Abstract
Motivation: Inferring the taxonomic profile of a microbial community from a large collection of anonymous DNA sequencing reads is a challenging task in metagenomics. Because existing methods for taxonomic profiling of metagenomes are all based on the assignment of fragmentary sequences to phylogenetic categories, the accuracy of results largely depends on fragment length. This dependence complicates comparative analysis of data originating from different sequencing platforms or resulting from different preprocessing pipelines.
Results: We here introduce a new method for taxonomic profiling based on mixture modeling of the overall oligonucleotide distribution of a sample. Our results indicate that the mixture-based profiles compare well with taxonomic profiles obtained with other methods. However, in contrast to the existing methods, our approach shows a nearly constant profiling accuracy across all kinds of read lengths and it operates at an unrivaled speed.
Availability: A platform-independent implementation of the mixture modeling approach is available in terms of a MATLAB/Octave toolbox at http://gobics.de/peter/taxy. In addition, a prototypical implementation within an easy-to-use interactive tool for Windows can be downloaded.
Contact: pmeinic@gwdg.de; thomas@gobics.de
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Metagenomics provides a holistic approach to the analysis of microbial communities that overcomes the necessity of isolating single organisms for cultivation (Beja et al., 2000; Rondon et al., 2000). Investigating a mixture of genetic material from the whole spectrum of organisms, researchers can now obtain comprehensive descriptions even of highly diverse communities. Further, comparative metagenomics offers new possibilities for studying the distinguishing characteristics of a wide range of ecosystems, which are shaped by specific combinations of microorganisms. In particular, research on the human microbiome has begun to elucidate the community structures associated with the human body. Important medical perspectives, for instance, arise from comparing gut microbiome profiles to differentiate between healthy and diseased states (Turnbaugh et al., 2009).
To investigate the taxonomic composition of metagenomes, many studies focus on sequencing the 16S rRNA gene (Hugenholtz, 2002), which currently provides the best resolution in terms of the available number of operational taxonomic units in the reference databases (Stach and Bull, 2005). Besides the selectivity of primers (Hong et al., 2009), another difficulty for quantitative analysis arises from the varying copy number of the 16S rRNA gene (Kunin et al., 2008). A pure 16S analysis, however, completely neglects the functional potential encoded in the metagenome. In contrast, whole metagenome sequencing allows simultaneous taxonomic and functional profiling, which provides a deeper insight into the structure of a microbial community.
The taxonomic profiling of whole metagenome sequencing reads is a challenging task, and several techniques have been developed to extract the phylogenetic signal encoded in the sequenced material. To date, all approaches rely on the classification of sequencing reads. In most cases, a supervised binning of sequences according to an assignment to predefined taxonomic categories is performed. Although an unsupervised binning is possible for longer contigs (Teeling et al., 2004), the comparison of metagenomes becomes difficult with the inclusion of unlabeled bins. Methods for the classification of sequencing reads have been based either on homology using sequence similarity or on genomic signatures in terms of oligonucleotide composition. Homology-based methods include the taxonomic evaluation of BLAST hits (Huson et al., 2007; Kosakovsky Pond et al., 2009; Meyer et al., 2008) and phylogenetic analyses of particular marker genes (von Mering et al., 2007; Wu and Eisen, 2008) or protein domains (Krause et al., 2008; Schreiber et al., 2010). Signature-based approaches have been realized based on the correlation of oligonucleotide frequencies (Teeling et al., 2004), machine learning techniques (Diaz et al., 2009; McHardy et al., 2007) and probabilistic models (Brady and Salzberg, 2009; Rosen et al., 2008).
All these methods are highly dependent on read length. For homology-based approaches, the number of significant similarity hits decreases considerably for shorter reads (Wommack et al., 2008). Additionally, the estimation of genomic signatures in sequencing reads becomes increasingly difficult for decreasing read lengths. For sequence lengths below 1000 bp, earlier approaches showed a sharp breakdown in accuracy (McHardy et al., 2007), which has been improved with recent tools (Brady and Salzberg, 2009). All methods require a minimum read length in order to be applicable. In many cases, this condition restricts the use of ultra-short read techniques for metagenome profiling. As a consequence of the length-dependent classification performance, the varying accuracy of taxonomic profiling methods particularly complicates the comparison of metagenomes.
With a rapidly increasing number of sequenced samples, comparative metagenomics faces the problem that many samples are difficult to compare due to different sequencing platforms with varying read lengths and platform-specific sequencing errors. In particular, the read length is highly variable across different platforms and generations of sequencing technologies. Another source of sequence length variability arises from different stages of assembly. Depending on the number of reads and the diversity of the community, for many samples, a varying number of assembled contigs exists. Although homology and signature-based methods perform significantly better on longer contigs, the sample-specific distribution of contig lengths introduces a bias towards more abundant species, which complicates the comparability of samples.
We here present a novel method for taxonomic profiling of metagenomes that is based on mixture modeling. Instead of a classification of sequencing reads based on a read-specific estimate of oligonucleotide frequencies, our method performs an analysis of the total oligonucleotide composition of a sample. The discrete distribution of oligonucleotides is modeled by a mixture of organism-specific oligonucleotide distributions as obtained from sequenced genomes. Taxonomic profiling then means to obtain the organism weights of that mixture from an approximation of the metagenomic distribution. We show that under a varying read length this mixture approach provides a more stable estimation of taxonomic composition than methods based on read classification. In particular, this advantage implies a better comparability of samples across different sequencing platforms. Another advantage of our approach is the computational speed; it is the first profiling approach that allows the analysis of large volumes of sequence data within a few minutes on a single laptop.
2 METHODS
Genomic signatures in terms of oligonucleotide distributions have widely been used for genome-based characterizations of microbial organisms (Bohlin et al., 2009). We here propose the analysis of the oligonucleotide distribution of a metagenome for the taxonomic characterization of the corresponding microbial community.
2.1 Computation of compositional parameters
We model the oligonucleotide distribution of a metagenome by a linear mixture of organism-specific genomic signatures from a reference database. Given the N oligonucleotide probabilities of the metagenomic and genomic signatures as N-dimensional vectors y and xi together with M positive organism weights wi, the metagenomic signature y arises from a convex combination of the database signature vectors xi:
| (1) |
The organism weights are the free parameters of the model and provide the basis for all kinds of taxonomic profiling tasks. To obtain a profile on a particular taxonomic level, all the weights of organisms belonging to the same category on that level are summed to yield the corresponding profile value.
A key question is how to determine the unknown mixture weights if only the metagenomic and genomic signatures are given. In general, it will be impossible to exactly reconstruct the metagenomic signature by some limited amount of genomic database signatures because a large fraction of organisms in the underlying community will not be covered by the available genomes. Therefore, the mixture weights have to be chosen to yield a close approximation of the metagenomic distribution according to some distance measure. The most common way would be to apply the EM algorithm (Dempster et al., 1977) to minimize the Kullback–Leibler divergence between the metagenomic distribution and the mixture approximation. However, the EM algorithm requires an initial estimate of the weights and only converges to a local optimum. Therefore, we here consider a weighted L2-distance measure which gives rise to a convex optimization problem. More specifically, in the N-dimensional space of oligonucleotide probabilities we minimize the normalized squared Euclidean distance between the metagenomic and model signatures. With xij and yj denoting the probability of oligonucleotide j for database organism i and the target (meta)genome, respectively, we use the following error function:
| (2) |
| (3) |
with weight vector w containing positive weights wi ≥ 0. The standard deviation σj of dimension j can be estimated from oligonucleotide frequencies observed for the M database genomes. Minimization of the above error gives rise to a convex quadratic programming problem (QP), which can be solved by standard optimization tools. We used a corresponding function from a MATLAB SVM toolbox (Canu et al., 2005).
In general, the solution is unique if the dimensionality of the signature vectors exceeds the number of database organisms used for the approximation. For uniqueness, the database signature vectors have to be linearly independent such that no organism-specific oligonucleotide signature can be perfectly (without error) reconstructed by a convex combination of the other signature vectors. This kind of non-redundancy condition geometrically means that all signature vectors have to be vertices of their convex hull. In practice, however, a redundancy elimination of signature vectors on the organism level is actually not necessary. For very close signatures, the solution is not unique only for the associated weights of the corresponding organisms. This ambiguity does not affect less specific phylogenetic levels, since here the organism-specific weights of closely related organisms are aggregated. In contrast to homology-based methods, the use of many closely related organisms does not imply a profiling bias toward these organisms.
2.2 FOU error and profile divergence
To map the value of the above approximation error E to an interpretable scale between 0 and 1, we compute an additional error measure which we refer to as the fraction of oligonucleotides unexplained (FOU). We define the FOU as the total one-sided error of predictions in the oligonucleotide frequency space
| (4) |
where yj is the relative frequency of oligonucleotide j from the observed metagenomic signature and ŷj is the corresponding prediction resulting from the estimated combination of genomic signatures. The FOU measures the fraction of metagenomic DNA that cannot be explained by the mixture of genomic signatures. It equals the sum of deviations resulting from ‘underpredicted’ relative frequencies that are lower than the corresponding observed metagenomic frequencies. The same error is necessarily obtained from the sum of deviations resulting from ‘overpredicted’ frequencies due to the unit sum of all probabilities of a signature.
In general, the FOU error of the Taxy method cannot be used for quantification of phylogenetic novelty because the divergence between the observed profile and the mixture model is forced to be minimized by the approximation method. Only in cases where even on higher phylogenetic levels no related organisms/signatures exist in the database, an increased FOU may indicate novel organisms. Analogously to analyses based on sequence similarity, this case will be hard to distinguish from a degradation of sequence quality. On the other hand, the unusual case where all metagenomic sequences refer to the known database organisms can easily be detected by a vanishing FOU.
Because the FOU error is defined as a distance between discrete distributions, also the divergence between two taxonomic profiles on the same phylogenetic level can be measured in that way. In this case, the absolute deviations in Equation (2) arise from the taxon-specific fractions on a particular level. In order to be able to compare discrete distributions, the corresponding categories have to be the same for all profiles and the weights of a profile have to sum up to some unique constant. In particular, when read classification methods are used, the comparison cannot include unclassified reads, i.e. reads that have not been assigned to some taxonomic category. In the following, we use the profile divergence to measure the deviation of a predicted profile from a reference profile. Similar to the FOU error, also the profile divergence can be interpreted in terms of a percental deviation.
2.3 Genomic and metagenomic signatures
The oligonucleotide probabilities can be estimated by counting exact DNA word matches of a certain length. For a word length k, the estimate comprises 4k relative frequencies according to the number of different DNA words. In the case of fragmented data, the estimate was established by summing up the oligonucleotide counts of all individual reads, contigs or chromosomes. To ensure that read orientation did not affect the taxonomic prediction, oligonucleotide counts of the reverse complement were added to every metagenomic and reference signature. This scheme implies a loss of information, which approximately halves the number of distinguished oligonucleotides. Finally, the oligonucleotide counts of each signature were normalized to relative frequencies.
For the genomic reference signatures, we chose the KEGG organism database (Kanehisa and Goto, 2000) as of March 2010, providing 1013 fully sequenced prokaryotic genomes. The NCBI taxonomy database was used for taxonomic annotation of the genomic signatures. As outlined in Section 2.1, the high number of reference organisms suggests a minimum word length of k = 6 to provide a unique solution for the mixture weights. In this case, the combination of two read orientations implies ~2000 non-redundant dimensions within a 4096-dimensional signature vector. For most metagenomic datasets, also longer words may be considered. Because the memory requirements for storing all reference signatures increase with k, we limited the word length to a maximum value of k = 8 to enable the computation on most of the current notebook and desktop architectures. For the experimental analyses in the following study, we used a medium word length of k = 7.
3 RESULTS
3.1 Profile comparison
First, we evaluated our method, which we refer to as ‘Taxy’ in the following, on the Northern Schneeferner glacial ice sample (Simon et al., 2009). This sample was used in Schreiber et al. (2010) to compare the predictions of the Treephyler tool with the results of CARMA (Krause et al., 2008), Phymm (Brady and Salzberg, 2009) and a classical 16S analysis. For the comparison with Taxy, we additionally used the homology-based web tool Galaxy (Kosakovsky Pond et al., 2009) for taxonomic profiling analysis. As suggested by the original study, we used an intermediate level between phylum and class rank for comparison. The sequence data from the glacier sample comprise 1 076 539 pyro-sequencing reads with an average 200 bp read length.
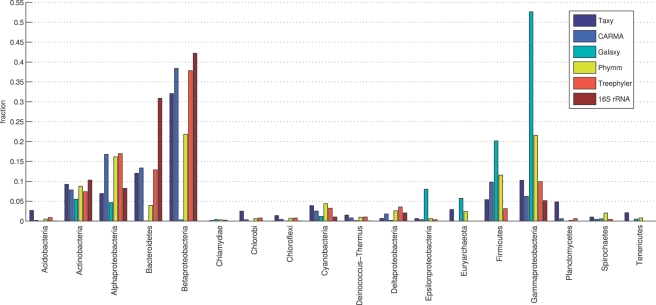
The approximation error (FOU error, see Section 2) of Taxy based on heptamer signatures was 0.021 and 0.02 for the QP and EM method, respectively. These low values indicate a good approximation of the metagenomic oligonucleotide distribution by the reference signatures. As shown in Figure 1, the taxonomic distribution predicted by Taxy was largely congruent with CARMA and Treephyler. For Alphaproteobacteria, Taxy showed a clearly lower level, which was closer to the 16S analysis. The profile divergence of the Taxy-based abundances from the 16S profile was 32.7 percentage points (p.p.). CARMA, Treephyler, Phymm and Galaxy diverged by 24.0, 25.3, 48.9 and 83.1 p.p., respectively, from the 16S-based prediction. Here, Galaxy showed large peaks for the Gammaproteobacteria and Firmicutes phyla, which can be explained by the overrepresentation of associated organisms in nucleotide databases. The maximum common difference between the tool-based predictions and the 16S profile occurred in the Bacteroidetes phylum. The Taxy prediction differed from the 16S result by 19.6 p.p. in this phylum, while CARMA, Treephyler, Phymm and Galaxy differed by 17.5, 18.0, 26.9 and 30.8 p.p., respectively. In many phyla, Taxy constituted a compromise between the Phymm and the CARMA/Treephyler-based predictions. While Taxy was not as close to the 16S profile as CARMA and Treephyler, it was closer to the 16S level than Phymm and Galaxy. However, the large Bacteroidetes prediction divergence of all tools with respect to the 16S analysis highlights the difficulty of establishing a gold standard for the taxonomic profiling of metagenomic data. Note that the comparison with a 16S analysis is not unquestionable due to the varying copy number of the corresponding marker gene. An additional problem arises from a possible bias of the 16S primers which has been reported to favor the amplification of Bacteroidetes 16S rRNA in human gut samples (Gill et al., 2006). Finally, marker gene counts measure organism frequency rather than genomic DNA content, which is measured by the above tools.
Fig. 1.
Phylum/class-level taxonomic profiles of the Norther Schneeferner metagenome as obtained from Taxy, CARMA, Galaxy, Phymm and Treephyler in comparison with a 16S rRNA profile.
In addition, we used the ‘simHC’ dataset introduced in Mavromatis et al. (2007) to measure the accuracy of taxonomic profiling methods on a simulated high-complexity community (see Supplementary Material). Besides the limited realism of a simulated metagenome, another problem arises from possible overlaps between reference or training organisms used by the profiling tools and the database organisms which have been used to construct the simulated metagenome sequences. To reduce this overlap, we removed all reference/training organisms belonging to genera which are present in the simHC data from the tools. Because we were not able to fully exclude these organisms from the CARMA and Treephyler prediction engines only Taxy, Phymm and, in addition, homology-based results from the Galaxy server (Kosakovsky Pond et al., 2009) were used in this evaluation. Besides the removal of all genus-level overlaps with simHC, we chose the remaining set of 654 reference organisms to be equal for all three tools to ensure comparability of the corresponding methods.
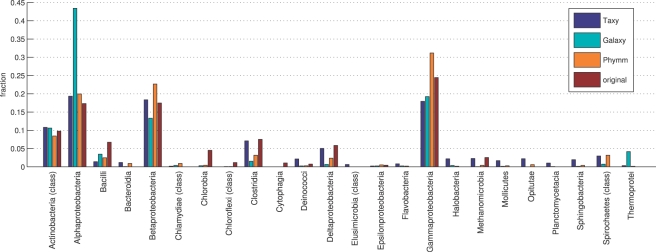
In Figure 2, the estimated profiles at class level for all three methods are shown together with the original profile according to the known composition of the simulated metagenome. In most taxonomic categories, the predictions agree well with the original profile. An exceptional deviation can be observed for the Galaxy predicted fraction of Alphaproteobacteria, which exceeds the original fraction by 26.1 p.p. To investigate whether this peak arises from the particular configuration of the homology search step within Galaxy, we used different BLAST parameter settings and repeated the analysis. However, the high deviation for the Alphaproteobacteria class was observed for all configurations. In contrast, the Taxy and Phymm predictions for Alphaproteobacteria were close to the original. In total, the profile divergences of Taxy, Phymm and Galaxy were 20.7, 22.5 and 33.84 p.p., respectively. We also analyzed the more general phylum level, where the corresponding values were 17.9, 18.3 and 17.2 p.p. The divergences for Taxy, Phymm and Galaxy on the more specific order level were 57.6, 46.8 and 56.4 p.p., which indicates that an estimation of the taxonomic distribution on this level is generally difficult within the chosen simHC setup.
Fig. 2.
Class-level taxonomic profiles of the simHC simulated metagenome as obtained from Taxy, Galaxy and Phymm in comparison with the original profile according to the known fractions of taxa.
3.2 Read length dependence
For comparative metagenome analyses, it is highly desirable that read length does not affect the estimation of taxonomic composition. Therefore, we also investigated the variation of profiling results with respect to a varying read length. For that purpose, we compared the read length dependence of Taxy, Galaxy and Phymm on the hypersaline microbial mat samples introduced in Kunin et al. (2008). The dataset includes 129 147 unassembled Sanger sequencing reads (~700 bp read length) from 10 samples according to different depth layers of the mat. For reasons of space, an analysis of the taxonomic profiles of all depth layers as estimated by the three methods on full-length reads can be found in the Supplementary Material.
We studied the effect of read length dependence by measuring the divergence between profiles obtained from full length data and different versions of fragmented data. We chose three different fragment lengths (350, 175 and 80 bp) to reflect the range of read lengths provided by current sequencing technologies. The fragmentation was implemented through a simple read splitting, which cut the original reads into fragments that approximately met the desired read lengths (see Supplementary Material). Because the fragments did not overlap, this scheme implied a loss of oligonucleotide information around the fragment border. For a word length of 7 bp and an average read length of 80 bp, about 10% of the original heptamers in the full-length reads were lost.
The results in Table 1 indicate that, on average, the phylum level divergence of the Taxy tool was at least one order of magnitude lower than the corresponding Galaxy and Phymm results. Thereby, the overall profile divergences of Galaxy and Phymm were rather similar. All methods exhibited a correlation between the fragment length and profile divergence. Both Galaxy and Phymm exhibited a stronger than average divergence in the top three layers, while Taxy did not diverge above average in these layers (see Supplementary Table S1). The Taxy method showed a very even and predictable small divergence due to the above-mentioned loss of oligonucleotide information in the fragments. In contrast, the Galaxy and Phymm profiles showed a large variation of the divergence with maximum deviations of 14.0 p.p. (Galaxy) and 19.3 p.p. (Phymm) for the 80 bp fragments in the first layer.
Table 1.
Profile divergence between results obtained from full and fragmented reads of the hypersaline microbial mat samples
| Method | Read length (bp) | Mean | Max. | Min. |
|---|---|---|---|---|
| 350 | 0.21 | 0.29 | 0.18 | |
| Taxy | 175 | 0.34 | 0.49 | 0.22 |
| 80 | 0.64 | 0.79 | 0.47 | |
| 350 | 5.51 | 7.95 | 3.54 | |
| Galaxy | 175 | 6.09 | 8.01 | 4.26 |
| 80 | 8.17 | 13.99 | 4.98 | |
| 350 | 2.55 | 4.32 | 2.00 | |
| Phymm | 175 | 6.30 | 10.91 | 5.04 |
| 80 | 10.32 | 19.32 | 5.29 |
Statistics in terms of the mean, maximum and minimum values over all 10 depth-specific samples for the Taxy, Galaxy and Phymm results.
The length dependence of fragment classification methods in particular can be problematic if partially assembled data have to be analyzed. With today's high-throughput sequencing technologies, even microbial communities with a medium complexity allow to assemble a large fraction of the original reads into longer contigs. On the one hand, the classification of longer contigs is more reliable than assigning the original short reads to taxonomic categories. On the other hand, a significant bias may arise from the fact that the probability that two reads can be assembled increases with the abundance of the corresponding organism. We analyzed the profile divergence between partly assembled data and simulated short read data for a human gut sample (Kurokawa et al., 2007) where we compared Taxy with WebCARMA (Gerlach et al., 2009) and the NBC web server tool (Rosen et al., 2011) on phylum level (see Supplementary Material). Because of the widely varying sequence length, for all methods, we compared the amount of DNA (bp) attributed to phylum level categories and not the number of sequences assigned to these categories. While WebCARMA and NBC showed a large deviation for the most abundant phyla, reaching 12.9 and 21.8 p.p. in the Bacteroidetes phylum, the deviations of our mixture approach were below 0.15 p.p. in all categories.
3.3 Run time
The Taxy runtime for the analysis of the 239.7 MB Northern Schneeferner dataset on a single core of an AMD Opteron (2.4 GHz) processor was 7.5 s. The single core run times for Treephyler, Phymm and CARMA were 12 h, 30 h and extrapolated 696 h, respectively. Considering the computational cost for analysis of the hypersaline microbial mat data, the Taxy run time on a single core of a 2.66 GHz Intel processor for the analysis of all 10 sets (84.35 MB) was 9 s, while Phymm and the Galaxy analyses required about 69 h (CPU time) and 95 min, respectively. The hardware requirements for a Taxy analysis are exceptionally low: we were able to process the complete 1.7 GB sequence file from the Sargasso Sea sample (Venter et al., 2004) on a notebook with a single core 1.4 GHz Pentium (M) CPU and 760 MB RAM under Octave 3.2.4 in 95 s.
3.4 Implementation
The taxonomic profiling algorithm described above was implemented using the MATLAB programming language. A MATLAB toolbox containing the computational routines, precomputed oligonucleotide signatures for 1013 reference organisms and documentation can be downloaded from http://gobics.de/peter/taxy. The toolbox allows the profiling of a given metagenome sample (in multiple FASTA format) on different taxonomic levels (phylum, class, order, family, genus). The output comprises histogram bar plots of the sample-specific taxonomic composition as well as comma-separated value (CSV) files for detailed analysis of the profiles with spreadsheet software such as Microsoft Excel. The toolbox code is also executable with recent versions of Octave (3.0 and above, http://www.gnu.org/software/octave/ http://www.gnu.org/software/octave/)), a freely available MATLAB-like software environment. The toolbox was tested under Microsoft Windows and Linux and can easily be used on other platforms.
In addition, we provide an implementation of the proposed method as part of the freely available Taxy tool for Windows. The Taxy tool prototype includes the precomputed taxonomic profiles of 256 metagenomes based on sequence data obtained from the CAMERA web site (Seshadri et al., 2007). Here, the 256 samples with the corresponding profiles can also be used for comparative analysis. Furthermore, the graphical user interface of the Taxy tool allows the user to inspect the sample metadata and the taxonomic profile as estimated by the mixture modeling method (see also Supplementary Material).
4 DISCUSSION
As do all other methods for taxonomic profiling of whole metagenome sequences, our method crucially depends on the range of microbial reference genomes available in current databases. The phylogenetic coverage of these genomes directly determines the limits for the achievable taxonomic resolution. Because genome databases still suffer from a significant bias toward certain culturable organisms, an important impact on profiling performance is expected from recent efforts to broaden the range of sequenced organisms (Wu et al., 2009). Obviously, all profiling methods will largely benefit from a more even sampling of the microbial world.
In several cases, it may be useful to include eukaryotic organisms in the analysis of the taxonomic composition. In particular, the inclusion of a known host genome provides a straightforward way to identify the fraction of host-specific DNA in a sample. In this context, the modular architecture of Taxy allows an easy integration of eukaryotic genomes in the database of signature vectors. Preliminary results with 28 fully sequenced eukaryotic organisms added to the database show that Taxy was able to benefit from eukaryotic signatures in the analysis of an insect herbivore microbiome dataset (Suen et al., 2010), which is characterized by a high proportion of eukaryotic DNA (see Supplementary Material).
The main advantage of the Taxy approach over all existing methods is the inherent read length invariance of the composition estimates. First of all, this property makes it possible to fully utilize ultra-short reads from all high-throughput sequencing technologies. Secondly, without losing comparability, it allows the use of datasets with heterogeneous sequence lengths, which for instance arise from a combination of raw reads and assembled contigs. In this case, the method is also robust with respect to erroneous assemblies because no taxonomic assignment of contigs is actually performed. Finally, Taxy facilitates the comparability of data obtained from different sequencing platforms. This advantage is of particular importance because the heterogeneity of sequencing technologies and the associated read lengths is still increasing. Read length invariance, however, cannot cope with the variability of metagenomic protocols, which affect the preparation of samples before sequencing and which can severely degrade the comparability of data. Other sources of variability, for instance, include the cloning bias of Sanger sequencing or particular sequencing errors.
Another consequence of the read length invariance is that the prediction performance cannot be assessed in terms of sensitivity and specificity as in sequence classification methods. Because no single read is actually assigned to a taxonomic category, it is impossible to measure the performance in terms of detection accuracy. Instead, the compositional parameters describing the abundance of taxonomic units are directly predicted from the overall oligonucleotide distribution. For many problems of quantitative metagenome analysis, direct predictions of the taxonomic composition will be sufficient, but in some cases, a more detailed investigation is necessary. For example, further analysis of reads from a particular phylogenetic group would require a sequence classification method for the identification of the corresponding reads. Therefore, Taxy complements the current range of profiling methods rather than replacing any of the existing methods. Furthermore, the organism-specific weights as obtained from a Taxy analysis can be used as priors in a probabilistic fragment classification framework such as the NBC approach (Rosen et al., 2008).
Currently, a number of web-based metagenome analysis systems exist, which provide the user with a comfortable platform for comparative metagenomics and taxonomic profiling: MG-RAST (Meyer et al., 2008), Galaxy (Kosakovsky Pond et al., 2009), IMG/M (Markowitz et al., 2008) and CAMERA (Seshadri et al., 2007). Although these platforms are of great value for metagenome analyses, they show the typical disadvantages of web-based tools, such as restrictions on user-supplied data or long response times. Several platforms are based on a BLAST (Altschul et al., 1990) engine, which matches the supplied sequence data against particular databases. BLAST analyses usually involve a number of parameters that have a measurable effect on the results. The optimal choice of BLAST parameters depends on the complexity and size of the sample and on the sequence length distribution. As a consequence, the specific adjustment of parameters such as E-value, word length, minimal alignment length and percent identity for each metagenomic dataset can complicate a BLAST-based comparative analysis. The same difficulties are encountered when using tools like MEGAN (Huson et al., 2007), which rely on prior results from a costly BLAST analysis. As our Galaxy results on the glacial ice sample and on the simHC data demonstrate, also the taxonomical distribution of the reference database may affect the estimation of profiles. On the other hand, read classification methods based on BLAST offer the adjustment of a similarity-based rejection criterion, which allows to exclude parts of the data from taxonomic profiling. This can be a great advantage if sequence quality is low and it suggests the combination of different methods rather than favoring one single approach.
The particular utility of Taxy arises from a quick overview of the taxonomic distribution of large datasets, which can be a good starting point for any kind of computational metagenome analysis. Besides the inherent read length independence of Taxy, which significantly simplifies comparative analysis, there is another striking advantage that qualifies the method as an excellent early stage data mining tool for metagenomics: the computational speed is orders of magnitude faster than that of any of the existing taxonomic profiling methods. Therefore, large amounts of data can be processed without having access to extensive computational facilities. All computations can be performed on a local standard PC requiring at most a few minutes for even the largest datasets. This efficiency makes it possible to already obtain a first estimate of the sample composition, long before extensive computations on external servers or computer clusters may provide a more detailed picture of the community structure.
Supplementary Material
ACKNOWLEDGEMENT
We would like to thank Peter Gumrich for programming the Taxy tool for Windows and Christian Opitz, Stefanie Mühlhausen and Alexander Kaever for additional technical support. We further thank two anonymous reviewers for their helpful comments.
Funding: Grants from the Deutsche Forschungsgemeinschaft (ME 3138, ‘Compositional descriptors for large scale comparative metagenome analysis’ to P.M. in part) and (LI 2050, ‘Development of machine learning methods for functional characterization of the peroxisome’ T.L. in part).
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beja O, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Bohlin J, et al. Analysis of genomic signatures in prokaryotes using multinomial regression and hierarchical clustering. BMC Genomics. 2009;10:487. doi: 10.1186/1471-2164-10-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat. Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canu S, et al. Perception Systèmes et Information. 2005. SVM and Kernel Methods Matlab Toolbox. INSA de Rouen, Rouen, France. [Google Scholar]
- Dempster AP, et al. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B. 1977;39:1–38. [Google Scholar]
- Diaz NN, et al. TACOA: taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformatics. 2009;10:56. doi: 10.1186/1471-2105-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach W, et al. WebCARMA: a web application for the functional and taxonomic classification of unassembled metagenomic reads. BMC Bioinformatics. 2009;10:430. doi: 10.1186/1471-2105-10-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, et al. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009;3:1365–1373. doi: 10.1038/ismej.2009.89. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-reviews0003. REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S, et al. Windshield splatter analysis with the Galaxy metagenomic pipeline. Genome Res. 2009;19:2144–2153. doi: 10.1101/gr.094508.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause L, et al. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36:2230–2239. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, et al. A bioinformatician's guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- McHardy AC, et al. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- Meyer F, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen G, et al. Metagenome fragment classification using N-mer frequency profiles. Adv. Bioinformatics. 2008;2008:205969. doi: 10.1155/2008/205969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen GL, et al. NBC: the Naive Bayes classification tool webserver for taxonomic classification of metagenomic reads. Bioinformatics. 2011;27:127–129. doi: 10.1093/bioinformatics/btq619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber F, et al. Treephyler: fast taxonomic profiling of metagenomes. Bioinformatics. 2010;26:960–961. doi: 10.1093/bioinformatics/btq070. [DOI] [PubMed] [Google Scholar]
- Seshadri R, et al. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, et al. Rapid identification of genes encoding DNA polymerases by function-based screening of metagenomic libraries derived from glacial ice. Appl. Environ. Microbiol. 2009;75:2964–2968. doi: 10.1128/AEM.02644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stach JE, Bull AT. Estimating and comparing the diversity of marine actinobacteria. Antonie Van Leeuwenhoek. 2005;87:3–9. doi: 10.1007/s10482-004-6524-1. [DOI] [PubMed] [Google Scholar]
- Suen G, et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, et al. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 2004;6:938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- von Mering C, et al. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- Wommack KE, et al. Metagenomics: read length matters. Appl. Environ. Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.