Abstract
Motivation: Well-annotated gene sets representing the universe of the biological processes are critical for meaningful and insightful interpretation of large-scale genomic data. The Molecular Signatures Database (MSigDB) is one of the most widely used repositories of such sets.
Results: We report the availability of a new version of the database, MSigDB 3.0, with over 6700 gene sets, a complete revision of the collection of canonical pathways and experimental signatures from publications, enhanced annotations and upgrades to the web site.
Availability and Implementation: MSigDB is freely available for non-commercial use at http://www.broadinstitute.org/msigdb.
Contact: gsea@broadinstitute.org
1 INTRODUCTION
Microarrays and other high-throughput genomic technologies typically produce long lists of potentially interesting genes, which are not always easily interpreted. Recognizing the importance of coordinately expressed sets of genes, our seminal paper (Mootha et al., 2003) introduced Gene Set Enrichment Analysis (GSEA) to discover metabolic pathways altered in human type 2 diabetes mellitus. GSEA and other analytical enrichment tools summarize genomic data in prioritized lists of higher-level biological features. As underscored by a recent survey of 68 enrichment tools, they critically depend on ‘backend annotation databases’ (Huang et al., 2009). Typically, such databases focus on a particular domain of knowledge or annotation procedure. For example, Gene Ontology (GO) (Ashburner et al., 2000) represents a hierarchy of controlled terms to describe individual gene products, while TRANSFAC (Matys et al., 2006) stores information about transcription factor binding sites. A growing number of databases obtain sets from gene expression signatures reported in the literature. These include SignatureDB (Shaffer et al., 2006), GeneSigDB (Culhane et al., 2009), CCancer (Dietmann et al., 2010) and L2L and LOLA (Cahan et al., 2007).
Molecular Signatures Database (MSigDB) differs from these resources in several distinguishing aspects. (i) MSigDB is explicitly designed to provide gene sets for enrichment analysis methods. As such, it is natively and seamlessly integrated with our GSEA software (Subramanian et al., 2005). (ii) MSigDB covers a substantially more diverse and wider range of gene set sources and types. These include signatures extracted from original research publications, and entire collections of sets derived from specialized resources such as GO, KEGG (Kanehisa and Goto, 2000), TRANSFAC and L2L. (iii) MSigDB gene sets are acquired both through manual curation and by automatic computational means, whereas other databases emphasize only one of these approaches. (iv) Finally, MSigDB contains the largest number of gene sets overall.
The initial MSigDB database, released in 2005 with GSEA software, contained 1325 sets. In contrast, MSigDB 3.0, released in September 2010, includes 6769 sets and a richer set of annotations. Here, we describe the MSigDB 3.0 sets in more detail and the accompanying online resource.
2 RESULTS
Gene set collections: gene sets in MSigDB 3.0 are organized into five collections according to their derivation:
C1: Genes located in the same chromosome or cytogenetic band.
C2: Gene sets representing canonical pathways from pathway resources [including 430 new sets contributed by Reactome (Matthews et al., 2009)], and sets corresponding to chemical and genetic perturbations from 786 scientific publications.
C3: Sets of genes sharing cis-regulatory motifs in their promoter (transcription factor targets) or 3′ UTR (micro-RNA targets) sequences.
C4: Clusters of coexpressed modules defined by computational analysis of large gene expression compendia.
C5: Gene sets corresponding to GO terms.
Table 1 shows the growth of the MSigDB collections and database since the initial release (see also online Release Notes).
Table 1.
MSigDB versions and changes in the number of gene sets
| Gene set category | 1.0 (2005) | 2.5 (2008) | 3.0 (2010) |
|---|---|---|---|
| C1: positional | 319 | 386 | 326a |
| C2: curated (total) | 522 | 1892 | 3272 |
| C2: chemical and genetic perturbations | 50 | 1186 | 2392 |
| C2: canonical pathways | 472 | 639 | 880 |
| C2: uncategorized | 0 | 66 | 0 |
| C3: motifs (total) | 57 | 837 | 836a |
| C3: transcription factor targets | 57 | 500 | 615 |
| C3: micro-RNA targets | 0 | 222 | 221a |
| C3: uncategorized | 0 | 115 | 0 |
| C4: computational | 427 | 883 | 881a |
| C5: GO terms | 0 | 1454 | 1454 |
| MSigDB total | 1325 | 5452 | 6769 |
aDecrease in number due to the removal of sets with too few genes to run GSEA.
Gene set annotations: each MSigDB gene set is a list of genes with relevant annotations and links to external resources. MSigDB focuses on human gene sets. However, we do include sets from some model organisms and gene set annotations include organism identification. We use HUGO gene symbols and, as of version 3.0, human Entrez Gene IDs serve as universal identifiers. These Entrez IDs are guaranteed to be unique and stable, can easily be mapped into a variety of other identifiers and are natively integrated with the GenBank resources of primary nucleic and protein sequences. We also preserve whatever original identifiers were used in the gene set source. All sets have unique database identifiers and names, and include brief and full descriptions. Other annotations depend on the type of gene set. Annotations linking to external resources are especially important as they allow researchers to place the sets in the context of a specific study and facilitate decisions on follow-up experiments.
Gene sets from publications are the most richly annotated. Their annotations include the PubMed ID of the publication, pointers to other gene sets from the same publication, and now also details on the exact table or figure from which the gene set was extracted. For version 3.0, we updated the names of these gene sets to make them more descriptive and standardized and the accompanying brief descriptions to follow a more uniform and consistent format. Other annotation features introduced with version 3.0 include links to source datasets in Gene Expression Omnibus (GEO) (Barrett et al., 2009) and ArrayExpress (Parkinson et al., 2009). Canonical pathway sets include links to the pathway at the source web site.
File formats: MSigDB gene set files are available for download in plain text and XML formats. The plain text files contain simple listings of gene set membership, while the XML files also include the annotations. To ensure reproducibility of GSEA results, older versions of the MSigDB files are always available. Note that users of our GSEA software do not need to download the MSigDB files as the tool directly and automatically retrieves the gene sets.
3 MSigDB ONLINE RESOURCE
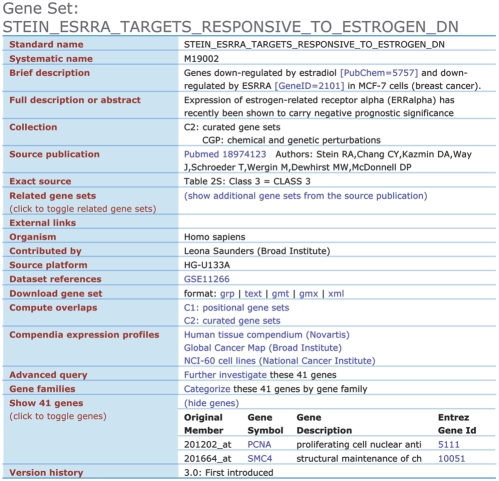
In version 3.0, we updated the MSigDB web site. First introduced in July 2007, the site allows users to view the annotated gene sets and perform simple search and analysis tasks. Each gene set and all of its annotations are presented on a separate web page (Fig. 1). Embedded hyperlinks connect annotations to corresponding external web resources, including PubMed, GEO and ArrayExpress, PubChem and Entrez Gene.
Fig. 1.
A typical gene set page on the MSigDB web site. The list of genes has been abbreviated from 41 to 2 for the purposes of this figure.
The MSigDB web site allows users to find gene sets by searching for keywords in the annotations. The online analysis tools allow users to: (i) compute overlaps between gene sets; (ii) view a heat map of a gene set in one of the reference expression compendia; and (iii) categorize the genes in a set by gene families. Gene families offer a quick view of a gene set by grouping its members into a small number of informative categories. We have updated the gene families and they now include: oncogenes, tumor suppressors, translocated cancer genes, transcription factors, protein kinases, homeodomain proteins, cell differentiation markers and cytokines/growth factors.
ACKNOWLEDGEMENTS
We thank J. Roberston, L. Saunders and L. Kazmierski for gene set collection; H. Kuehn and J. McLaughlin for documentation; and M. Wrobel for web site development.
Funding: National Cancer Institute (5R01CA121941).
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;30:D5–D15. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, et al. Meta-analysis of microarray results: challenges, opportunities, and recommendations for standardization. Gene. 2007;401:12–18. doi: 10.1016/j.gene.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane A, et al. GeneSigDB – a curated database of gene expression signatures. Nucleic Acids Res. 2009;38:D716–D725. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmann S, et al. CCancer: a bird's eye view on gene lists reported in cancer-related studies. Nucleic Acids Res. 2010;38(Suppl):W118–W123. doi: 10.1093/nar/gkq515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Huang W, et al. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Parkinson H, et al. ArrayExpress update – from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]