Abstract
Summary: OTUbase is an R package designed to facilitate the analysis of operational taxonomic unit (OTU) data and sequence classification (taxonomic) data. Currently there are programs that will cluster sequence data into OTUs and/or classify sequence data into known taxonomies. However, there is a need for software that can take the summarized output of these programs and organize it into easily accessed and manipulated formats. OTUbase provides this structure and organization within R, to allow researchers to easily manipulate the data with the rich library of R packages currently available for additional analysis.
Availability: OTUbase is an R package available through Bioconductor. It can be found at http://www.bioconductor.org/packages/release/bioc/html/OTUbase.html.
Contact: msettles@uidaho.edu
1 INTRODUCTION
New sequencing technologies, such as Roches 454 pyrosequencing, have made it possible to sequence large amounts of DNA quickly. These new capabilities are being used to sequence portions of marker genes that allow investigators to determine which organisms are present within a sample. This is especially useful for the study of microbes. Microbes are often difficult to culture. Therefore, culture-independent methods are used to determine the composition of microbial communities. The 454 16s amplicon sequencing is one such culture-independent method (Hugenholtz, 2002).
There are currently two primary approaches used to analyze environmental community amplicon data. The first approach uses a database of identified sequences to train an algorithm to classify a set of unknown sequences, such as 454 sequence reads. A widely used example of this is the RDP classifier (Wang et al., 2007). This type of analysis labels each sequence with a taxonomic classification. A classification approach has the advantage of producing taxonomic names for each amplicon. However, it is dependent on the accuracy of the algorithm and the quality of the database used to train the algorithm.
A second approach to the analysis of amplicon data is to cluster the sequences based on overall sequence similarity (Schloss et al., 2009). These clusters represent operational taxonomic units, (OTUs). The OTU approach potentially decreases the biases of using an incomplete database to classify the sequences. However, the number of OTUs may be inflated by sequencing error and the taxonomic identity of the microbes in the sample remains unresolved (Kunin et al., 2010).
Programs are available to perform both of these types of data analysis. In particular, some programs or pipelines are able to both cluster and classify sequences. These include Mothur (Schloss et al., 2009), the RDP pipeline (Cole et al., 2009), QIIME (Caporaso et al., 2010), and PANGEA (Giongo et al., 2010). These programs are also able to perform some limited downstream analysis of the data.
OTUbase is an R infrastructure package that imports OTU or classification files produced by these programs along with the experiment metadata and sequence data. The imported data is stored in a structure that is easy to access and manipulate. The researcher is able to quickly summarize and visualize the data. More importantly, the data is abstracted from the raw data. This abstraction allows for the development of novel analysis techniques using the power of the R statistical programming environment. The structure of OTUbase is general enough to be applicable to all amplicon types, including 16s, 18s and IGS regions, or other targeted genes. Additionally, OTUbase is not restricted to any specific sequencing technique. Any data that can be clustered or classified into OTUs may be explored using OTUbase. This may include data from older Sanger sequencing and newer aproaches such as Illumina sequencing.
2 FEATURES
OTUbase is an R package available through Bioconductor (http://www.bioconductor.org/). The package includes two types of S4 classes, OTUset and TAXset, that organize and structure OTU and classification (taxonomic) data, respectively. S4 classes are a versatile data structure available in R. These classes include slots, or compartments within the class that hold distinct parts of the data. There are many types of data associated with typical community amplicon experiments. These data may include sample metadata, sequencing reads and quality, sequence memberships in OTUs, and sequence classifications. The classes provided by OTUbase organize these data into arrays.
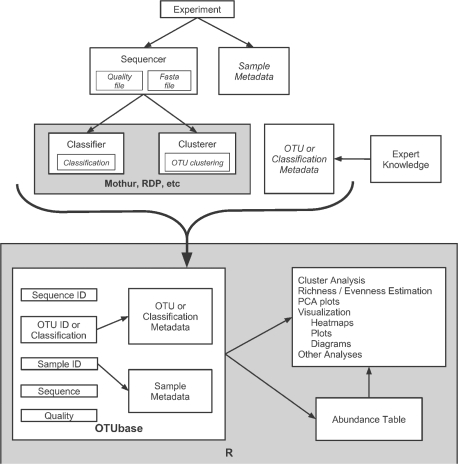
The OTUset class type in OTUbase is designed for OTU related analyses. It includes slots for: the read identifier, the read's DNA sequence, the DNA sequence quality of the read, the sample identifier the read comes from, and the OTU identifier the read belongs to (Fig. 1). These slots are linked together by position. For example, the first sequence ID is linked to the first sequence in the sequence slot and the first sample ID in the sample ID slot. In addition to these slots, two slots hold metadata. The first metadata slot is for sample metadata that is linked to a corresponding sample identifier. The last slot holds OTU metadata and is linked to the OTU identifier slot. The OTU metadata slot can be used to hold OTU classification information, for example.
Fig. 1.
Organization of data within OTUbase. Data analysis starts by either clustering or classifying the reads obtained from the sequencer with an extermal application. The clustering or classification results are collected by OTUbase along with read and quality data and sample metadata. OTUbase then organizes the data into an R object. The library of R tools can then be used to analyze and visualize the data.
The TAXset class type provided in OTUbase is designed for classification data. This class is similar to the OTU class with the key difference being that the slot for the OTU identifier is replaced by a slot for the classification. The OTU metadata is replaced by a taxonomy metadata slot, which may hold additional information about specific taxonomic categories, such as family, order, etc.
In addition to providing structure and organization for amplicon data, OTUbase makes it easy to import data into OTUbase objects. OTUbase is able to read the output files produced by both the RDP classifier and Mothur. In general OTUbase is able to import large datasets quickly. Memory and computation limitations that arise in the analysis of amplicon data are much more important during the classification or clustering of sequences into OTUs then in the downstream analysis. Consequently, OTUbase can handle large datasets quickly and efficiently when the optional sequence and quality data is not included. It is expected that the majority of users will not find sequence data necessary. This capability is included, however, due to the possible use of this data in the development of new analysis techniques.
The key strength of OTUbase is that it interfaces easily with the rich library of packages already available in R for data exploration, visualization and statistical analyses. For example, OTUbase can produce abundance data that is accepted as input to packages that provide ecological data analyses [e.g. vegan from the CRAN project (Oksanen et al., 2010)]. By providing a structure and organization for amplicon data within R, OTUbase enables the efficient development of new analyses and visualization techniques.
A detailed example workflow can be found in the vignette included with the OTUbase package. This example uses data originally presented in Sogin et al., 2005. A typical workflow will include data import and the generation of an abundance table using the functions readOTUset and abundance. This abundance table can then be used in many analyses such as richness calculations (estimateR from the vegan package) and cluster analyses (clusterSamples). The data may be visualized using R commands such as heatmap. OTUbase allows an abundance table to be generated based on any column in the metadata. This makes OTUbase a powerful tool for data exploration. OTUbase also includes functions to manipulate OTU data. The subsetting function subOTUset is able to quickly generate subsets of the complete dataset. This allows the user to focus analyses on interesting subsets of the data.
3 CONCLUSION
OTUbase organizes and structures data associated with community amplicon experiments into R classes, which allows the researcher to easily visualize and manipulate the experimental data. OTUbase has a number of functions that allow OTUbase to import data from the commonly used Mothur package and the RDP classifier. It also includes functions that enable the user to easily use existing R packages to perform complex analyses and visualizations.
Funding: Bioinformatics facilities and some student support were provided by National Institutes of Health/National Center for Research Resources (P20RR16448, P20RR016454).
Conflict of Interest: none declared.
REFERENCES
- Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giongo A, et al. PANGEA: pipeline for analysis of next generation amplicons. The ISME Journal. 2010;4:852–861. doi: 10.1038/ismej.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:reviews0003.1–reviews0003.8. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, et al. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]