Abstract
Objective
Certain class II alleles are associated with susceptibility to develop arthritis. However, some individuals carrying non-RA associated alleles develop arthritis is still unexplained. An individual heterozygous for both the DQA1 and DQB1 genes, can express the DQ molecule in cis or trans heterodimers. In a cis-heterodimer the alpha chain interacts with the beta chain coded by the same chromosome, while in a trans-heterodimer it interacts with the beta chain on the other chromosome. In this study we tried to find out if trans-heterodimer of 2 non-associated alleles, DQB1*0601 and DQB1*0604, can predispose to arthritis using a humanized mouse model of arthritis.
Methods
DQB1*0601 and *0604 occur in linkage with DQA1*0103 and *0102 respectively. To understand the role of trans-heterodimer, we generated DQB1*0604/DQA1* 0103 transgenic mice lacking endogenous class II molecules.
Results
The DQB1*0604/A1*0103 mice developed severe arthritis and in vitro generated antigen-specific response. The DQB1*0604/DQA1*0103 could present type II collagen (CII)-derived peptides that are not presented by arthritis- resistant DQB1*0601 allele, suggesting that trans-heterodimer molecules between two DQB1 and DQA1 molecules may result in presentation of unique antigens and susceptibility to develop arthritis. Molecular modeling of the CII peptides showed that DQB1*0604/DQA1*0103 shares p4 pocket with arthritis-susceptible DQB1*0302 allele and further a critical role of p4 and p9 pockets is suggested with susceptibility to arthritis.
Conclusions
The present data provides a possible explanation for the parental inheritance of non-susceptible alleles in some patients with rheumatoid arthritis and a mechanism by which they can predispose to develop arthritis.
Rheumatoid arthritis (RA) is an autoimmune disease of unknown etiology that leads to destruction of the joints. The presence of certain Human leukocyte antigen (HLA) class II alleles predispose to develop arthritis. Most of the studies in various ethnic populations show an association between RA and DRB1 alleles sharing the 3rd hypervariable region 67–74 with the DRB1*0401 molecule while DRB1*0402 and DR2 have been shown to be protective. Even though there is no clear cut association with DQ alleles in RA, DQB1*0301 and DQB1*0302 that occur in linkage with DRB1*0401 have been associated with arthritis in various populations (1, 2) while DQB1*0601 in linkage with DR2 is protective (3). In humans it is difficult to interpret the association as there is a strong linkage between DR and DQ loci. In arthritis, heterozygous genotype such as DRB1*01-DQB1*05/DRB1*04-DQB1*03 have higher relative risk of disease compared to DRB1*04-DQB1*03 homozygous genotypes indicating an epistatic or synergistic effect. This suggests that the relative risk of HLA genotypes is not the additive effect of the single haplotype relative risks. Similarly in diabetes, DRB1*03-DQB1*02/DRB1*04-DQB1*03 shows a higher risk than individual homozygous genotype (4). The human HLA-DQ molecule constitutes of two polymorphic chains, DQα and DQβ. An individual heterozygous for both the DQA1 (α) and DQB1 (β) genes, can express DQ molecule in cis or trans heterodimers. In a cis heterodimer α chain interacts with the β chain coded by the same chromosome, while in a trans heterodimer it interacts with the β chain on the other chromosome (5). Epistatic effect of highly susceptible trans-coded DQ heterodimers has been hypothesized for predisposition to type 1 diabetes and celiac disease (6, 7). Genetic predisposition to RA is mainly focused on the presence or absence of shared epitope in humans, although animal models using HLA-DQ transgenic mice have brought into focus the role of DQ alleles in arthritis (8–11).
Collagen induced arthritis (CIA) is an experimental model of autoimmune inflammatory polyarthritis sharing clinical and pathological features to RA (11, 12). Our previous studies have reported that Aβo transgenic mice expressing HLA-DQA1*0301/DQB1*0302 (DQ8) are highly susceptible to CIA (9–11) and can present multiple human type II collagen (CII) derived peptides (13). On the other hand, DQA1*0103\DQB1*0601 (DQ6) mice are resistant to develop arthritis (8). It has been hypothesized that both alpha and beta chain influence peptide binding. DQB1*0604 occurs in linkage with DQA1*0102. To determine the role of trans-heterodimer DQ molecules in arthritis, we generated transgenic mice expressing DQB1*0604/DQA1*0103 molecules. Our observations show that DQB1*0604/DQA1*0103 predisposes transgenic mice to develop CIA. Our studies provide a possible explanation for the parental inheritance of non-susceptible alleles in some patients and mechanism by which they can predispose to develop arthritis.
MATERIAL AND METHODS
Mice
Transgenic mice expressing functional HLA-DQ8 (DQA1*0301/DQB1*0302) and DQ6 (DQB1*0601/DQA1*0103) molecules were generated as described before (8, 9). Cosmids pPKQ5121 and pAKQ 4116 containing DQA1*0103 and DQB1*0604 gene respectively were a kind gift of Dr. H. Inoko. A 40-kb construct containing DQB1*0604, was microinjected into FVB (F1) embryos, producing DQ6.4β transgenic mice, and a construct containing the DQA1*0103 gene was microinjected into (B6/J X B10.M)F2 embryos, resulting in B10.M-DQ6α transgenic mice. These mice were bred on FVB background. Each strain was then bred onto a mouse class II–deficient (Aβo) background. The DQ6a (Aβo) and DQ6.4β (Aβo) mice were then mated to obtain a DQ*0604 αβ strain (DQ6.4). The Aβo mice express H2-Aα and H2-Eβ in the cytoplasm, but these chains do not contribute to the expression of functional heterodimers on the cell surface (14). For convenience DQB1*0604/DQA1*0103 mice are referred to as DQ6.4 and DQB1*0601/DQA1*0103 as DQ6.1 in the entire manuscript.
For all the groups, parental mice and negative littermates were included as controls. Mice of both sexes (8–12 weeks of age) used in this study were bred and maintained in the pathogen free Immunogenetics Mouse Colony of Mayo Clinic. All experimental procedures were done with the approval of the Institutional Animal Use and Care Committee.
Induction and evaluation of CIA
Pure native chick type II collagen (CII) was obtained by multiple step purification described previously (15). Eight to twelve weeks old transgenic mice along with negative littermates were immunized with 100 μg chick CII emulsified 1:1 with complete Freund’s adjuvant H37Ra (Difco Laboratories, Detroit, MI) as previously described (10). The onset and progression of CIA was monitored from 3–12 weeks postimmunization. The arthritic severity of mice was evaluated as described previously with a grading system for each paw of 0–3 (12). The mean arthritic score was determined using arthritic animals only.
Anti collagen antibodies
Mice were bled on day 35 postimmunization and the level of anti-mouse CII, and anti chick CII in serum were determined using a standard ELISA technique as described previously (16). Serial dilutions of a known high titer anti-CII IgG-positive serum were run as control. Anti-CII IgG levels for samples were calculated from the OD of the high titer standard sera, arbitrarily determined to equal 100Ab units (AU/ml).
Flow cytometry
Expression of HLA -DQ, H2A, CD3, CD4, CD8, and T-cell receptor Vβ chains molecules on peripheral blood lymphocytes (PBLs) was analyzed by flow cytometry using FACS IV (Beckton Dickinson) as described earlier (10). Antibodies used for staining were L227 (anti-DQB1*0601), IVD12 (anti DQB1), HB163 (anti-H2-Ab), GK1.5 (anti-CD4), 53.6.8 (anti CD8), MR9-4( anti-Vβ5.1), MR9-8 (anti-Vβ5.1.2), 44-22-1(anti-Vβ6), F23.1 (anti-Vβ8.1.2.3), KJ-16 (anti-Vβ8.1.2), F23.2 (anti-Vβ8.2), KT11 (anti-Vβ11).
In vitro T cell proliferation
Mice were immunized with 200 μg of CII or peptide emulsified 1:1 with CFA (Difco) injected intradermally at the base of the tail. Ten days postimmunization, draining popliteal, caudal and lumbar lymph nodes were removed and prepared for in vitro culture. LNCs (1×106) were challenged in vitro with or without native collagen. Inhibition experiments were done by adding mAb GK1.5 (anti-CD4), IVD12 (anti HLA-DQ), or Lyt2 (anti-CD8) to the cells challenged in vitro with CII at 50 μg/ml. During the last 18 h the cells were pulsed with 3H-thymidine (1μCi/well). At the end of the assay, the cells were harvested using a plate harvester and incorporated radioactivity was determined using an automated counter (Micrebeta, Perkin Elmer Wallac).
Mice were also tested for T cell response to human collagen II (HII)-derived peptides synthesized and purified at Mayo Clinic Peptide Facility. The DQ8-specific CII-derived peptides have been characterized (13). The mice were primed with 200 μg of peptide and challenged in vitro with 100 μg/ml of the peptide as described previously (13).
Cloning and genomic sequence of T cell receptor (TCR) Vα11.1
DNA was isolated from the spleens of transgenic mice using a kit. PCR fragments were generated by using oligos published previously (17, 18) ). The amplified fragment was cloned using pCR2.1 vector using TOPO TA cloning kit for subcloning (Invitrogen) according to manufacturer’s instructions and sequenced with 377 DNA Sequencer (ABI PRISM).
Molecular Modeling
Models of DQ8, DQ6.1 and DQ6.4 with the various collagen peptides were prepared on a Silicon Graphics Octane and Fuel work station using the program Insight II-version 2005 (Accelrys Inc., San Diego, CA), essentially as previously described (19, 20). The crystal structure of DQ*0602 complexed to the hypocretin 1–13 peptide was used as a base-molecule for all the simulation studies (21), while the modeled structure of the DQ8 allele with the collagen peptide was based on the crystal structure of the same Major Histocompatibility Complex (MHC) II molecule with the Insulin B11-23 peptide (22). In all cases possible, a 13-mer peptide was used, containing the core nonamer and two residues on each of the amino- and carboxy-termini. Where the core nonamer was at the beginning or the end of a sensitizing peptide, minimization was performed using the core nonamer flanked by at most two Ala residues extending only to the beginning or the end of the sensitizing peptide. The numbering scheme developed by Fremont and colleagues (23) for mouse H2-A MHC class II molecules, and modified by Bondinas and coworkers for human MHC II alleles (24), was used here. Essentially, it yields identical numbering in equivalent structural positions in all MHC class II alleles.
Statistical analysis
Difference in incidence of arthritis between groups was analyzed using chi-square test with Yates’ correction. Antibody levels and means scores for arthritic mice were compared using Student’s t test.
RESULTS
DQB1*0604/DQA1*0103 (DQ6.4) mice develop arthritis
All RA patients do not carry RA-susceptible HLA genes, while some carry HLA genes thought to be protective. We hypothesized that the presence of trans-heterodimer HLA molecules may render transgenic mice susceptible to disease, similar to the known HLA susceptible molecules. Although most associations are reported with DQB1 alleles, our hypothesis suggests that both alpha and beta chain are required for presenting antigens and thus contribute to development of arthritis. We tested our hypothesis by generating DQ6.4 mice; DQB1*0604 generally occurs in linkage with DQA1*0102. All transgenic mice developed normally and expressed transgenes (Figure 1). Aβo mice have less than 2 % of CD4+ T cells. Introduction of DQA1*0103 and DQB1*0604 transgenes rescued CD4+ T cells to levels of FVB/N mice (data not shown). FVB/N mice have deletion of Vβ8.2 which has been suggested as one of the reasons for resistance to develop arthritis (17). Transgenic mice showed positive selection of Vβ8.2 T cells (8±1.4 in DQ6.4 compared to 3±2.8 in FVB) suggesting transgenes can positively select TCR profile in thymus.
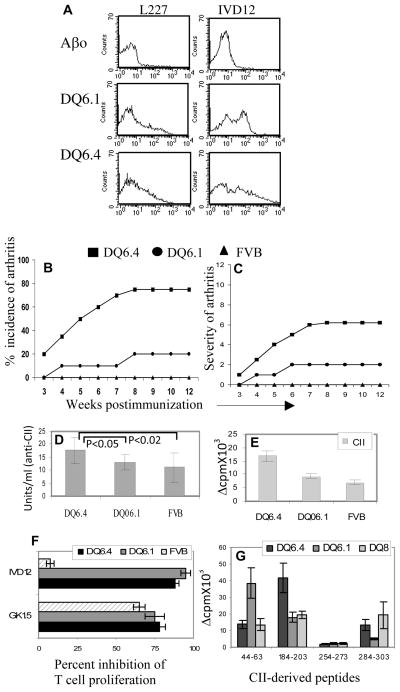
Figure 1.
DQ6.4 mice are susceptible to develop collagen-induced arthritis. A) Expression of DQ in transgenic DQ6.1 and DQ6.4 mice. Both DQ6.1 and DQ6.4 mice showed positive staining with conjugated antibodies IVD12 (anti-DQ) and L227 (anti-DQ6) antibodies while Aβo mice were negative. N=4 B) Incidence and C) severity of arthritis in transgenic and control FVB mice (10–12 mice in each group). D) Anti-collagen antibodies in mice immunized with type II collagen, measured by ELISA. E) DQ6.4 mice generate a more robust in vitro T cell response to CII compared to DQ6.1 mice (3–4 mice in each group). In vitro T cell response was measured in LNCs isolated from CII-primed mice. F) The CII-specific response was CD4-mediated and DQ-restricted as tested by inhibition studies using anti-DQ (IVD12) and anti-CD4 (GK1.5) antibodies. G) In vitro recall response to CII-derived peptides in transgenic mice primed with individual peptides.
Both DQB1*0604 and *0601 have been associated with resistance from arthritis (25, 26). To determine the contribution of DQA1 chain in development of arthritis, we tested Aβo.DQ6.4 mice in CIA protocol. Immunization of DQ6.4 mice with type II collagen led to development of severe arthritis with an incidence of 75 % (Figure 1B, 1C). On the other hand, DQ6.1 and FVB mice were completely resistant to arthritis. All of the immunized strains, DQ6.1, DQ6.4 and FVB, produced anti CII antibodies (Figure 1D).
DQA1*0103/DQB1*0604 mice have a deletion in Vα11.1
Studies have suggested that even though FVB/N mice are H2Aq, they are resistant to CIA (17, 18). This resistance to develop arthritis has been attributed in part to TCRα-V11.1 coding sequence polymorphism. We sequenced TCR Vα11 in DQ6.4 mice (Table 1). DQ6.4 mice showed deletion at position 68 as known for FVB/N mice suggesting this deletion may not be a critical factor for resistance to arthritis in FVB/N mice.
Table 1.
TCR/Vα 11.1 sequences in DQB1*0604 and DQA1*0103 mice compared to FVB/N and DBA/1 mice
| Mice/Vα 11.1 position | 18 | 29 | 31 | 37 | 45 | 68 |
|---|---|---|---|---|---|---|
| DQA1*0103 | G | M | S | Q | N | - |
| DQB1*0604 | S | T | S | Q | N | - |
| DBA/1Vα11.1d | D | M | S | Q | S | R |
| FVB/N Vα11.1c | G | M | S | Q | S | - |
| FVB/N Vα11.2c | S | T | S | Q | N | - |
DQ6.4 mice present CII peptides similar to CIA susceptible DQ8 mice
Mice expressing DQ6.4 generated a CII-specific cellular immune response that was significantly higher than the control DQ6.1, p<0.03 (Figure 1E). The response was mediated by CD4+ T cells and restricted by DQ molecules as observed by HLA-DQ-antibody inhibition studies (Figure 1F). All mice produced anti-CII antibodies although antibodies were significantly higher in the susceptible mice compared to CIA resistant DQ6.1 mice. To determine if DQ6.4 mice present CII-derived peptides similar to DQ8 mice, we carried out in vitro T cell proliferation to various CII-derived peptides. DQ8 and DQ6.1 restricted epitopes of CII have been characterized (13). There are few CII-derived peptides that are not presented by CIA resistant DQ6.1 mice but are presented by CIA susceptible DQ8 mice suggesting they might be important in pathogenesis. As shown in Figure 1G, DQ6.4 mice generated response to the CII-derived peptides that DQ6.1 mice do not, but DQ8 mice do.
DQB1*0604 and DQB1*0302 share peptide binding pockets
As DQ6.1 gene confers protection from arthritis, we first compared differences in sequences between DQ6.1 and DQ6.4. Both molecules differ at 12 positions in beta chains, of which 9 positions are involved in various peptide binding pockets. The DQA1*0102 chain which is generally associated with DQB1*0604 differs from the DQA1*0103 chain at 5 positions (Table 2). To understand the molecular basis of arthritis and T cell response to CII-derived peptides in DQ6.4 mice, we compared the peptide binding pockets among CIA susceptible DQ8 and DQ6.4 and CIA resistant DQ6.1. As shown in Table 2, DQ6.4 and DQ8 molecules share P9 pocket except for two semi-conservative differences at α73 (Met vs. Val) and β57 (Val vs. Ala). On the other hand, p9 pocket of DQ6.1 has two acidic residues (β37Asp and β57Asp) and differs substantially from both its counterparts in DQ6.4 and DQ8. P1 pocket shows the maximum difference among these three alleles, yet all of them share a preference for aliphatic residues and Phe, while DQ8 also shows a distinct preference for acidic residues (22, 27–33) (Table 3). We modeled the binding of those CII peptides to the respective HLA-DQ alleles that generated a T cell response according to the published predicted peptide binding pockets (Tables 3 and 4). T cell responses corresponded with the peptide binding even though all proposed epitopes have at least one weak anchor. This is to be expected, as they are obtained after active immunization, a process that boosts weak epitope antigenicity.
Table 2.
Polymorphism in DQ6 molecules and peptide binding pockets in various DQ haplotypes
| A. Amino Acid Polymorphisms outside anchoring pockets* | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpha chain | 15 | 22+ | 23 | 38 | 42 | 44 | 45 | 47 | 49 | 50 | 53 | 58 | 61 | 63 | 77 | 126 | 127 | 172 | 184 | 204 |
| DQA1*0102 | F | Y | T | R | A | R | W | E | S | K | G | G | R | M | Y | Q | S | Q | A | M |
| DQA1*0103 | F | F | T | K | A | R | W | E | S | K | G | G | R | M | Y | H | A | Q | A | V |
| DQA1*0301 | S | Y | S | R | V | Q | L | L | R | R | R | F | T | I | S | H | S | E | T | V |
| Beta chain | 3 | 53 | 55 | 66 | 84 | 87 | 125 | 130 | 140 | 167 | ||||||||||
| DQB1*0302 | S | L | P | E | Q | L | A | R | T | R | ||||||||||
| DQB1*0601 | P | Q | R | D | E | F | G | R | A | H | ||||||||||
| DQB1*0604 | S | Q | R | E | E | Y | G | Q | A | R | ||||||||||
| B. Amino Acid Polymorphisms within anchoring Pockets | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DQA1/DQB1 | P1 | P4 | P6 | P7 | P9 | ||||||||||
| α9, α31, α52, β86 | β13, β26, | α66, β9, β30 | β67 | α73, β9, β37, β38, β57 | |||||||||||
| *0301/*0302 | Y | E | R | E | G | L | L | Y | Y | V | V | Y | Y | A | A |
| *0103/*0601 | C | Q | G | A | A | Y | A | L | Y | I | M | L | D | V | D |
| *0103/*0604 | C | Q | G | G | G | L | A | Y | H | V | M | Y | Y | A | V |
Table 3.
Amino acid residue preferences (motifs) at given pockets for alleles used in the study*
| Allele | Pocket 1 | Shelf p3 | Pocket 4 | Pocket 6 | Pocket 7 | Pocket 9 |
|---|---|---|---|---|---|---|
| DQA1*0301/B1*0302 | Acidic, Aromatic (F, Y) Aliphatic (L, M, I, V) Polar (Q, N) 10X less No R, K, H |
NA** | Aromatic (F, Y) Aliphatic (L, M, I, V) Acidic 10X less No R, K, H No Q, N |
Aliphatic (L, M, I, V) Proline D, not E No R, K, H No Q, N No aromatic (F, Y, W) |
Wide preferences | Acidic, Q, N 10X less Aliphatic (L, M, I, V) 10X less No R, K, H No Aromatic (F, Y, W) |
| DQA1*0103/B1*0601 | Aliphatic (L, M, I, V) Aromatic (F) Polar (Q, N) Hydroxyl (S, T) Acidic ( D, E) No R, K, H No G, P |
All, incl. P and G, but No R, K, H | G, A, S only Pro 10X less No F, Y, W No V, M, I, L No Q, N No D, E No R, K, H |
Aliphatic (L, M, I, V) A, P, T, S, N, Q G 10X less No D, E No F, Y, W No R, K, H |
Wide preferences | Aliphatic (L, M, I, V, A) G Aromatic 10X less Q, N ca. 10X less S, T ca. 10 X less P 10X less No R, K, H No D, E |
| DQA1*0103/B1*0604 | Aliphatic (L, M, I, V) Aromatic (F) Polar (Q, N) No D, E No R, K, H No G, P |
All, incl. G, except italicized below No P No T, S No R, K, H |
Hydroxyl (S, T) Aliphatic (A, V, L, M, I) Aromatic (F, Y) No Q, N No D, E No R, K, H No G, P |
Aliphatic (L, M, I, V) A 10X less Acidic (D, E) No G, P No Q, N No F, Y, W No R, K, H |
Wide preferences | A, Aromatic (Y>F>W) Aliphatic (L, M, I, V) 10X less Acidic 5-10X less Q, N ca. 10X less S, T ca. 10 X less G 10X less No P No R, K, H |
Table 4.
Peptide binding and in vitro T cell response to CII- derived peptides
| CII | peptides P1–P9 | Binding | |
|---|---|---|---|
| 1. DQ8(A1*0301/B1*0302) | |||
| 44–63 | PPGPPGKPGDDGEAGKPGKA | + | weak, p9G weak |
| 184–203 | GPEGAQGPRGEPGTPGSPGP | + | weak, p4Q poor |
| 254–273 | TGGKPGIAGFKGEQGPKGEP | − | no register fits |
| 284–303 | PAGEEGKRGARGEPGGVGPI | + | weak, p4R poor |
| 2. DQ6.1 (A1*103/B1*601) | |||
| 44–63 | PPGPPGKPGDDGEAGKPGKA | + | gly anchors weak |
| 184–203 | GPEGAQGPRGEPGTPGSPGP | + | “ ““ |
| 254–273 | TGGKPGIAGFKGEQGPKGEP | − | no register fits |
| 284–303 | PAGEEGKRGARGEPGGVGPI | − | no register fits |
| 3. DQ6.4 (A1*103/B1*604) | |||
| 44–63 | PPGPPGKPGDDGEAGKPGKA | + | weak |
| 184–203 | GPEGAQGPRGEPGTPGSPGP | + | weak |
| 254–273 | TGGKPGIAGFKGEQGPKGEP | − | none fits |
| 284–303 | PAGEEGKRGARGEPGGVGPI | + | no single clear register |
Bold letters indicate peptide binding anchor positions P1–P9.
Of the four CII epitopes tested in the DQ8 transgenic mice, 184–203 and 284–303 showed the strongest sensitization, while appreciable responses were observed with the 44–63 CII-derived peptides, but hardly any response is seen with the 254–273 CII peptide (Figure 1G). It was possible to identify a single unique register in the cases of the three sensitized peptides, while no obvious epitope that could even remotely fulfill the DQ8 motif could be identified in the non-sensitized peptide (Table 4). It should be stressed however, that all three sensitized epitopes have at least one weak anchor residue: the epitope in the 44–63 peptide has weak p4E, p6G and p9G anchors, the 184–203 epitope has a weak p4Q anchor, while the 284–303 epitope has a very weak p4K anchor (Table 4, AFigure 2A). Likewise, in the DQ6.1 mice only the 44–63 and 184–203 CII epitopes exhibit one binding register each, albeit with weak p1G and p7G anchors in the former, and weak p6G and medium p9G in the latter (Table 4, Figure 2B). The remaining two epitopes lack any register in which they could bind to the DQ6.1 molecule (Table 4). Lastly, in the DQ6.4 mice there is sensitization to the peptide epitopes 44–63, 184–203, and 284–303, but none to the 254–273 peptide (Figure 1G, 2C). In this case as well, the p1G, p4G and p7G anchors in the first peptide are very weak, while in the second peptide the p4R anchor is extremely weak. No register is evident in the 254–273 peptide, as there is no apparent nonamer fulfilling the motif. Quite surprisingly, no register is evident in the 284–303 peptide as well for the DQA1*0103/DQB1*0604 molecule, even though there is sensitization of CII-immunized spleen cells to this peptide. A number of candidate core nonamers appear in this peptide with several extremely weak anchors (e.g. AGEEGKRGA, EGKRGARGE, GEPGGVGPI, putative anchors in bold). Yet molecular simulation of the respective structures does not yield any clear-cut favored peptide, so we do not consider it proper to propose any such peptide as the most likely one.
Figure 2.
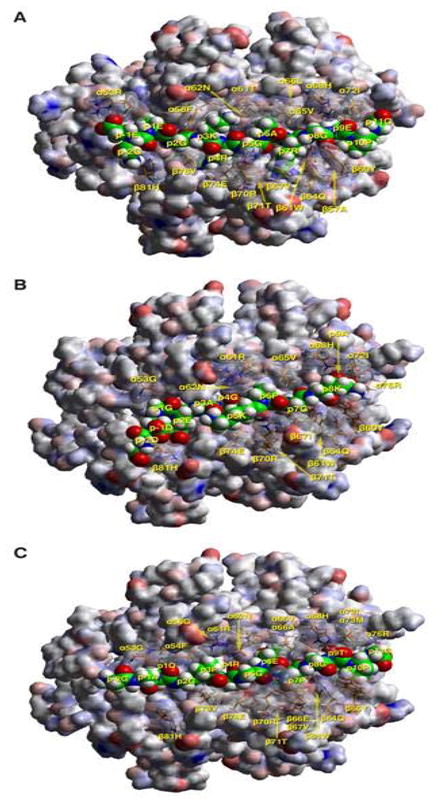
Molecular simulation complexes of CII peptides with respective human MHC II alleles. A) TCR view of the chick CII peptide 286–298 (GEEGKRGARGEPG, anchors in bold, core nonamer underlined) in the groove of HLA-DQ8, after energy minimization at pH 5.4 based on the crystal structure of HLA-DQ8 (22). The α1β1 domain of the DQ302 molecule is depicted in van der Waals surface representation, with the surface atomic charges color-coded (blue, positive; grey, neutral; red, negative), and the antigenic peptide is shown in space filling form (color code: carbon, green; oxygen, red; nitrogen, blue; hydrogen, white; sulfur, yellow). Several DQ302 residues that have particular interactions with the antigenic peptide or the cognate TCR are shown in stick form with transparent van der Waals surfaces, and with the same color-code as the antigenic peptide with the exception of carbon (orange). Figure was drawn with the aid of Web Lab Viewer version 3.5 and DS Viewer Pro version 6.0, of Accelrys. TCR view of the chick CII peptides B) 53–63 (DDGEAGKPGKA, anchors in bold, core nonamer underlined) in the groove of HLA-DQ6.1, and C) 187–199 (GAQGPRGEPGTPG, anchors in bold, core nonamer underlined) in the groove of HLA-DQ6.4, after energy minimization at pH 5.4 based on the crystal structure of HLA-DQA1*0102/B1*0602 (21) Mode of depiction, colors and conventions as in 2A.
As seen in Table 4, even in the cases of CII peptides 44–63 and 184–203, where all three transgenes show a T cell response upon immunization, the individual binding registers of the nonamer core are quite different, depending on the DQ allele. This is to be expected, as the affinity of a peptide for an HLA-DQ allele is the sum total of the individual affinities of the specific anchor residues for each individual pocket. More so, as in HLA-DQ alleles there does not seem to be any single affinity-determinant pocket, as pocket 1 is for HLA-DR alleles (33, 34).
DISCUSSION
Among all the genetic factors studied to date, genes encoded by MHC class II loci have shown the strongest association with RA. While DR4/DQ3 haplotype is associated with susceptibility to arthritis, DR2/DQ6 is associated with resistance (3, 26). Although development of arthritis is associated with shared epitope (SE), not all patients carry the shared epitope haplotype. Some patients carry DR2/DQ6 that has been shown to be associated with protection from RA in some ethnic groups (25) suggesting that protection is incomplete. Disease in SE-negative patients has been attributed to the presence of various non-MHC alleles and environmental factors. The resistance of FVB/NJ mouse to arthritis induction is suggested to be due to polymorphism at codon 18 and genomic deletion in coding sequences TCR Vα11.1(18). DBA/1 mice have Vα11.1d that carries Asp at position 18 while FVB/N mice have Vα11.1c or Vα11.2c that carries serine or glycine at that position. DQ6.4 mice showed Vα11 characteristic of FVB/N mice suggesting this region may not determine susceptibility to CIA in this model.
There are no studies on the possible role of trans-heterodimers of RA-resistant alleles in predisposition to RA. We show here that trans-heterodimer, DQB1*0604/DQA1*0103, between 2 resistant DQ6 alleles can predispose to CIA. The molecular basis of disease development and antigen presentation by DQ6.4 can be explained by the peptide binding characteristics of the P4, P6 and P9 pockets. Both *0604 and *0302 molecules share similarities, albeit with some distinct differences, in these pockets as shown in Tables 2 and Table 3. In the P9 pocket, β57 is Val in DQ6.4 and Ala in DQ8 compared to Asp in DQ6.1, an allele associated with resistance to arthritis. As suggested before, β57 Asp forms a salt bridge with α76 Arg but the potential to form this salt bridge is absent in DQ6.4 and DQ8 molecules. As this is a critical region promoting inter-chain contacts between α-, β-chains and the peptide backbone, the residues occupying pocket 9 in case of β57 non-Asp residues, must promote such contacts, either via salt bridges, as in the case of p9Asp—α76Arg in the complex of DQ8—InsB13-21 or van der Waals contacts as in the case of p9Trp, in the complex of MHC I αchain peptide with DQ2 (32). DQ6.4 and DQ8 differ at residues 55–57 of the beta chain (P-P-A versus R-P-V). However, β57Val in DQ6.4 shapes a far different set of preferences for pocket 9 of DQ6.4, than β57Ala does for DQ8 (35). Pocket 9 also sharesβ9 r residue in DQB1*0604 and *0302, Tyr, compared to Leu in DQ6.1 thus leading to a bigger and more hydrophobic area at the bottom of this pocket in the latter allele. DQ6.1 has an overall negative electrostatic surface potential in the P9 pocket thus favoring binding of basic and polar amino acids while DQ8 favors acidic residues and DQ6.4 favors hydrophobic amino acids but also has low affinity for acidic amino acids. The effect of polymorphisms of both DQα- and β-chains on peptide binding is expected, as both chains contribute residues for the binding pockets of the class II molecules. In type 1 diabetes, DQB1*0602 provides protection while closely related DQB1*0604 does not (35). The protection offered by DQB1*0602 is explained by the presence of β57Asp that forms a highly favored salt bridge toα76Arg leading to protein stability (36). Substitution of β57 alters the MHC II–peptide binding activity (37, 38). However as observed in the present results, most of the peptides bind with weak anchors yet generate an immune response. These results are reminiscent of earlier reports with native Ac1-9 MBP peptide that binds fleetingly to H2-Au, yet active immunization with the peptide leads to experimental autoimmune encephalomyelitis (39, 40). Besides the fact that the binding of most of these CII peptides to the respective MHC II molecules are weak and forced by Freund’s complete adjuvant mediated immunization, we also must consider the possibility that we may be dealing with more than one possible registers, all of them weak ones. This was recently shown, with not so weakly binding registers, in the case of peptides and the MHC II allele (41). The p4 pocket of DQ8 and DQ6.4 shows similarities with the residues of β13Gly and β26Leu, compared to Ala and Tyr, respectively, in DQ6.1 at these positions. The p4 pocket in DQ8 and DQ6.4 is deep while DQ6.1, associated with protection, probably has a smaller pocket that is partially filled because of the substitutions of Tyr for Leu at β26, and Ala for Gly at β13, predisposing to small aliphatics and not large aliphatics or Phe/Tyr. DQ6.4 differs from DQ8 at pockets p6 and p1. This is similar to the difference observed in this pocket 4 between DQ7 and DQ8, where identical residue substitutions lead to sharply decreased volume in this pocket in the case of DQ7 (29, 30). These two alleles differing in four residues in the β1 antigen-binding domain, three of which (β13, 26, 57) are in pocket 4 and 9, show indeed distinct preferences there, but identical preferences in the other three pockets. By contrast, DQ8 and DQ2, despite considerable allelic variation, also exhibit remarkable overlap in peptide binding specificity (27, 29–32, 35, 42). The present study suggests a critical role of p4 and p9 pockets of these two DQ molecules in susceptibility to arthritis. In addition, the presence of α22Phe in DQA1*0103 further destabilizes the DQ6.4—peptide complex in comparison with a complex of the same peptide with DQA1*0102/DQB1*0604 because the former complex has one hydrogen bond less between MHCII protein and peptide. A similar situation has been observed in comparing peptide complexes of DQA1*0501/B1*0201 with those of DQA1*0201/B1*0202 (43). Previous studies have shown that allelic variation in DQ6 alleles in peptide binding pockets may result in difference in protein stability that can change TCR recognition and T cell selection thus resulting in association or non-association with type 1 diabetes (35, 44, 45).
Results of this study suggest that HLA alleles thought to be protective for arthritis can render hosts susceptible if a DQA1/DQB1 trans-heterodimer is formed between the 2 protective DQ alleles. DQA1*0103/DQB1*0601 and DQA1*0102/DQ*0604 are associated with protection from arthritis (25). In general, DQB1*0604 occurs in linkage with DQA1*0102 while DQB1*0601 occurs in linkage with DQA1*0103. Our data suggest that DQB1*0604 when paired with DQA1*0103 in transgenic mice can present arthritogenic peptides of CII and lead to development of disease. Trans-heterodimers have been shown to impair peptide binding for some although trans-encoded dimers have been suggested to be more effective in presenting diabetogenic epitopes (6, 46).
DQA1*0103 also occurs in linkage with DQB1*05 and DQB1*0603. This would imply that an individual who is positive for the known protective HLA alleles, subtypes of DQ5 or DQ6, may express trans-heterodimers that can predispose to arthritis. In heterozygous individuals, products of both parental DQ haplotypes interact and have a potential to form DQ trans-heterodimers. Formation of trans-heterodimers between DQA1 and DQB1 has been shown although the stability of trans-heterodimers varies depending on the haplotype (47). DQ1 trans-heterodimers are more stable than between DQA1*01 and beta chains of DQ2 and DQ3 which has been shown to be due to the steric incompatibility resulting from structural differences (21). Our results are reminiscent of studies in diabetes and celiac disease where trans-heterodimers are associated with susceptibility to develop disease Recent studies using DQA1*0103/DQB1*0302 trans-heterodimer expressing transgenic mice have shown that they develop relapsing polychondritis while DQA1*0301/DQB1*0302 mice do not (48). Together with the available data, this study suggests that non-RA—associated HLA alleles present in some patients may predispose to disease by the generation of trans-heterodimers.
Acknowledgments
Authors thank Julie Hanson for breeding and maintaining transgenic mice and, Dr. George Bondinas and Mr. Demetrios Kyrkas for help with the molecular simulation.
Grant support for the study- This study was supported by NIH grants AI075262 and AR30752, An institutional grant to the Epirus Institute of Technology by the Regional Development Project of the Region of Epirus, under the 3rd Community Support Framework of the EU (80% EU funds, 20% Hellenic State funds).
Footnotes
No Financial conflicts. No financial support or benefits from commercial sources were obtained for this study.
References
- 1.Taneja V, Mehra NK, Chandershekaran AN, Ahuja RK, Singh YN, Malaviya AN. HLA-DR4-DQw8, but not DR4-DQw7 haplotypes occur in Indian patients with rheumatoid arthritis. Rheumatol Int. 1992;11:251–5. doi: 10.1007/BF00301502. [DOI] [PubMed] [Google Scholar]
- 2.Singal DP, D’Souza M, Reid B, Bensen WG, Kassam YB, Adachi JD. HLA-DQ beta-chain polymorphism in HLA-DR4 haplotypes associated with rheumatoid arthritis. Lancet. 1987;2:1118–20. doi: 10.1016/s0140-6736(87)91548-0. [DOI] [PubMed] [Google Scholar]
- 3.Taneja V, Mehra NK, Kailash S, Anand C, Malaviya AN. Protective & risk DR phenotypes in Asian Indian patients with rheumatoid arthritis. Indian J Med Res. 1992;96:16–23. [PubMed] [Google Scholar]
- 4.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–92. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trowsdale J. The gentle art of gene arrangement: the meaning of gene clusters. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-comment2002. COMMENT2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tollefsen S, Arentz-Hansen H, Fleckenstein B, Molberg O, Raki M, Kwok WW, et al. HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J Clin Invest. 2006;116:2226–36. doi: 10.1172/JCI27620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koeleman BP, Lie BA, Undlien DE, Dudbridge F, Thorsby E, de Vries RR, et al. Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans dimer associations with disease. Genes Immun. 2004;5:381–8. doi: 10.1038/sj.gene.6364106. [DOI] [PubMed] [Google Scholar]
- 8.Bradley DS, Nabozny GH, Cheng S, Zhou P, Griffiths MM, Luthra HS, et al. HLA-DQB1 polymorphism determines incidence, onset, and severity of collagen-induced arthritis in transgenic mice. Implications in human rheumatoid arthritis. J Clin Invest. 1997;100:2227–34. doi: 10.1172/JCI119760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nabozny GH, Baisch JM, Cheng S, Cosgrove D, Griffiths MM, Luthra HS, et al. HLA-DQ8 transgenic mice are highly susceptible to collagen-induced arthritis: a novel model for human polyarthritis. J Exp Med. 1996;183:27–37. doi: 10.1084/jem.183.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taneja V, Taneja N, Paisansinsup T, Behrens M, Griffiths M, Luthra H, et al. CD4 and CD8 T cells in susceptibility/protection to collagen-induced arthritis in HLA-DQ8-transgenic mice: implications for rheumatoid arthritis. J Immunol. 2002;168:5867–75. doi: 10.4049/jimmunol.168.11.5867. [DOI] [PubMed] [Google Scholar]
- 11.Taneja V, Taneja N, Behrens M, Griffiths MM, Luthra HS, David CS. Requirement for CD28 may not be absolute for collagen-induced arthritis: study with HLA-DQ8 transgenic mice. J Immunol. 2005;174:1118–25. doi: 10.4049/jimmunol.174.2.1118. [DOI] [PubMed] [Google Scholar]
- 12.Wooley PH, Luthra HS, Stuart JM, David CS. Type II collagen-induced arthritis in mice. I. Major histocompatibility complex (I region) linkage and antibody correlates. J Exp Med. 1981;154:688–700. doi: 10.1084/jem.154.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krco CJ, Pawelski J, Harders J, McCormick D, Griffiths M, Luthra HS, et al. Characterization of the antigenic structure of human type II collagen. J Immunol. 1996;156:2761–8. [PubMed] [Google Scholar]
- 14.Cosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, et al. Mice lacking MHC class II molecules. Cell. 1991;66:1051–66. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths MM, Eichwald EJ, Martin JH, Smith CB, DeWitt CW. Immunogenetic control of experimental type II collagen-induced arthritis. I. Susceptibility and resistance among inbred strains of rats. Arthritis Rheum. 1981;24:781–9. doi: 10.1002/art.1780240605. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths MM, Nabozny GH, Hanson J, Harper DS, McCall S, Moder KG, et al. Collagen-induced arthritis and TCRs in SWR and B10. Q mice expressing an Ek alpha transgene. J Immunol. 1994;153:2758–68. [PubMed] [Google Scholar]
- 17.Osman GE, Hannibal MC, Anderson JP, Lasky SR, Ladiges WC, Hood L. FVB/N (H2(q)) mouse is resistant to arthritis induction and exhibits a genomic deletion of T-cell receptor V beta gene segments. Immunogenetics. 1999;49:851–9. doi: 10.1007/s002510050564. [DOI] [PubMed] [Google Scholar]
- 18.Osman GE, Hannibal MC, Anderson JP, Cheunsuk S, Lasky SR, Liggitt HD, et al. T-cell receptor vbeta deletion and valpha polymorphism are responsible for the resistance of SWR mouse to arthritis induction. Immunogenetics. 1999;49:764–72. doi: 10.1007/s002510050550. [DOI] [PubMed] [Google Scholar]
- 19.Moustakas AK, Routsias J, Papadopoulos GK. Modelling of the MHC II allele I-A(g7) of NOD mouse: pH-dependent changes in specificity at pockets 9 and 6 explain several of the unique properties of this molecule. Diabetologia. 2000;43:609–24. doi: 10.1007/s001250051350. [DOI] [PubMed] [Google Scholar]
- 20.Reichstetter S, Papadopoulos GK, Moustakas AK, Swanson E, Liu AW, Beheray S, et al. Mutational analysis of critical residues determining antigen presentation and activation of HLA-DQ0602 restricted T-cell clones. Hum Immunol. 2002;63:185–93. doi: 10.1016/s0198-8859(01)00377-9. [DOI] [PubMed] [Google Scholar]
- 21.Siebold C, Hansen BE, Wyer JR, Harlos K, Esnouf RE, Svejgaard A, et al. Crystal structure of HLA-DQ0602 that protects against type 1 diabetes and confers strong susceptibility to narcolepsy. Proc Natl Acad Sci U S A. 2004;101:1999–2004. doi: 10.1073/pnas.0308458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol. 2001;2:501–7. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 23.Fremont DH, Monnaie D, Nelson CA, Hendrickson WA, Unanue ER. Crystal structure of I-Ak in complex with a dominant epitope of lysozyme. Immunity. 1998;8:305–17. doi: 10.1016/s1074-7613(00)80536-1. [DOI] [PubMed] [Google Scholar]
- 24.Bondinas GP, Moustakas AK, Papadopoulos GK. The spectrum of HLA-DQ and HLA-DR alleles, 2006: a listing correlating sequence and structure with function. Immunogenetics. 2007;59:539–53. doi: 10.1007/s00251-007-0224-8. [DOI] [PubMed] [Google Scholar]
- 25.Laivoranta-Nyman S, Mottonen T, Hermann R, Tuokko J, Luukkainen R, Hakala M, et al. HLA-DR-DQ haplotypes and genotypes in Finnish patients with rheumatoid arthritis. Ann Rheum Dis. 2004;63:1406–12. doi: 10.1136/ard.2003.009969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taneja V, Giphart MJ, Verduijn W, Naipal A, Malaviya AN, Mehra NK. Polymorphism of HLA-DRB, -DQA1, and -DQB1 in rheumatoid arthritis in Asian Indians: association with DRB1*0405 and DRB1*1001. Hum Immunol. 1996;46:35–41. doi: 10.1016/0198-8859(95)00165-4. [DOI] [PubMed] [Google Scholar]
- 27.Godkin A, Friede T, Davenport M, Stevanovic S, Willis A, Jewell D, et al. Use of eluted peptide sequence data to identify the binding characteristics of peptides to the insulin-dependent diabetes susceptibility allele HLA-DQ8 (DQ 3.2) Int Immunol. 1997;9:905–11. doi: 10.1093/intimm/9.6.905. [DOI] [PubMed] [Google Scholar]
- 28.Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci U S A. 2004;101:4175–9. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwok WW, Domeier ML, Raymond FC, Byers P, Nepom GT. Allele-specific motifs characterize HLA-DQ interactions with a diabetes-associated peptide derived from glutamic acid decarboxylase. J Immunol. 1996;156:2171–7. [PubMed] [Google Scholar]
- 30.Kwok WW, Nepom GT, Raymond FC. HLA-DQ polymorphisms are highly selective for peptide binding interactions. J Immunol. 1995;155:2468–76. [PubMed] [Google Scholar]
- 31.Stepniak D, Wiesner M, de Ru AH, Moustakas AK, Drijfhout JW, Papadopoulos GK, et al. Large-scale characterization of natural ligands explains the unique gluten-binding properties of HLA-DQ2. J Immunol. 2008;180:3268–78. doi: 10.4049/jimmunol.180.5.3268. [DOI] [PubMed] [Google Scholar]
- 32.van de Wal Y, Kooy YM, Drijfhout JW, Amons R, Papadopoulos GK, Koning F. Unique peptide binding characteristics of the disease-associated DQ(alpha 1*0501, beta 1*0201) vs the non-disease-associated DQ(alpha 1*0201, beta 1*0202) molecule. Immunogenetics. 1997;46:484–92. doi: 10.1007/s002510050309. [DOI] [PubMed] [Google Scholar]
- 33.Zarutskie JA, Sato AK, Rushe MM, Chan IC, Lomakin A, Benedek GB, et al. A conformational change in the human major histocompatibility complex protein HLA-DR1 induced by peptide binding. Biochemistry. 1999;38:5878–87. doi: 10.1021/bi983048m. [DOI] [PubMed] [Google Scholar]
- 34.Natarajan SK, Stern LJ, Sadegh-Nasseri S. Sodium dodecyl sulfate stability of HLA-DR1 complexes correlates with burial of hydrophobic residues in pocket 1. J Immunol. 1999;162:3463–70. [PubMed] [Google Scholar]
- 35.Ettinger RA, Papadopoulos GK, Moustakas AK, Nepom GT, Kwok WW. Allelic variation in key peptide-binding pockets discriminates between closely related diabetes-protective and diabetes-susceptible HLA-DQB1*06 alleles. J Immunol. 2006;176:1988–98. doi: 10.4049/jimmunol.176.3.1988. [DOI] [PubMed] [Google Scholar]
- 36.Ettinger RA, Liu AW, Nepom GT, Kwok WW. Beta 57-Asp plays an essential role in the unique SDS stability of HLA-DQA1*0102/DQB1*0602 alpha beta protein dimer, the class II MHC allele associated with protection from insulin-dependent diabetes mellitus. J Immunol. 2000;165:3232–8. doi: 10.4049/jimmunol.165.6.3232. [DOI] [PubMed] [Google Scholar]
- 37.Sato AK, Sturniolo T, Sinigaglia F, Stern LJ. Substitution of aspartic acid at beta57 with alanine alters MHC class II peptide binding activity but not protein stability: HLA-DQ (alpha1*0201, beta1*0302) and (alpha1*0201, beta1*0303) Hum Immunol. 1999;60:1227–36. doi: 10.1016/s0198-8859(99)00120-2. [DOI] [PubMed] [Google Scholar]
- 38.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–21. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 39.Liu GY, Wraith DC. Affinity for class II MHC determines the extent to which soluble peptides tolerize autoreactive T cells in naive and primed adult mice--implications for autoimmunity. Int Immunol. 1995;7:1255–63. doi: 10.1093/intimm/7.8.1255. [DOI] [PubMed] [Google Scholar]
- 40.Fugger L, Liang J, Gautam A, Rothbard JB, McDevitt HO. Quantitative analysis of peptides from myelin basic protein binding to the MHC class II protein, I-Au, which confers susceptibility to experimental allergic encephalomyelitis. Mol Med. 1996;2:181–8. [PMC free article] [PubMed] [Google Scholar]
- 41.Landais E, Romagnoli PA, Corper AL, Shires J, Altman JD, Wilson IA, et al. New design of MHC class II tetramers to accommodate fundamental principles of antigen presentation. J Immunol. 2009;183:7949–57. doi: 10.4049/jimmunol.0902493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidney J, del Guercio MF, Southwood S, Sette A. The HLA molecules DQA1*0501/B1*0201 and DQA1*0301/B1*0302 share an extensive overlap in peptide binding specificity. J Immunol. 2002;169:5098–108. doi: 10.4049/jimmunol.169.9.5098. [DOI] [PubMed] [Google Scholar]
- 43.Fallang LE, Bergseng E, Hotta K, Berg-Larsen A, Kim CY, Sollid LM. Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat Immunol. 2009;10:1096–101. doi: 10.1038/ni.1780. [DOI] [PubMed] [Google Scholar]
- 44.Routsias J, Papadopoulos GK. Polymorphic structural features of modelled HLA-DQ molecules segregate according to susceptibility or resistance to IDDM. Diabetologia. 1995;38:1251–61. doi: 10.1007/BF00401756. [DOI] [PubMed] [Google Scholar]
- 45.Ettinger RA, Kwok WW. A peptide binding motif for HLA-DQA1*0102/DQB1*0602, the class II MHC molecule associated with dominant protection in insulin-dependent diabetes mellitus. J Immunol. 1998;160:2365–73. [PubMed] [Google Scholar]
- 46.Reichstetter S, Kwok WW, Nepom GT. Impaired binding of a DQ2 and DQ8-binding HSV VP16 peptide to a DQA1*0501/DQB1*0302 trans class II heterodimer. Tissue Antigens. 1999;53:101–5. doi: 10.1034/j.1399-0039.1999.530111.x. [DOI] [PubMed] [Google Scholar]
- 47.Kwok WW, Kovats S, Thurtle P, Nepom GT. HLA-DQ allelic polymorphisms constrain patterns of class II heterodimer formation. J Immunol. 1993;150:2263–72. [PubMed] [Google Scholar]
- 48.Lamoureux JL, Buckner JH, David CS, Bradley DS. Mice expressing HLA-DQ6alpha8beta transgenes develop polychondritis spontaneously. Arthritis Res Ther. 2006;8:R134. doi: 10.1186/ar2023. [DOI] [PMC free article] [PubMed] [Google Scholar]