Abstract
Summary: The Signaling Gateway Molecule Pages (SGMP) database provides highly structured data on proteins which exist in different functional states participating in signal transduction pathways. A molecule page starts with a state of a native protein, without any modification and/or interactions. New states are formed with every post-translational modification or interaction with one or more proteins, small molecules or class molecules and with each change in cellular location. State transitions are caused by a combination of one or more modifications, interactions and translocations which then might be associated with one or more biological processes. In a characterized biological state, a molecule can function as one of several entities or their combinations, including channel, receptor, enzyme, transcription factor and transporter. We have also exported SGMP data to the Biological Pathway Exchange (BioPAX) and Systems Biology Markup Language (SBML) as well as in our custom XML.
Availability: SGMP is available at www.signaling-gateway.org/molecule.
Contact: shankar@ucsd.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 BACKGROUND
Signal transduction in mammalian systems can be simple, like transfer of ions, which results in change in the electrical potential of the cell that, in turn, propagates the signal in the cell or more complex signal transduction involving many intracellular protein cascades (Gutkind, 1998). All these signaling processes are driven by complex systems of functionally interacting molecules inside the cell. Thus, understanding functional states of signaling molecules and their interactions is essential to explain normal or pathological biological function. During the past few years, there has been a steady development of multiple resources on cellular signaling, e.g. Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa, 2002) and Reactome (Joshi-Tope et al., 2005). All these databases contain overall information on molecules involved in specific signal transduction processes while some databases like the Database of Quantitative Cellular Signaling (DOQCS) (Hariharaputran et al., 2003) have quantitative modeling data on cellular signaling. In an effort to provide a holistic view of cellular signaling, we created a repository derived from a comprehensive signaling protein ontology (Li et al., 2002) which covers functional states of a protein, the transitions between those states and the defined functions of a protein in a given cellular context (Table 1). Further, we provide data on quantitative parameters associated with each signaling function. The SGMP database contains over 4000 mammalian signaling proteins (see Supplementary Table S1 and S2) deposited as molecule pages which are available either as published (peer reviewed via Nature Publishing Group) or unpublished (awaiting peer review or awaiting authoring).
Table 1.
Molecule pages data statistics
| S. no | Molecule page event category | No. of events | No. of PubMed % citations used |
|---|---|---|---|
| 1 | Summary | 655 | 26 313 |
| 2 | State | 10 601 | 10 500 |
| 3 | State transition | 10 817 | 3069 |
| 4 | State function | ||
| Channel | 62 | 80 | |
| Receptor | 732 | 891 | |
| Enzyme | 3172 | 2694 | |
| Transcription factor | 83 | 163 | |
| Transporter | 182 | 113 | |
| 5 | Class moleculea | 394 | 779 |
All presently published (655) molecule pages have the above data as of March 4, 2011.
aClass molecule (also called Protein Class) is represented by a group of two or more proteins with sequence and/or functional homology (see ‘Class molecule’ section in Supplementary Material), which is used as one of the state constituents.
A molecule page is defined by a specific protein sequence and as a consequence by its biophysical properties. All molecule pages are augmented by sequence analysis and database-driven automated annotation, providing details on sequence, domains and motifs, interactions, pathways, sequence homology and structural information where available. Each published molecule page contains an abstract, a text summary and structured data on protein states, state transitions and state functions. To the best of our knowledge, no other resource provides details on experimentally characterized functional states of signaling proteins.
2 DATA MODEL
The data model was developed by defining states of a given protein, biological processes associated with state transitions and functions for each state.
2.1 State
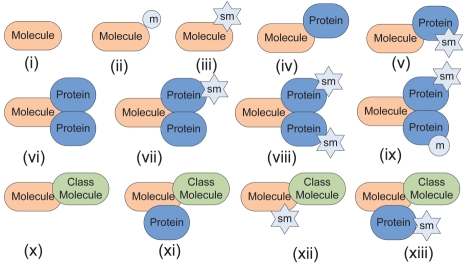
A protein, which is represented by a molecule page, has at least one state i.e. the native protein. This state represents a molecule immediately following protein synthesis, before addition of any post-translational modifications, and is the primary building block for all other states. The new states are created based on protein localization (Ashburner et al., 2000) and interaction with other proteins, small molecules and/or class molecules (Fig. 1). Examples of processes which create a state include phosphorylation, proteolysis, Guanosine triphosphate binding, membrane localization, etc. Transient or fleeting interactions like non-specific binding are not included in state creation. Each state has at least one reference, except a native state, showing experimental evidence for its existence with a specific role in cellular processes. In molecule pages, a state name follows a rigid set of rules (see ‘States’ section in Supplementary Material). Interacting proteins in a state are separated by slashes (‘/’), bound ligands are connected by hyphens and subcellular locations are included in parentheses. For example: ‘ProA/ProB-P2 (nuc)’ represents Protein A bound to Protein B, which has been phosphorylated twice and the state exists in the nucleus.
Fig. 1.
Schematic representation of some states. A new state is formed (i) with a native protein itself (ii) with covalent modification(s) (iii) with binding to small molecule (iv–ix) with binding to one or more other proteins, small molecules, modifications, etc. and (x–xiii) above changes with a class molecule. Molecule, molecule page protein; m, covalent modification; protein, other binding protein; loc, cellular location; sm, small molecule.
2.2 State transitions
State transition occurs when a new state is created from the native or previously created state by addition or removal of a covalent modification, association or dissociation of a protein, a small molecule or a change in location (see ‘State transitions’ section in Supplementary Material). One or more transitions can occur within a pair of states and there can be multistep transitions between states. Some transitions are associated with biological processes which are caused by one or more catalysts, where a catalyst could be a state, class molecule or a starting state with intrinsic enzyme activity (see ‘Enzyme function’ section in Supplementary Material). If, however, it is caused by ligand binding, then the ligand-receptor details (see ‘Receptor function’ section in Supplementary Material) are included.
2.3 State functions
A state may associate with one or more of five different functions depending on the protein functional category (see ‘State functions’ section in Supplementary Material). The channel function is defined with the channel ions, ionic conductance, channel blockers and thermodynamic data on channel gating. Receptor function is defined with bound ligand and dissociation rates. For enzymes, functions are represented for the catalysis with balanced chemical equations. If known, kinetic data of the species for which data were measured in vivo or in vitro are included. For transcription factors, bound DNA sequences with target genes are provided. These functions may overlap for a given protein state—for example, nuclear receptor may function as receptor and transcription factor, receptor tyrosine kinase may function as an enzyme and as a receptor and some ligand-gated channels are also associated with receptor function.
3 DATA EXPORT
The comparison of data availability between databases (Table 2) highlights the necessity to integrate data through a common platform. SGMP provides data export in various community standardized formats such as SBML (Hucka et al., 2003) and BioPAX (Demir et al., 2010) to facilitate integration of data from disparate databases while using systems biology tools for analysis. One such common use-case is to load a BioPAX file into Cytoscape (Shannon et al., 2003) for analysis (see ‘Data export’ section in Supplementary Material). The data can also be exported in PDF (see ‘PDF export’ section in Supplementary Material).
Table 2.
Comparison of data availability at different databases
| S. No | Data category | SGMP | Mousecyca | KEGG | Reactome | DOQCSb |
|---|---|---|---|---|---|---|
| 1 | Interaction details | Yes | No | No | Yes | No |
| 2 | Signaling reactions | Yes | No | No | Yes | No |
| 3 | Quantitative datac | Yes | No | No | No | Yes |
| 4 | Gene regulation | Yes | No | No | No | No |
| 5 | Biochemical pathways | Yesd | Yes | Yes | Yes | Yes |
| 6 | Publicatione | Yes | No | No | Yes | No |
| 7 | Exports | |||||
| BioPAX | Yes | Yes | Yes | Yes | No | |
| SBML | Yes | No | No | Yes | Yes |
aMousecyc (Evsikov et al., 2009) is representing metabolic pathways.
bDOQCS includes quantitative modeling data.
cKinetic/quantitative data is associated with reactions, if available in published papers.
dTransitions associated with a given protein are displayed as a network map.
ePublished data is assigned with Digital Object Identifier.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the present and past SGMP editorial and advisory board members and previous application developers.
Funding: SGMP is funded by NIH/NIGMS Grants (GM078005-05) to UC San Diego.
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov AV, et al. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkind JS. Cell growth control by G protein-coupled receptors: from signal transduction to signal integration. Oncogene. 1998;17:1331–1342. doi: 10.1038/sj.onc.1202186. [DOI] [PubMed] [Google Scholar]
- Hariharaputran S, et al. The Database of Quantitative Cellular Signaling: management and analysis of chemical kinetic models of signaling networks. Bioinformatics. 2003;19:408–415. doi: 10.1093/bioinformatics/btf860. [DOI] [PubMed] [Google Scholar]
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Joshi-Tope G, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- Li J, et al. The Molecule Pages database. Nature. 2002;420:716–717. doi: 10.1038/nature01307. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.