Abstract
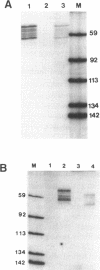
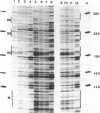
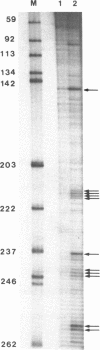
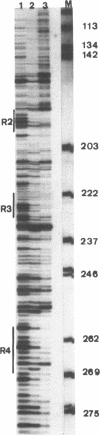
Two distinct regions in the replication origin, oriC, of Escherichia coli are separately distorted upon initiation complex formation by the initiator protein DnaA. The AT-rich region in the left part of oriC and the start site region in the right part of oriC. Chemical modification of single-stranded DNA was observed at both regions whereas endonuclease recognition of DNA mini-bulges specifically occurred in the start site region. We show that the helical phasing of binding sites for DnaA protein in oriC is important for origin function. An insertion or deletion of one helical turn between the two rightmost binding sites does not alter the efficiency of replication initiation, whereas all modifications of distance by less or more than one helical turn result in inactivation of oriC. DnaA binding and helical distortions in the AT-rich region as well as in the start site region are not affected in the distance mutants irrespective of their functionality in vivo. We propose a specific compact nucleoprotein structure for the initiation complex.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asada K., Sugimoto K., Oka A., Takanami M., Hirota Y. Structure of replication origin of the Escherichia coli K-12 chromosome: the presence of spacer sequences in the ori region carrying information for autonomous replication. Nucleic Acids Res. 1982 Jun 25;10(12):3745–3754. doi: 10.1093/nar/10.12.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., von Kitzing E., Diekmann S., Kemper B., Lilley D. M. Model for the interaction of DNA junctions and resolving enzymes. J Mol Biol. 1991 Oct 20;221(4):1191–1207. doi: 10.1016/0022-2836(91)90928-y. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988 Oct;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Buhk H. J., Messer W. The replication origin region of Escherichia coli: nucleotide sequence and functional units. Gene. 1983 Oct;24(2-3):265–279. doi: 10.1016/0378-1119(83)90087-2. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. Structural junctions in DNA: the influence of flanking sequence on nuclease digestion specificities. Nucleic Acids Res. 1985 Jun 25;13(12):4445–4467. doi: 10.1093/nar/13.12.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., Roll J. The requirement of IHF protein for extrachromosomal replication of the Escherichia coli oriC in a mutant deficient in DNA polymerase I activity. New Biol. 1990 Sep;2(9):818–827. [PubMed] [Google Scholar]
- Filutowicz M., Ross W., Wild J., Gourse R. L. Involvement of Fis protein in replication of the Escherichia coli chromosome. J Bacteriol. 1992 Jan;174(2):398–407. doi: 10.1128/jb.174.2.398-407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kaguni J. M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987 Jul 25;262(21):10327–10334. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H., Egan J. B., Roth A., Messer W. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 1991 Aug 11;19(15):4167–4172. doi: 10.1093/nar/19.15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H., Messer W. Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 1991 Jun;10(6):1579–1584. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz A., Schaefer C., Gille H., Jueterbock W. R., Messer W. Mutations in the DnaA binding sites of the replication origin of Escherichia coli. Mol Gen Genet. 1992 May;233(1-2):81–88. doi: 10.1007/BF00587564. [DOI] [PubMed] [Google Scholar]
- Hwang D. S., Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem. 1992 Nov 15;267(32):23083–23086. [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Eddy M. J. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989 Dec 20;8(13):4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M., Oka A., Takanami M., Yasuda S., Hirota Y. Sites of dnaA protein-binding in the replication origin of the Escherichia coli K-12 chromosome. J Mol Biol. 1985 Aug 5;184(3):529–533. doi: 10.1016/0022-2836(85)90299-2. [DOI] [PubMed] [Google Scholar]
- Messer W., Egan B., Gille H., Holz A., Schaefer C., Woelker B. The complex of oriC DNA with the DnaA initiator protein. Res Microbiol. 1991 Feb-Apr;142(2-3):119–125. doi: 10.1016/0923-2508(91)90018-6. [DOI] [PubMed] [Google Scholar]
- Messer W., Hartmann-Kühlein H., Langer U., Mahlow E., Roth A., Schaper S., Urmoneit B., Woelker B. The complex for replication initiation of Escherichia coli. Chromosoma. 1992;102(1 Suppl):S1–S6. doi: 10.1007/BF02451779. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Oka A., Sasaki H., Sugimoto K., Takanami M. Sequence organization of replication origin of the Escherichia coli K-12 chromosome. J Mol Biol. 1984 Jul 15;176(4):443–458. doi: 10.1016/0022-2836(84)90171-2. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugimoto K., Takanami M., Hirota Y. Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol Gen Genet. 1980 Apr;178(1):9–20. doi: 10.1007/BF00267207. [DOI] [PubMed] [Google Scholar]
- Parsons R., Tegtmeyer P. Spacing is crucial for coordination of domain functions within the simian virus 40 core origin of replication. J Virol. 1992 Apr;66(4):1933–1942. doi: 10.1128/jvi.66.4.1933-1942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaczek P. Bending of the origin of replication of E. coli by binding of IHF at a specific site. New Biol. 1990 Mar;2(3):265–271. [PubMed] [Google Scholar]
- Pottmeyer S., Kemper B. T4 endonuclease VII resolves cruciform DNA with nick and counter-nick and its activity is directed by local nucleotide sequence. J Mol Biol. 1992 Feb 5;223(3):607–615. doi: 10.1016/0022-2836(92)90977-r. [DOI] [PubMed] [Google Scholar]
- Pottmeyer S., Kemper B. T4 endonuclease VII resolves cruciform DNA with nick and counter-nick and its activity is directed by local nucleotide sequence. J Mol Biol. 1992 Feb 5;223(3):607–615. doi: 10.1016/0022-2836(92)90977-r. [DOI] [PubMed] [Google Scholar]
- Quiñones A., Jüterbock W. R., Messer W. Expression of the dnaA gene of Escherichia coli is inducible by DNA damage. Mol Gen Genet. 1991 May;227(1):9–16. doi: 10.1007/BF00260699. [DOI] [PubMed] [Google Scholar]
- Samitt C. E., Hansen F. G., Miller J. F., Schaechter M. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989 Mar;8(3):989–993. doi: 10.1002/j.1460-2075.1989.tb03462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Schauzu M. A., Kücherer C., Kölling R., Messer W., Lother H. Transcripts within the replication origin, oriC, of Escherichia coli. Nucleic Acids Res. 1987 Mar 25;15(6):2479–2497. doi: 10.1093/nar/15.6.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu K., Yung B. Y., Kornberg A. The dnaA protein of Escherichia coli. Abundance, improved purification, and membrane binding. J Biol Chem. 1988 May 25;263(15):7136–7140. [PubMed] [Google Scholar]
- Seufert W., Messer W. Initiation of Escherichia coli minichromosome replication at oriC and at protein n' recognition sites. Two modes for initiating DNA synthesis in vitro. EMBO J. 1986 Dec 1;5(12):3401–3406. doi: 10.1002/j.1460-2075.1986.tb04656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., Messer W. Start sites for bidirectional in vitro DNA replication inside the replication origin, oriC, of Escherichia coli. EMBO J. 1987 Aug;6(8):2469–2472. doi: 10.1002/j.1460-2075.1987.tb02527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaro P. C., Birkenkamp K., Pfeiffer P., Kemper B. Endonuclease VII of phage T4 triggers mismatch correction in vitro. J Mol Biol. 1993 Apr 5;230(3):868–877. doi: 10.1006/jmbi.1993.1207. [DOI] [PubMed] [Google Scholar]
- Yung B. Y., Kornberg A. The dnaA initiator protein binds separate domains in the replication origin of Escherichia coli. J Biol Chem. 1989 Apr 15;264(11):6146–6150. [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]