Abstract
Summary: We built a web server named APOLLO, which can evaluate the absolute global and local qualities of a single protein model using machine learning methods or the global and local qualities of a pool of models using a pair-wise comparison approach. Based on our evaluations on 107 CASP9 (Critical Assessment of Techniques for Protein Structure Prediction) targets, the predicted quality scores generated from our machine learning and pair-wise methods have an average per-target correlation of 0.671 and 0.917, respectively, with the true model quality scores. Based on our test on 92 CASP9 targets, our predicted absolute local qualities have an average difference of 2.60 Å with the actual distances to native structure.
Availability: http://sysbio.rnet.missouri.edu/apollo/. Single and pair-wise global quality assessment software is also available at the site.
Contact: chengji@missouri.edu
1 INTRODUCTION
Protein model quality assessment plays an important role in protein structure prediction and application. Assessing the quality of protein models is essential for ranking models, refining models and using models (Cheng, 2008). Model Quality Assessment Programs (MQAPs) predict model qualities from two perspectives: the global quality of the entire model and the residue-specific local qualities. The techniques often used by MQAPs include multiple-model (clustering) methods (Ginalski et al., 2003; McGuffin, 2007, 2008; Paluszewski and Karplus, 2008; Wallner and Elofsson, 2007; Zhang and Skolnick, 2004a), single model methods (Archie and Karplus, 2009; Benkert et al., 2008; Cline et al., 2002; Qiu et al., 2008; Wallner and Elofsson, 2003; Wang et al., 2008) and hybrid methods (Cheng et al., 2009; McGuffin, 2009).
According to the CASP experiments, multiple-model clustering methods are currently more accurate than single model methods. However, they cannot work well if only a small number of models are available. A hybrid quality assessment method (Cheng et al., 2009) was recently developed to combine the two approaches and integrate their respective strengths. Here, we build a web server to provide the community with access to all three model quality assessment approaches (i.e. single, clustering and hybrid).
2 METHODS
2.1 Input and output
Users only need to upload or paste a single model file in Protein Data Bank (PDB) format or a zipped file containing multiple models.
If a single model is submitted, APOLLO predicts the absolute global and local qualities. If multiple models are submitted, APOLLO outputs the absolute global qualities, average pair-wise GDT-TS scores, refined average pair-wise Q-scores, refined absolute scores and pair-wise local qualities. All the global qualities range between (0, 1), where 1 indicates a perfect model and 0 indicates the worst case.
2.2 Algorithms
The absolute global quality score is generated based on our single model QA predictor—ModelEvaluator (Wang et al., 2008). Given a single model, ModelEvaluator (as MULTICOM-NOVEL server in CASP9) extracts secondary structure, solvent accessibility, beta-sheet topology and a contact map from the model, and then compares these items with those predicted from the primary sequence using the SCRATCH program (Cheng et al., 2005). These comparisons generate match scores which are then fed into an SVM model trained on CASP6 and CASP7 data to predict the absolute global quality of the model in terms of GDT-TS scores. To predict absolute local quality score of a residue, the secondary structure and solvent accessibility predicted from the sequence are compared with the ones parsed from the model in a 15-residue window around the residue. For each residue in the window, we also gather its contact residues that are ≥ 6 residues away in sequence and have an Euclidean distance ≤ 8 Å in the model. Their probabilities of being in contact according to the predicted contact probability map are averaged. The averaged contact probabilities, the match scores of secondary structure and solvent accessibility comparison and the residue encoding are fed into an SVM to predict local quality. The SVM are trained on the models of 30 CASP8 single domain targets.
The average pair-wise GDT-TS score is generated using our latest implementation (as MULTICOM-CLUSTER server in CASP9) of the widely used pair-wise comparison approach (Larsson et al., 2009). Taking a pool of models as an input, it first filters out illegal characters and chain-break characters in their corresponding PDB files. It then uses TM-Score (Zhang and Skolnick, 2004b) to perform a full pair-wise comparison between these models. The average GDT-TS score between a model and all other models is used as the predicted GDT-TS score of the model. One caveat is that the GDT-TS score of a partial model is scaled down by the ratio of its length divided by the full target length.
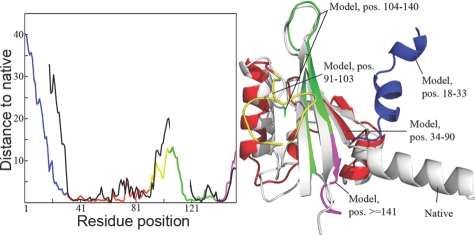
The refined global and local quality scores are generated using a hybrid approach (as MULTICOM-REFINE server in CASP9) (Cheng et al., 2009) that integrates single model ranking methods with structural comparison-based methods. It first selects several top models (i.e. top five or top ten) as reference models. Each model in the ranking list is superposed with the reference models by the TM-Score. The average GDT-TS score of these superposition is considered as the predicted quality score. The superposition with the reference models is also used to calculate Euclidean distances between the same residues in the superposed models. The average distance is used as the predicted pair-wise local quality of the residue (Fig. 1). Higher distances correspond to poorer local quality.
Fig. 1.
A local quality example for CASP9 target T0563. On the left is a plot of predicted local quality scores (colorful line) and actual distance (black line) against residue positions. On the right is the superposition between native structure (grey) and the model. The regions of the model with different local quality are visualized in different colors corresponding to the color of line segments in the plot on the left. Disordered regions are not plotted in the actual distance line.
The refined average pair-wise Q-scores are generated using a consensus approach (as MULTICOM-CONSTRUCT server in CASP9). APOLLO first uses the average pair-wise similarity scores, calculated in terms of Q-score (Ben-David et al., 2009; McGuffin and Roche, 2010), to generate an initial ranking of all the models. The Q-score between a pair of residues (i, j) in the two models is computed as: Qij = exp[ − (rija − rijb )2], where rija and rijb are the distance between Cα atoms at residue positions i and j in models a and b, respectively. The overall Q-score between models a and b is equal to the average of all Qij scores of all residue pairs in the entire model. The average Q-score between a model and all other models is used as the predicted quality score of the model. The initial quality scores are refined by the same refinement process used by our hybrid method in MULTICOM-REFINE.
3 RESULTS
We assessed most of the methods used by APOLLO on 107 valid CASP9 targets. We downloaded all the CASP9 models from CASP9 (http://predictioncenter.org/download_area/CASP9/) and the experimental structures from the PDB (Berman et al., 2000). These PDB files were preprocessed in order to select correct chains and residues that match the CASP9 target sequences. TM-Score was used to align each model with the corresponding native structure and generate its real quality score (GDT-TS). The CASP9 QA predictions made by our methods were evaluated against the actual quality scores by four criteria: average per-target correlation (Cozzetto et al., 2009), the average sum of the GDT-TS scores of the top one ranked models, the overall correlation on all targets and the average loss—the difference in GDT-TS score between the top ranked model and the best model (Cozzetto et al., 2009) (Table 1). The results show that the average correlation can be as high as 0.92 (respectively, 0.67) and the average loss can be as low as 0.057 (respectively, 0.095) for multiple model (respectively, single model). Our multiple- and single-model global QA methods were ranked among the most accurate QA methods of their respective kind according to the CASP9 official assessment (http://www.predictioncenter.org/casp9/doc/presentations/CASP9_QA.pdf). The average per-target correlation of our pair-wise local quality predictions is ~ 0.53, which is also among the top local quality predictors in CASP9. We also conducted a blind test of the absolute local quality predictor (trained on the CASP8 dataset) on the CASP9 models of 92 CASP9 single domain proteins. On the residues whose actual distances to the native are ≤ 10 and 20 Å, the average absolute difference between our predicted distances and the actual distances is 2.60 and 3.18 Å, respectively.
Table 1.
Results of global quality assessment methods used by APOLLO server on 107 CASP9 targets
| Methods | Average correlation | Average top 1 | Overall correlation | Average % loss |
|---|---|---|---|---|
| Absolute score | 0.671 | 0.552 | 0.767 | 0.095 |
| Average pair-wise GDT-TS | 0.917 | 0.591 | 0.943 | 0.057 |
| Refined absolute score | 0.870 | 0.567 | 0.928 | 0.081 |
| Refined pair-wise Q-score | 0.835 | 0.572 | 0.904 | 0.076 |
Funding: A National Institutes of Health (NIH) (grant 1R01GM093123 to J.C.).
Conflict of Interest: none declared.
REFERENCES
- Archie J, Karplus K. Applying undertaker cost functions to model quality assessment. Proteins. 2009;75:550–555. doi: 10.1002/prot.22288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David M, et al. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;77:50–65. doi: 10.1002/prot.22591. [DOI] [PubMed] [Google Scholar]
- Benkert P, et al. QMEAN: a comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, et al. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. A multi-template combination algorithm for protein comparative modeling. BMC Struct. Biol. 2008;8:18. doi: 10.1186/1472-6807-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, et al. Prediction of global and local quality of CASP8 models by MULTICOM series. Proteins. 2009;77:181–184. doi: 10.1002/prot.22487. [DOI] [PubMed] [Google Scholar]
- Cline M, et al. Predicting reliable regions in protein sequence alignments. Bioinformatics. 2002;18:306–314. doi: 10.1093/bioinformatics/18.2.306. [DOI] [PubMed] [Google Scholar]
- Cozzetto D, et al. Evaluation of CASP8 model quality predictions. Proteins. 2009;77:157–166. doi: 10.1002/prot.22534. [DOI] [PubMed] [Google Scholar]
- Ginalski K, et al. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Larsson P, et al. Assessment of global and local model quality in CASP8 using Pcons and ProQ. Proteins. 2009;77:167–172. doi: 10.1002/prot.22476. [DOI] [PubMed] [Google Scholar]
- McGuffin L. Benchmarking consensus model quality assessment for protein fold recognition. BMC Bioinformatics. 2007;8:345. doi: 10.1186/1471-2105-8-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008;24:586. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ. Prediction of global and local model quality in CASP8 using the ModFOLD server. Proteins. 2009;77(Suppl. 9):185–190. doi: 10.1002/prot.22491. [DOI] [PubMed] [Google Scholar]
- McGuffin L, Roche D. Rapid model quality assessment for protein structure predictions using the comparison of multiple models without structural alignments. Bioinformatics. 2010;26:182–188. doi: 10.1093/bioinformatics/btp629. [DOI] [PubMed] [Google Scholar]
- Paluszewski M, Karplus K. Model quality assessment using distance constraints from alignments. Proteins. 2008;75:540–549. doi: 10.1002/prot.22262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, et al. Ranking predicted protein structures with support vector regression. Proteins. 2008;71:1175. doi: 10.1002/prot.21809. [DOI] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Prediction of global and local model quality in CASP7 using Pcons and ProQ. Proteins. 2007;69:184–193. doi: 10.1002/prot.21774. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. Evaluating the absolute quality of a single protein model using structural features and support vector machines. Proteins. 2008;75:638–647. doi: 10.1002/prot.22275. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein folds. J. Comput. Chem. 2004a;25:865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004b;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]