 1 pigs inoculated with PBS buffer (Table 1).
Table 1.
Detection of fecal virus shedding by RT-PCR in pigs inoculated with a genotype 3 strain of swine HEV and subsequently challenged at 12 weeks post-inoculation (wpi) with different genotypes of human and swine HEV
| Groups | Pig ID# |
Inoculum | Weeks post-inoculation (wpi) | Challenge inoculum |
Weeks post- challenge (wpc) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 1 | 2 | 3 | 4 | ||||
| 1 | PBS buffer | PBS buffer | |||||||||||||||||
| 1737 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1739 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1746 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 2 |
Genotype 3
swine HEV |
Genotype 3
human HEV |
|||||||||||||||||
| 1644 | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | |||
| 1647 | − | + | + | − | + | + | + | + | + | − | − | − | − | − | − | − | |||
| 1738 | + | + | + | − | + | + | − | + | + | − | − | − | − | − | − | − | |||
| 1741 | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | |||
| 1744 | + | + | + | − | + | + | + | + | + | − | − | − | − | − | − | − | |||
| 1748 | + | + | + | − | + | + | + | + | + | − | − | − | − | − | − | − | |||
| 3 |
Genotype 3
swine HEV |
Genotype 3
swine HEV |
|||||||||||||||||
| 1645 | − | + | − | − | − | − | − | − | − | − | − | − | +* | − | − | − | |||
| 1648 | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1650 | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | +* | |||
| 1743 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1749 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1750 | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 4 |
Genotype 3
swine HEV |
Genotype 4
human HEV |
|||||||||||||||||
| 1640 | − | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||
| 1641 | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||
| 1646 | − | + | − | − | + | + | + | + | + | + | + | + | − | − | − | − | |||
| 1649 | − | + | − | + | + | + | + | + | + | + | + | + | +* | − | − | − | |||
| 1740 | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||
Positive for genotype 3 swine HEV RNA but negative for genotype 4 human HEV RNA
3.2. Viremia in pigs inoculated with genotype 3 swine HEV prior to challenge
During the 12 weeks prior to challenge, viremia was detected variably in 16 of 17 pigs (except for pig # 1644) inoculated with the genotype 3 swine HEV in groups 2, 3, and 4 (Table 2). Viremia began at 1 wpi in 4/6 pigs in group 2, 4/6 pigs in group 3, and 2/5 pigs in group 4 (Table 2). Negative control pigs in group 1 inoculated with PBS buffer remained negative (Table 2). The viremia in most pigs is transient lasting 1-3 weeks (Table 2), however some animals such as pigs #1738, #1744 and #1748 in group 2, and pig #1640 in group 4 had a prolonged period of viremia (Table 2).
Table 2.
Detection of viremia by RT-PCR in pigs inoculated with a genotype 3 strain of swine HEV and subsequently challenged at 12 weeks post-inoculation (wpi) with different genotypes of human and swine HEV
| Groups | Pig ID# |
Inoculum | Weeks post-inoculation (wpi) | Challenge inoculum |
Weeks post- challenge (wpc) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 1 | 2 | 3 | 4 | ||||
| 1 | PBS buffer | PBS buffer | |||||||||||||||||
| 1737 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1739 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1746 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 2 |
Genotype 3
swine HEV |
Genotype 3
human HEV |
|||||||||||||||||
| 1644 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1647 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1738 | − | + | + | + | + | − | + | + | + | − | − | − | − | − | − | − | |||
| 741 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1744 | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | |||
| 1748 | + | + | + | + | + | + | − | − | + | + | − | − | − | − | − | − | |||
| 3 |
Genotype 3
swine HEV |
Genotype 3
swine HEV |
|||||||||||||||||
| 1645 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1648 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1650 | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1743 | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1749 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1750 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 4 |
Genotype 3
swine HEV |
Genotype 4
human HEV |
|||||||||||||||||
| 1640 | + | − | + | + | + | + | + | + | + | + | + | + | − | − | − | − | |||
| 1641 | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1646 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 1649 | − | + | + | − | + | + | − | − | − | − | − | − | − | − | − | − | |||
| 1740 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |||
3.3. Seroconversion to IgG anti-HEV in pigs inoculated with genotype 3 swine HEV prior to challenge
Seroconversion to IgG anti-HEV started at 1 wpi in some of the groups 2 and 4 pigs, and at 2 wpi in some of the group 3 pigs (Fig. 1). During the first 12 weeks prior to challenge, pigs in all three experimental groups seroconverted to IgG anti-HEV antibody except for pig 1645 in group 3 and pig #1740 in group 4 (Fig. 1). Negative control pigs (ID# 1739 and #1746) remained seronegative throughout the study, although pig #1737 had an elevated OD reading that was at the borderline of the cutoff (Fig. 1). Serum and fecal samples from pig#1737 are tested negative by RT-PCR for HEV RNA throughout the study.
Figure 1.
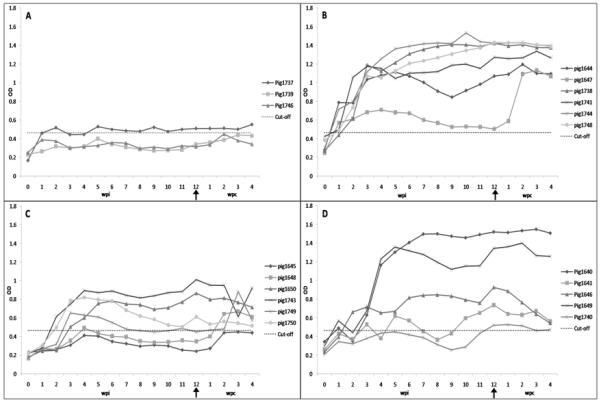
IgG anti-HEV response in pigs inoculated with a genotype 3 swine HEV and subsequently challenged, at 12 weeks post-inoculation (wpi), with different strains of human and swine HEV. A: Pigs inoculated and challenged with PBS buffer. B: Pigs inoculated with a genotype 3 swine HEV and challenged at 12 wpi with a genotype 3 human HEV (US-2 strain). C: Pigs inoculated with a genotype 3 swine HEV and challenged at 12 wpi with the same genotype 3 swine HEV. D: Pigs inoculated with a genotype 3 swine HEV and challenged at 12 wpi with a genotype 4 human HEV (Taiwanese strain). Arrow in the X-axis indicates the time of challenge at 12 wpi. All pig were necropsied at 4 weeks post-challenge (wpc)
3.4. Protection and cross-protection of HEV infections in pigs challenged with homologous and heterologous genotypes of HEV
At 12 wpi, pigs in groups 1, 2, 3, and 4 were challenged with sterile PBS buffer (control), a genotype 3 US-2 strain of human HEV, the same genotype 3 strain of swine HEV as used in the inoculum, and a genotype 4 Taiwanese strain of human HEV, respectively. After challenge, weekly serum and fecal samples were collected from each pig, and all pigs were necropsed at 4 wpc. Fecal virus shedding and viremia were tested by nested RT-PCR assays specific for each challenge strain of HEV. At no time point after challenge, was there any HEV RNA detected in the sera of pigs in any group (Table 2). Likewise, HEV RNA specific to the challenge virus was not detected in the feces of pigs in group 2 or group 4. Pig 1649 in group 4 continued to shed genotype 3 swine HEV from the original inoculation as confirmed by sequence analysis in feces for one week after challenge (Table 1). In group 3 pigs challenged with the homologous same genotype 3 strain of swine HEV, pig 1645 shed the same genotype 3 swine HEV in feces at 1 wpc while pig 1650 shed the same genotype 3 swine HEV in feces at 4 wpc (Table 1). After challenge, the levels of IgG anti-HEV antibodies maintained in similar levels with a slight decrease in titers in some pigs towards the end of the study. A booster effect on IgG anti-HEV level after challenge was not evident, which is expected since the prior infection protected against the challenge (Fig. 1). As expected, viremia, fecal virus shedding and seroconversion were not detected in the negative control pigs in group 1 (Fig. 1).
4. Discussion
A vaccine against HEV has not yet been available, largely due to the inability to efficiently propagate HEV in cell culture. The recent adaptation of several strains of HEV to replicate more efficiently in established cell lines (Okamoto, 2011; Shukla et al., 2011) may aid the future development of an affordable HEV vaccine. The current experimental HEV vaccines, which are all based on the recombinant ORF2 capsid protein of a single strain of HEV, are promising in clinical trials (Arankalle et al., 2009; Shrestha et al., 2007; Zhu et al., 2010), however their efficacies against the diversified field strains of HEV from different genotypes, especially against the emerging and zoonotic strains of HEV, need to be evaluated. In a given geographic region, multiple genotypes of HEV are co-circulating in the same population. For example, genotypes 1, 3 and 4 HEV strains are all circulating in China (Zhang et al., 2010). Thus, it is possible that an individual may be exposed to more than one genotype of HEV. It will be important to know if prior exposure to one genotype will induce sufficient protective immunity against subsequent exposure to a different genotype. In addition, the high prevalence of swine HEV infection in pigs increases the risk of zoonotic human infections by swine HEV (Meng et al., 2003). For example, individuals from the major swine-producing state of Minnesota are at least 5 times more likely to be seropositive for IgG anti-HEV than individuals from traditionally non-major swine state such as Alabama (Meng et al., 2003). It is unclear if the individuals infected by the zoonotic swine HEV will confer protection against subsequent exposure to human strains of HEV of a different genotype. This study was designed to partially address these questions by evaluating the cross-protection of HEV infection in pigs inoculated with a genotype 3 swine HEV and then subsequently challenged with homologous genotype 3 swine HEV and heterologous human HEV strains from a different genotype.
All experimental pigs inoculated with the genotype 3 swine HEV became infected as evidenced by seroconversion to IgG anti-HEV (Fig. 1), fecal virus shedding (Table 1) and viremia (Table 2). Consistent with previous infection studies in pigs, the inoculated animals remained clinically healthy but began to shed virus in feces and become viremic within the first two weeks after inoculation. As was the case in previous studies, pig-to-pig variations in the onset and duration of fecal virus shedding, viremia and seroconversion were observed in inoculated pigs. Some inoculated animals such as pigs #1738 and #1748 in group 2 and pigs #1640 and #1649 in group 4 had extended fecal virus shedding and prolonged viremia. The exact reason for the observed long period of fecal virus shedding and viremia is not known, but similar observations have been reported in some of HEV-infected chickens and pigs in our previous studies (Billam et al., 2005; Feagins et al., 2007, 2008). Also, cases of chronic HEV infections and prolonged virus shedding have been reported in transplant patients as well HIV-infected individuals (Legrand-Abravanel et al., 2010; Renou et al., 2010). In addition, long-term shedding of genotype 3 swine HEV for up to 12 weeks in naturally-infected pigs has also been reported (Kanai et al., 2010). It is possible that pigs infected by HEV may develop persistent infection more often than what we originally thought. While this study is not designed to determine the reason for the observed pig-to-pig variation in virus shedding or viremia, it is possible that some behavioral characteristics such as stress or altered sleep patterns in a certain group or animal may affect the course of HEV infection. There is also a possibility that some animals with extended fecal virus shedding and viremia may become re-infected before the animal mounts an effective immune response from the initial infection.
Importantly for this study, pigs inoculated with the genotype 3 swine HEV seroconverted to IgG anti-HEV antibodies (Fig. 1). Pig-to-pig variations in IgG anti-HEV antibody response are evident within each group. A few pigs such as pig #1645 in group 3 and pig #1740 in group 4 had a very low level of IgG anti-HEV response below the cutoff before challenge. However, overall the majority of pigs inoculated with swine HEV developed IgG anti-HEV prior to challenge at 12 wpi, and the results indicated that pigs inoculated with the swine HEV became infected and elicited humoral immune response prior to challenge.
At 12 wpi, the pigs were challenged with two different genotypes of human HEV (genotypes 3 and 4) as well as with the homologous same genotype 3 strain of swine HEV. After challenge, fecal virus shedding and viremia were undetectable in the pigs that had been previously infected with the genotype 3 swine HEV. Two pigs in group 3 (ID# 1645, # 1650) and one pig in group 4 (ID# 1649) had transient fecal virus shedding of one week after challenge, and subsequent sequence analyses of the excreted viruses recovered from the fecal samples of the three pigs indicated that the virus belongs to the genotype 3 swine HEV from the original inoculation. Thus, the transient one-week fecal virus shedding after challenge in pig #1649 in group 4 is due to the persistent infection of the genotype 3 swine HEV from the original inoculation rather than from the challenge genotype 4 human HEV. However, the source of genotype 3 swine HEV detected after challenge in pigs #1645 and #1650 in group 3 could not be determined since the pigs in this group were challenged with the same genotype 3 swine HEV. It is possible that the transient one-week fecal virus shedding in these two pigs may be due to incomplete protection against the homologous challenge virus, since fecal virus shedding from the initial inoculation were fully cleared in both pigs by 6 weeks post-inoculation. Overall, the results from this study clearly demonstrated that prior infection with the genotype 3 swine HEV prevented pigs from developing viremia and fecal virus shedding upon challenges with homologous swine HEV and heterologous human HEV strains. Our result is consistent with a non-human primate study in which rhesus macaques initially infected with genotypes 1 or 4 human HEV are protected from subsequent challenge with heterologous genotypes of HEV (Huang et al., 2008). The results from this study suggest that a vaccine derived from one genotype of HEV or from an animal strain of HEV is sufficient to elicit protective immunity against other HEV genotypes, and this finding is important when designing future HEV vaccine strategies.
Acknowledgements
We thank Dr. Suzanne U. Emerson and Dr. Robert H. Purcell from the National Institutes of Health, Bethesda, MD for providing the US-2 strain of genotype 3 human HEV and the Taiwanese strain of genotype 4 human HEV. This study is supported by grants from the National Institutes of Health (R01 AI074667 and R01 AI050611).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aikawa T, Kojima M, Takahashi M, Nishizawa T, Okamoto H. Identification of indigenous hepatitis E virus from a Japanese patient who contracted sporadic acute hepatitis in 1982. J. Infect. Dis. 2002;18(10):1535–1536. doi: 10.1086/344349. [DOI] [PubMed] [Google Scholar]
- Arankalle VA, Chobe LP, Chadha MS. Type-IV Indian swine HEV infects rhesus monkeys. J. Viral. Hepat. 2006;13(11):742–745. doi: 10.1111/j.1365-2893.2006.00759.x. [DOI] [PubMed] [Google Scholar]
- Arankalle VA, Lole KS, Deshmukh TM, Srivastava S, Shaligram US. Challenge studies in Rhesus monkeys immunized with candidate hepatitis E vaccines: DNA, DNA-prime-protein-boost and DNA-protein encapsulated in liposomes. Vaccine. 2009;27(7):1032–1039. doi: 10.1016/j.vaccine.2008.11.097. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Singhal A, Panda SK, Acharya SK. A 20-year single-center experience with acute liver failure during pregnancy: is the prognosis really worse? Hepatology. 2008;48(5):1577–1585. doi: 10.1002/hep.22493. [DOI] [PubMed] [Google Scholar]
- Bilic I, Jaskulska B, Basic A, Morrow CJ, Hess M. Sequence analysis and comparison of avian hepatitis E viruses from Australia and Europe indicate the existence of different genotypes. J. Gen. Virol. 2009;90(Pt 4):863–873. doi: 10.1099/vir.0.007179-0. [DOI] [PubMed] [Google Scholar]
- Billam P, Huang FF, Sun ZF, Pierson FW, Duncan RB, Elvinger F, Guenette DK, Toth TE, Meng XJ. Systematic pathogenesis and replication of avian hepatitis E virus in specific-pathogen-free adult chickens. J. Virol. 2005;79(6):3429–3437. doi: 10.1128/JVI.79.6.3429-3437.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwknegt M, Frankena K, Rutjes SA, Wellenberg GJ, de Roda Husman AM, van der Poel WH, de Jong MC. Estimation of hepatitis E virus transmission among pigs due to contact-exposure. Vet. Res. 2008;39(5):40. doi: 10.1051/vetres:2008017. [DOI] [PubMed] [Google Scholar]
- Casas M, Pujols J, Rosell R, de Deus N, Peralta B, Pina S, Casal J, Martín M. Retrospective serological study on hepatitis E infection in pigs from 1985 to 1997 in Spain. Vet. Microbiol. 2009;135(3-4):248–252. doi: 10.1016/j.vetmic.2008.09.075. [DOI] [PubMed] [Google Scholar]
- Colson P, Borentain P, Queyriaux B, Kaba M, Moal V, Gallian P, Heyries L, Raoult D, Gerolami R. Pig liver sausage as a source of hepatitis E virus transmission to humans. J. Infect. Dis. 2010;202(6):825–834. doi: 10.1086/655898. [DOI] [PubMed] [Google Scholar]
- de Deus N, Casas M, Peralta B, Nofrarías M, Pina S, Martín M, Segalés J. Hepatitis E virus infection dynamics and organic distribution in naturally infected pigs in a farrow-to-finish farm. Vet. Microbiol. 2008a;132(1-2):19–28. doi: 10.1016/j.vetmic.2008.04.036. [DOI] [PubMed] [Google Scholar]
- de Deus N, Peralta B, Pina S, Allepuz A, Mateu E, Vidal D, Ruiz-Fons F, Martín M, Gortázar C, Segalés J. Epidemiological study of hepatitis E virus infection in European wild boars (Sus scrofa) in Spain. Vet. Microbiol. 2008b;129(1-2):163–170. doi: 10.1016/j.vetmic.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Drobeniuc J, Favorov MO, Shapiro CN, Bell BP, Mast EE, Dadu A, Culver D, Iarovoi P, Robertson BH, Margolis HS. Hepatitis E virus antibody prevalence among persons who work with swine. J. Infect. Dis. 2001;184(12):1594–1597. doi: 10.1086/324566. [DOI] [PubMed] [Google Scholar]
- Emerson SU, Purcell RH. Hepatitis E virus. Rev. Med. Virol. 2003;13(3):145–54. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- Feagins AR, Opriessnig T, Guenette DK, Halbur PG, Meng XJ. Detection and characterization of infectious Hepatitis E virus from commercial pig livers sold in local grocery stores in the USA. J. Gen. Virol. 2007;88(Pt 3):912–917. doi: 10.1099/vir.0.82613-0. [DOI] [PubMed] [Google Scholar]
- Feagins AR, Opriessnig T, Huang YW, Halbur PG, Meng XJ. Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J. Med. Virol. 2008;80(8):1379–1386. doi: 10.1002/jmv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqshenas G, Shivaprasad HL, Woolcock PR, Read DH, Meng XJ. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 2001;82(Pt 10):2449–1462. doi: 10.1099/0022-1317-82-10-2449. [DOI] [PubMed] [Google Scholar]
- Haqshenas G, Huang FF, Fenaux M, Guenette DK, Pierson FW, Larsen CT, Shivaprasad HL, Toth TE, Meng XJ. The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. J. Gen. Virol. 2002;83(Pt 9):2201–2209. doi: 10.1099/0022-1317-83-9-2201. [DOI] [PubMed] [Google Scholar]
- Halbur PG, Kasorndorkbua C, Gilbert C, Guenette D, Potters MB, Purcell RH, Emerson SU, Toth TE, Meng XJ. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 2001;39(3):918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang H, Harrison TJ, Lang S, Huang G, Wang Y. Cross-protection of hepatitis E virus genotypes 1 and 4 in rhesus macaques. J. Med. Virol. 2008;80(5):824–832. doi: 10.1002/jmv.21140. [DOI] [PubMed] [Google Scholar]
- Jameel S. Molecular biology and pathogenesis of hepatitis E virus. Expert Rev. Mol. Med. 1999:1–16. doi: 10.1017/S1462399499001271. 1999. [DOI] [PubMed] [Google Scholar]
- Jilani N, Das BC, Husain SA, Baweja UK, Chattopadhya D, Gupta RK, Sardana S, Kar P. Hepatitis E virus infection and fulminant hepatic failure during pregnancy. J. Gastroenterol. Hepatol. 2007;22(5):676–682. doi: 10.1111/j.1440-1746.2007.04913.x. [DOI] [PubMed] [Google Scholar]
- Johne R, Plenge-Bönig A, Hess M, Ulrich RG, Reetz J, Schielke A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol. 2010;91(Pt 3):750–758. doi: 10.1099/vir.0.016584-0. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Tsujikawa M, Yunoki M, Nishiyama S, Ikuta K, Hagiwara K. Long-term shedding of hepatitis E virus in the feces of pigs infected naturally, born to sows with and without maternal antibodies. J. Med. Virol. 2010;82(1):69–76. doi: 10.1002/jmv.21647. [DOI] [PubMed] [Google Scholar]
- Legrand-Abravanel F, Kamar N, Sandres-Saune K, Garrouste C, Dubois M, Mansuy JM, Muscari F, Sallusto F, Rostaing L, Izopet J. Characteristics of autochthonous hepatitis E virus infection in solid-organ transplant recipients in France. J. Infect. Dis. 2010;202(6):835–844. doi: 10.1086/655899. [DOI] [PubMed] [Google Scholar]
- Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, Haynes JS, Thacker BJ, Emerson SU. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA. 1997;94(18):9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ, Halbur PG, Shapiro MS, Govindarajan S, Bruna JD, Mushahwar IK, Purcell RH, Emerson SU. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J .Virol. 1998;72(12):9714–9721. doi: 10.1128/jvi.72.12.9714-9721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ, Wiseman B, Elvinger F, Guenette DK, Toth TE, Engle RE, Emerson SU, Purcell RH. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J. Clin. Microbiol. 2003;40(1):117–122. doi: 10.1128/JCM.40.1.117-122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 2010a;140(3-4):256–265. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ. Recent advances in Hepatitis E virus. J. Viral Hepat. 2010b;17(3):153–161. doi: 10.1111/j.1365-2893.2009.01257.x. [DOI] [PubMed] [Google Scholar]
- Mizuo H, Yazaki Y, Sugawara K, Tsuda F, Takahashi M, Nishizawa T, Okamoto H. Possible risk factors for the transmission of hepatitis E virus and for the severe form of hepatitis E acquired locally in Hokkaido, Japan. J. Med. Virol. 2005;76(3):341–349. doi: 10.1002/jmv.20364. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Takahashi K, Taira K, Taira M, Ohno A, Sakugawa H, Arai M, Mishiro S. Hepatitis E virus infection in wild mongooses of Okinawa, Japan: Demonstration of anti-HEV antibodies and a full-genome nucleotide sequence. Hepatol. Res. 2006;34(3):137–140. doi: 10.1016/j.hepres.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Okamoto H. Genetic variability and evolution of hepatitis E virus. Virus Res. 2007;127(2):216–228. doi: 10.1016/j.virusres.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Okamoto H. Hepatitis E virus cell culture models. Virus Res. 2011 Feb 10; doi: 10.1016/j.virusres.2011.01.015. 2011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Pavio N, Meng XJ, Renou C. Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet. Res. 2010;41(6):46. doi: 10.1051/vetres/2010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J. Hepatol. 2008;48(3):494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Renou C, Lafeuillade A, Cadranel JF, Pavio N, Pariente A, Allègre T, Poggi C, Pénaranda G, Cordier F, Nicand E. Hepatitis E virus in HIV-infected patients. AIDS. 2010;24(10):1493–1499. doi: 10.1097/QAD.0b013e32833a29ab. [DOI] [PubMed] [Google Scholar]
- Rolfe KJ, Curran MD, Mangrolia N, Gelson W, Alexander GJ, L′estrange M, Vivek R, Tedder R, Ijaz S. First case of genotype 4 human hepatitis E virus infection acquired in India. J. Clin. Virol. 2010;48(1):58–61. doi: 10.1016/j.jcv.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Shrestha MP, Scott RM, Joshi DM, Mammen MP, Jr, Thapa GB, Thapa N, Myint KS, Fourneau M, Kuschner RA, Shrestha SK, David MP, Seriwatana J, Vaughn DW, Safary A, Endy TP, Innis BL. Safety and efficacy of a recombinant hepatitis E vaccine. N. Engl. J. Med. 2007;356(9):895–903. doi: 10.1056/NEJMoa061847. [DOI] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. USA. 2011;108(6):2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda H, Abe M, Sugimoto T, Sato Y, Bando M, Fukui E, Mizuo H, Takahashi M, Nishizawa T, Okamoto H. Prevalence of hepatitis E virus (HEV) Infection in wild boars and deer and genetic identification of a genotype 3 HEV from a boar in Japan. J. Clin. Microbiol. 2004;42(11):5371–5374. doi: 10.1128/JCM.42.11.5371-5374.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Kitajima N, Abe N, Mishiro S. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology. 2004;330(2):501–505. doi: 10.1016/j.virol.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Nishizawa T, Sato H, Sato Y, Jirintai D, Nagashima S, Okamoto H. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J. Gen. Virol. 2011 Jan 12; doi: 10.1099/vir.0.029470-0. 2011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Tei S, Kitajima N, Takahashi K, Mishiro S. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet. 2003;362(9381):371–373. doi: 10.1016/S0140-6736(03)14025-1. [DOI] [PubMed] [Google Scholar]
- van der Poel WH, Verschoor F, van der Heide R, Herrera MI, Vivo A, Kooreman M, de Roda Husman AM. Hepatitis E virus sequences in swine related to sequences in humans, The Netherlands. Emerg. Infect. Dis. 2001;7(6):970–976. doi: 10.3201/eid0706.010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JC, Chen CM, Chiang TY, Sheen IJ, Chen JY, Tsai WH, Huang YH, Lee SD. Clinical and epidemiological implications of swine hepatitis E virus infection. J. Med. Virol. 2000;60(2):166–171. [PubMed] [Google Scholar]
- Yazaki Y, Mizuo H, Takahashi M, Nishizawa T, Sasaki N, Gotanda Y, Okamoto H. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 2003;84(Pt 9):2351–2357. doi: 10.1099/vir.0.19242-0. [DOI] [PubMed] [Google Scholar]
- Zhang W, He Y, Wang H, Shen Q, Cui L, Wang X, Shao S, Hua X. Hepatitis E virus genotype diversity in eastern China. Emerg. Infect. Dis. 2010;16(10):1630–1632. doi: 10.3201/eid1610.100873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, Wang Y. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J. Med. Virol. 2009;81(8):1371–1379. doi: 10.1002/jmv.21536. [DOI] [PubMed] [Google Scholar]
- Zhu FC, Zhang J, Zhang XF, Zhou C, Wang ZZ, Huang SJ, Wang H, Yang CL, Jiang HM, Cai JP, Wang YJ, Ai X, Hu YM, Tang Q, Yao X, Yan Q, Xian YL, Wu T, Li YM, Miao J, Ng MH, Shih JW, Xia NS. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet. 2010;376(9744):895–902. doi: 10.1016/S0140-6736(10)61030-6. [DOI] [PubMed] [Google Scholar]


