Abstract
Summary: Rule-based modeling (RBM) is a powerful and increasingly popular approach to modeling intracellular biochemistry. Current interfaces for RBM are predominantly text-based and command-line driven. Better visual tools are needed to make RBM accessible to a broad range of users, to make specification of models less error prone and to improve workflows. We present RULEBENDER, an open-source visual interface that facilitates interactive debugging, simulation and analysis of RBMs.
Availability: RULEBENDER is freely available for Mac, Windows and Linux at http://rulebender.org.
Contact: faeder@pitt.edu; marai@cs.pitt.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Rule-based modeling (RBM) enables precise and concise specification of biochemical models by representing molecules as structured objects and molecular interactions as rules for transforming the attributes of these objects. The two most common languages for specifying RBMs, κ (Feret et al., 2009) and the BioNetGen language (Faeder et al., 2009), have nearly identical syntax and semantics based on graphs and graph rewriting. Each language and associated modeling framework has been used to construct a wide variety of models of biochemical processes. In this article, we focus on the BioNetGen framework.
While the potential benefits of RBM to biology are outstanding, building an RBM and detecting and correcting modeling errors (i.e. debugging) can be tedious and frustrating. RBMs are typically defined by the user via a text file. The user defines a set of molecules and proceeds to write rules governing their interactions derived from specific knowledge about the system. Although individual rules are easy to write, it is often difficult to fully grasp the implications of a set of rules; the challenge in grasping the global perspective is particularly acute when trying to understand models written by different researchers. This problem complicates debugging and reduces the accessibility of RBM, especially for users with limited programming experience.
Simulating and analyzing RBMs pose additional challenges. Once a working model has been defined, an RBM can be simulated using a number of different approaches including ordinary differential equations, stochastic simulations or particle-based stochastic simulations (Colvin et al., 2010; Danos et al., 2007; Sneddon et al., 2011). The typical workflow relies on an external plain text editor, command console and external plotting tools for displaying simulation results, which is inconvenient because it requires modelers to switch between different tools over repeated cycles of model editing and simulation. The process gets further complicated when exploring alternative simulations and models.
Here we present RuleBender, an integrated development environment that aims to facilitate the construction, simulation and analysis of RBMs through interactive editing, syntax checking, visualization and integrated execution.
2 RULEBENDER
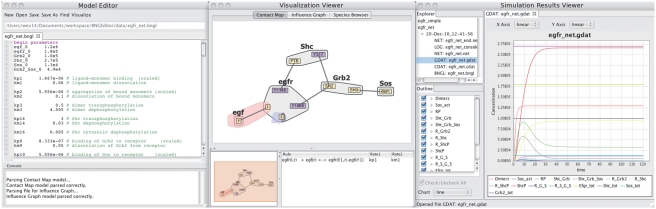
RuleBender provides a complete development environment for RBM. RuleBender is freely available and open source [Java with SWT, Prefuse (http://prefuse.org) and JFreeChart (http://www.jfree.org/jfreechart) libraries.] RuleBender's main features include syntax checking, syntax highlighting, visual global model exploration, integrated execution of simulations, support for multiple simulation modules, simulation journaling, interactive plotting including comparison of multiple datasets and parameter scanning. The three main interface windows provide support for RBM construction and simulation through interactive editing, visual model exploration and results analysis capabilities, respectively, as shown in Figure 1.
Fig. 1.
The RULEBENDER interface with Model Editor window including Console for simulation (left), a Visualization Viewer window (middle) and a Simulation Results Viewer window (right). RULEBENDER's main features include syntax checking, syntax highlighting, visual global model exploration, integrated execution, support for multiple simulation modules, simulation journaling, interactive plotting including comparison of multiple datasets and parameter scanning.
Model browsing and editing: the Model Editor window (Fig. 1, left panel) in RuleBender consists of a toolbar, a tabbed editor pane and a console that displays output of simulations and other tasks performed on the model. In addition to standard text editor capabilities, the editor provides a number of useful features for creating and editing RBMs in the BioNetGen Language (BNGL) format. These features include syntax highlighting, code folding, error highlighting and tabbing to allow multiple files to be open at the same time. The editor also provides a BNGL model template that expedites model construction. A detailed description of the BNGL language is given in Faeder et al. (2009); an example rule is shown in Supplementary Figure S1. The editor window could easily be adapted to support other RBM languages.
Model exploration and debugging: to further support model understanding and debugging, the Visualization Viewer window (Fig. 1, middle panel) provides both global and local views of the model currently loaded in the Model Editor window. The views are automatically generated from models.
The first global view is the Contact Map, which is a graph representation of the molecules and rules in the model. Molecules (gray shapes in Fig. 1) are the basic units of a BioNetGen model and may contain components (yellow and purple rectangles in Fig. 1), which represent internal and binding states of the molecule. Edges in the map show that two components may be bound through the action of a rule. Selecting an edge brings up a list of all rules that create or destroy bonds between the linked components. Selecting one of these rules brings up bubble sets (Collins et al., 2009) that highlight separately the components modified by the rule (called the reaction center) and the components whose states are required to have specified values for the rule to be applied (called the reaction context), as shown in Figure 1 and Supplementary Figure S2. The Contact Map provides global organization for a rule set and enables the user to browse the rules in a systematic fashion.
An alternative global view is the Influence Graph, which shows interactions between the rules. A rule can activate (inhibit) another rule by producing (consuming) a reactant required by the other rule. Selecting a rule node displays the rule text in the lower right information pane of the visualization window (Supplementary Fig. S3). Both visualizations enable quick identification of ‘orphan’ molecules or rules that do not interact with other molecules/rules, thus further supporting understanding and debugging of the models.
Simulation and results analysis: BioNetGen simulator commands can be performed on the model currently loaded in the Editor window. If the model is valid, simulation results are displayed in the Simulation Results Viewer. Otherwise, syntax errors and other output are reported via the console in the lower section of the editor window. Parameter scanning (Supplementary Fig. S4), which allows the researcher to interactively see the effects of varying the value of a single model parameter, is also supported.
The simulation results, along with the model files, parameters scan results, etc. are stored and managed by RuleBender through a journaling system that allows users to distinguish simulation results. The Simulation Results Viewer contains an explorer in the upper left quadrant of the window (Fig. 1, right panel), which makes it easy to retrieve the exact version of the model that produced a specific set of results.
Network files generated from a model may be displayed with syntax highlighting (Supplementary Fig. S5). Charts in linear or log scale show the time series for concentrations of chemical species and observables as seen in the far right window of Figure 1. Any number of species and observables can be compared in the same chart (Supplementary Fig. S6). Furthermore, multiple simulation runs can be compared in order to analyze the effects of changing the model (Supplementary Fig. S7).
A species and observables browser is also provided (lower left of the results viewer in Fig. 1). Graph-based models of species and observables, similar to the Contact Map, are displayed by selecting an observable in the Simulation Results Viewer (Supplementary Fig. S8).
3 CONCLUSION
RuleBender is a powerful tool for the development of RBMs that makes RBM accessible to users with a wide range of computational experience, while providing a uniform interface across computing platforms. The support of RBM creation, debugging and interactive visualization expedites the RBM learning process and reduces model construction time; while built-in model simulation and analysis with multiple linked views streamline the execution and analysis of newly created models and generated networks. A development cycle that includes close interaction with expert users allows RuleBender to better serve the needs of the systems biology community.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks are also due to Yao Sun, John Sekar and Bill Hlavacek.
Funding: National Science Foundation (grant numbers CCF-0829788, IIS-0952720); National Institutes of Health (grant numbers GM-076570, UL1-RR024153).
Conflict of Interest: none declared.
REFERENCES
- Collins C, et al. Bubble sets: revealing set relations with isocontours over existing visualizations. IEEE Trans. Vis. CG. 2009;15:1009–1016. doi: 10.1109/TVCG.2009.122. [DOI] [PubMed] [Google Scholar]
- Colvin J, et al. RuleMonkey: software for stochastic simulation of rule-based models. BMC Bioinformatics. 2010;11:404. doi: 10.1186/1471-2105-11-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danos V, et al. Scalable simulation of cellular signaling networks. Lect. Notes Comput. Sci. 2007;4807:139–157. [Google Scholar]
- Faeder JR, et al. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol. Biol. 2009;500:113–168. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- Feret J, et al. Internal coarse-graining of molecular systems. Proc. Natl Acad. Sci. USA. 2009;106:6453–6458. doi: 10.1073/pnas.0809908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon MW, et al. Efficient modeling, simulation and coarse-graining of biological complexity with NFsim. Nat. Methods. 2011;8:177–183. doi: 10.1038/nmeth.1546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.